Abstract
Interventions such as dietary restriction (DR) have been reported to ameliorate age-related proteasome inhibition in some tissues, suggesting that there may be plasticity in the proteasome proteolytic pathway which contributes to the preservation of proteasome function. Currently it is not known what effects aging and DR have on proteasome plasticity and proteasome biogenesis in the liver and brain, nor have previous studies identified the links between changes in proteasome plasticity, biogenesis, and activity in the aging brain and liver. In the present study we demonstrate that the brain and liver exhibit age-dependent decreases in 26S and 20S proteasome activity. Additionally, our studies demonstrate that the brain and liver undergo selective changes in proteasome plasticity, and increases in proteasome biogenesis in response to aging and DR, with the liver exhibit more robust plasticity as compared to the brain. Lastly, studies demonstrated that aging and DR alter the interaction of Hsp90 with the 20S proteasome complex in the brain and liver. These studies affirm the dynamic nature of the proteasome complexes in both the liver and brain following aging and DR, and indicate that the relationship between proteasome plasticity/biogenesis and proteasome activity in tissues is extremely complex.
Keywords: Hepatocyte, Neuron, Oxidative stress, Ubiquitin Proteasome Pathway
1. Introduction
Protein degradation is essential for life and not surprisingly compromises in intracellular proteolysis are believed to mediate deleterious effects on cellular homeostasis (Sullivan et al., 2004; Cecarini et al., 2007; Van Tijn et al., 2008; Petropoulos et al., 2006). Compromises in the proteasome proteolytic pathway are known to occur during the aging of many tissues (Widmer et al., 2006; Carrad et al., 2003; Kapphahn et al., 2007) and numerous lines of evidence suggest that proteasome-mediated protein degradation may potentially contribute to the progression of both aging and age-related disease (Keller et al., 2000b; 2006; Breusing and Gurne, 2008; de Vrij et al., 2004). Additionally, studies have demonstrated that dietary restriction (DR) is capable of ameliorating age-related declines in proteasome-mediated protein degradation in some tissues, although it is clear that DR does not preserve proteasome function in all tissues and experimental conditions (Aksensova et al., 1998; Goto et al., 2007; Radak et al., 2002; Li et al., 2008; Zhang et al., 2007).
Proteasome-mediated protein degradation is achieved by the activity of the 26S and 20S proteasome complexes, which are large protein complexes of ∼2500 kDa and ∼700 kDa, respectively (Peters et al., 1994; Coux et al., 1996; Dahlmann et al., 1995; Hilt and wolf, 1996). The 20S proteasome is the core unit of the proteasome degradation system, and is responsible for all proteolytic activity of the proteasome. The 19S/PA700 regulator and the 11S/PA28 complexes can bind to the 20S proteasome to form a 26S proteasome complex, which mediates the degradation of proteins tagged with ubiquitin in an ATP dependent pathway (Ciechanover et al., 2000; Pickart., 2001), where as oxidized proteins and ornithine decarboxylase appear to be degraded in an ATP independent pathway by the 20S proteasome (Grune et al., 1996; 2003; Shrigarpure et al., 2003).
Proteasome biogenesis in eukaryotes is a multi step process, which requires several chaperones and accessory proteins including the chaperone UMP1 in yeast (m/hUMP1, POMP1, proteoassemblin) (Ramos et al., 1998; Witt et al., 2004) and the Proteasome Assembling Chaperones 1-4 (PAC1-4) (Le Tallec et al., 2007; Hirano et al., 2005; 2006). Despite the tremendous progresses which have been made in regards to our understanding of the molecular and cellular basis of proteasome biogenesis, it currently is unclear how proteasome biogenesis may be altered in tissues such as the brain and liver during aging and DR. However, recent studies have suggested that age-related changes in proteasome function in the heart and spleen may be due in part to modulation of proteasome biogenesis (Li et al., 2008; Zhang et al., 2007).
The majority of work thus far on the regulation of proteasome activity in vivo has focused on the role of post-translational modifications, in particular oxidative damage (Amici et al., 2003; Friguet and Szweda, 1997; Stadtman 1992) as modulators of proteasome function. However, recent studies have demonstrated that interactions between Hsp90 and the proteasome may be another mechanism by which proteasome function is regulated in response to aging and DR. For example, addition of Hsp90 to 20S proteasomes was found to protect the trypsin-like activity of the proteasome from oxidative inactivation by metal catalyzed oxidation and 4-hydroxy-2-nonenal treatment (Conconi et al., 1996; 1998).
Understanding how aging and DR alter the expression, composition, biogenesis, and activity of the proteasome complexes is important to understanding the basis of aging and DR effects on tissues. In the current study, we demonstrate that both the brain and liver undergo changes in proteasome activity, plasticity, and biogenesis. Our findings indicate that the liver undergoes more robust age-related changes in multiple aspects of proteasome plasticity, as compared to the brain, with the 26S proteasome more affected by aging and DR than the 20S proteasome. Cumulatively our studies indicate that the effects of aging and DR on proteasome plasticity, proteasome biogenesis, and proteasome activity appear to be extremely complex.
2. Materials and Methods
2.1. Materials
The proteasome substrate Suc-Leu-Leu-Val-Tyr-AMC (for measurement of chymotrypsin-like activity) was purchased from Bachem Inc., (Torrance, CA, USA). The antibodies used in this study were antibodies against 20S proteasome core subunits, Rbt (EMD Chemicals, Gibbstown, NJ, USA), Alpha 7 or C8 (clone MCP72) subunit 20S proteasome- mouse mAb, 19S regulator ATPase subunit Rpt3 (S6b,TBP7) -mouse mAb, Subunit β5 for 20S proteasome-Rbt (BIOMOL International, Plymouth meeting, PA, USA), Anti-Hsp90 mouse antibody (Assay Designs, Ann Arbor, MI, USA), 11S regulator subunit PA28 alpha rabbit polyclonal antibody (BIOMOL international Plymouth meeting, PA, USA), and DSCR2 (PAC1) mouse monoclonal antibody (ABNOVA, Walnut, PA, USA). All the HRP conjugated secondary antibodies were purchased from Vector Laboratories, USA. MG132 was purchased from EMD Chemicals (Gibbstown, NJ, USA). The Superose-6 HR 10/300 (Cat# 17517201) gel filtration column was purchased from GE Life Sciences (Piscataway, NJ, USA). The BCA (Cat# 23223) and micro-BCA (Cat#23235) reagents were purchased from Thermo Scientific, Inc. (IL, USA). All electrophoresis and immunoblot reagents were purchased from Bio-Rad Laboratories (Hercules, CA, USA). All other items and chemicals including protein A-agarose beads were purchased from Sigma-Aldrich, Corp. (St. Louis, MO, USA).
2.2. Animal tissues
Male Helicobacter-free F344/Brown Norway (F344 × BN F1) rats were obtained from the NIA Dietary Restriction (DR) colony. The rats in this study consisted of 6 three month-old ad libitum (AL), 6 twenty five month-old AL, and 6 twenty five month-old DR rodents. All animals were handled and euthanatized in accordance with IACUC approved protocols at the Pennington Biomedical Research Center.
2.3. Preparation of liver homogenates and separation of 20S and 26S proteasomes by gel filtration chromatography
The separation of proteasomes from liver lysates using Superose 6 Gel filtration column was done as described previously (Brooks et al., 2000) with some minor modifications. Liver tissues were homogenized in buffer containing 20 mM Tris-HCl pH-7.4, 5 mM ATP, 10% Glycerol and 100 mM NaCl. Lysates were centrifuged at 19,000 × g for 20 minutes. All the steps were performed either on ice or below 10°C. One hundred microliters of the resulting solution (∼2 mg) was loaded on to Superpose 6 HR 10/300 Gel filtration column (GE Life sciences) equilibrated with the buffer containing 100mM NaCl, 20mM Tris-HCl pH-7.4, 5mM ATP and 10% Glycerol which are the buffer conditions for separating the different proteasome complexes (Brooks et al., 2000) and eluted with same buffer. All the lysates belonging to different age groups were run under similar conditions with equal amounts (2mg) of protein. The 500 μl fractions were collected at a flow rate of 0.3mL/min. The fractions were numbered as per their elution from the column. The peak fractions corresponding to 26S and 20S proteasomes were determined by specific activity assays and characterized by Western blotting analysis using antibodies against 20S proteasome subunits (alpha 7 and beta 5), 19S complex (S6b subunit) and also to alpha subunits of 11S complex (PA28) as described in previous studies (Brooks et al., 2000; Bose et al., 2001).
2.4. Protein estimation
Protein concentration of the crude lysates or FPLC separated fractions of liver and brain was estimated using either BCA or micro BCA reagent (Thermo Scientific) as described by the manufacturer.
2.5. Assays for proteasome activity
Assays for Chymotrypsin like activity of proteasomes were performed using fluorogenic Succinyl-Leu-Leu-Val-Tyr-AMC as a substrate. All assays were carried out in 250 μl of activity assay buffer in a 96 well black assay plate with clear bottom as described previously by our laboratory (Dasuri et al., 2009). In each well we added 50 μl of each Superose -6 fraction, and 40 μM of substrate. The 26S proteasome activity assay was carried out in the presence of 2mM ATP where as 20S proteasome activity was carried out in the presence of 0.02%SDS which activates 20S proteasomes and partially inhibits the 26S proteasome activity as described previously (Brooks et al., 2000). The reaction mixture was incubated for 3 hours at 37°C and the fluorescence of the released AMC product was measured in Perkins-Elmer plate reader at an emission wavelength of 355 nm and an excitation wavelength of 460 nm. The background fluorescence values obtained by incubating the fractions with MG132 were subtracted from activity values and the difference in activity was considered as the actual activity of the proteasome. Relative proteasome activities were represented as μM AMC formed/hour per mg of protein.
2.6. Immunoprecipitation with Hsp90
Equal amounts of Brain and Liver lysates in PBS buffer containing 0.2%Triton X-100 and Proteasome inhibitor cocktail were incubated overnight with 20S proteasome antibodies against the core subunits. The precipitated protein-antibody complexes were pulled down using Protein A-agarose beads and then the beads were washed for 4X times with PBS buffer containing 0.2%Triton X-100. Complexes were separated by SDS-PAGE and then immunoblotted with anti-HSP90 antibodies. Control experiment was done, where the lysates were incubated with beads alone without antibody, to establish the background signal for the immunoprecipitation procedure.
2.7. Western blotting
The protein samples were boiled in Laemmlli sample buffer and loaded on to BioRad Precast Criterion gels and immunoblotted with specified antibodies as described previously by our laboratory (Dasuri et al., 2009). All blots presented in individual figures were developed simultaneously on the same film to ensure equal exposure.
3. Results
3.1. Analysis of Proteasome Activity
In order to conduct analysis of proteasome complexes from liver lysates, we have used Fast Protein Liquid Chromatography (FPLC), using Superose-6 HR 10/300 gel filtration column as described previously (Brooks et al., 2000). Analysis of brain and liver proteasome complexes revealed that aging induces robust declines in both 20S and 26S proteasome activities in both tissues (Figure 1). Dietary restriction (DR) was observed to not have significant effects on age-related proteasome inhibition in the liver, but did promote the amelioration of age-related inhibition of the 26S proteasome in the brain (Figure 1). No significant effect of DR on 20S proteasome activity was observed in either tissue (Figure 1). It should be noted that the activity and levels of proteasome complexes is much lower in the brain compared to liver (Figure 1), consistent with previous studies (Dasuri et al., 2009).
Figure1. Aging decreases proteasome activity in brain and liver.
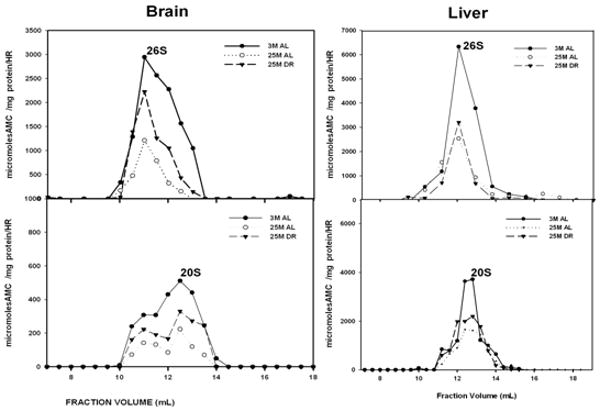
Rat liver (A) and brain (B) lysates were fractionated on Superose- 6 HR 10/300 gel filtration chromatography column and 500 μl fractions were collected as described in methods section. Fractions were then assayed for proteasome activity using Succinyl-Leu-leu-Val-Tyr-7-amido-4-methylcoumarin as substrate under conditions optimized for 26S proteasomes (+ATP) and for 20S proteasomes (+0.02% SDS). The peak fractions showing 26S and 20S proteasome activity are indicated in the figure. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent the mean of three sets of experiments done under similar conditions.
3.2. Brain and liver differ in composition of proteasome subunits following aging and DR
In our next set of experimentation we sought to determine whether the composition of core proteasome subunits is altered in response to aging and DR, and to elucidate whether the liver and brain differ in regards to potential changes in proteasome composition. In this and all Western blot analysis the blots depicted in each figure were all developed simultaneously on the same film to ensure the same duration of film exposure. In our analysis we observed that in the brain the levels of the alpha-7/C8 proteasome subunit are decreased in the 26S proteasome of the aged brain, which is reversed by DR (Figure 2). In contrast in the liver there was an age-related increase in alpha-7/C8 proteasome subunits within the 26S proteasome complex (Figure 2), which was also partially reversed by DR. The levels of the 19S/S6b proteasome subunit were increased in the 26S proteasome complex in both the brain and liver of aged rodents (Figure 3), which was reversed by DR in both tissues. The levels of beta-5 proteasome subunit were observed to be relatively unchanged in all experimental conditions in the brain (Figure 4), while the liver exhibited a significant decline in beta-5 expression in the 20S proteasome complex with age (Figure 4), that was reversible by DR (Figure 4). Taken together, these data are consistent with the 20S and 26S proteasome complexes undergoing changes in composition, and potentially even changes in the level of proteasome complexes, in response to aging and DR, with the liver showing a greater degree of plasticity as compared to the brain.
Figure 2. Expression of Alpha-7/C8 proteasome subunits of proteasome are altered by aging and dietary restriction.
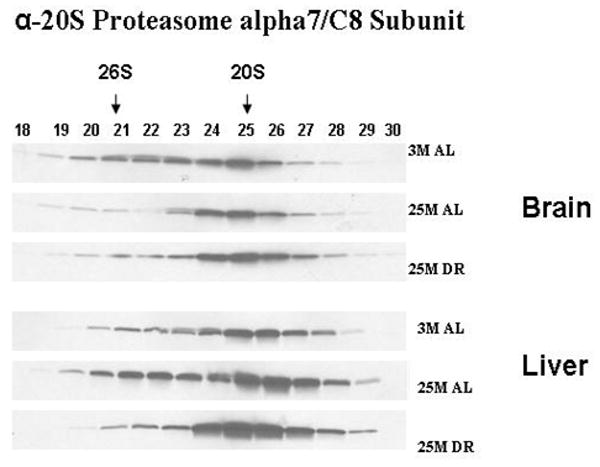
A 30 μl aliquot of each of the chromatography column fraction obtained after gel filtration of liver and brain lysates on Superose-6 HR10/300 column, were separated by SDS-PAGE and immunoblotted using 20S proteasome antibodies specific to α7 subunit. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent two sets of experiments done under similar conditions.
Figure 3. Expression of S6b proteasome subunits of 19S proteasome regulator are altered by aging and dietary restriction.
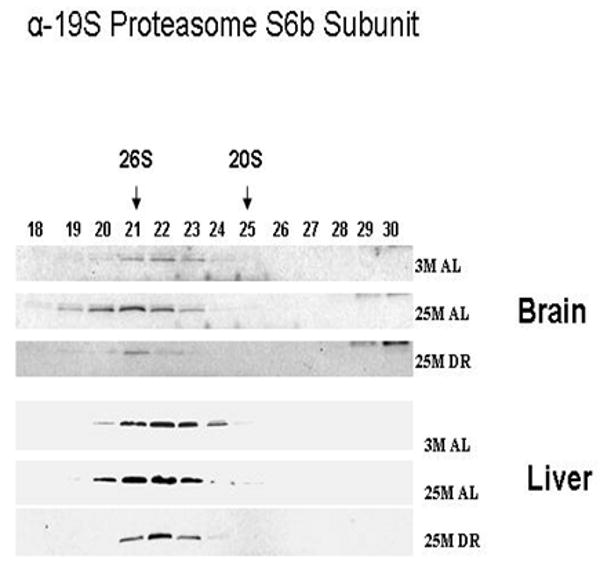
A 30 μl aliquot of each of the chromatography column fraction obtained after gel filtration of liver and brain lysates on Superose-6 HR10/300 column were separated by SDS-PAGE and immunoblotted using 19S proteasome antibodies specific to S6b subunit. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent two sets of experiments done under similar conditions.
Figure 4. Expression of Beta 5 proteasome subunits of proteasome are altered by aging and dietary restriction.
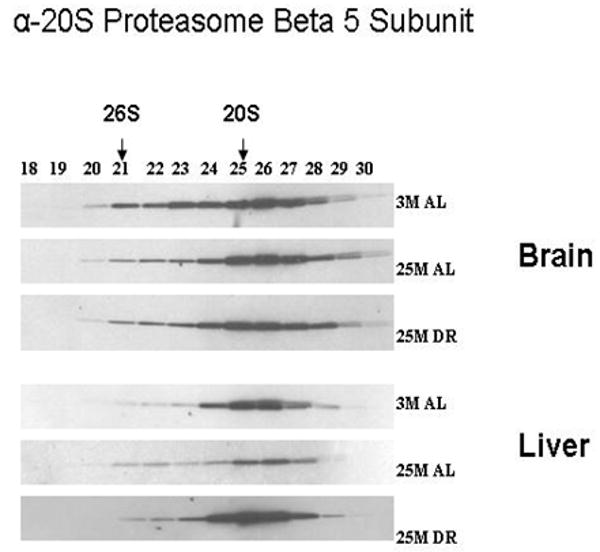
A 30 μl aliquot of each of the chromatography column fraction obtained after gel filtration of liver and brain lysates on Superose-6 HR10/300 column, were separated by SDS-PAGE and immunoblotted using 20S proteasome antibodies specific to Beta5 subunit. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent two sets of experiments done under similar conditions.
3.3. The brain and liver exhibit differences in regards to the age and DR induced changes in 11S/PA28 proteasome regulator and proteasome assembly chaperone, PAC1
In the next set of experimentation we sought to examine the effects of aging and DR on the levels of 11S/PA28 caps of the proteasome complexes, as well as the levels of proteasome biogenesis. In these studies we observed that the levels of 11S/PA28 were significantly increased by aging and DR in the liver (Figure 5), while the brain exhibited little difference in any of the experimental paradigms. We next conducted analysis of PAC1, an essential regulator of proteasome biogenesis (Hirano et al., 2005; 2006; Le Tallec et al., 2007), in fractions containing complexes of proteasome precursors. These studies revealed that in the aging brain and liver there was a significant increase in proteasome biogenesis (Figure 6), which remained elevated in DR rodents. The levels of biogenesis appeared to be dramatically more robust in the liver as compared to the brain (Figure 6).
Figure 5. 11S/PA28 proteasome regulator levels are altered by aging and dietary restriction.
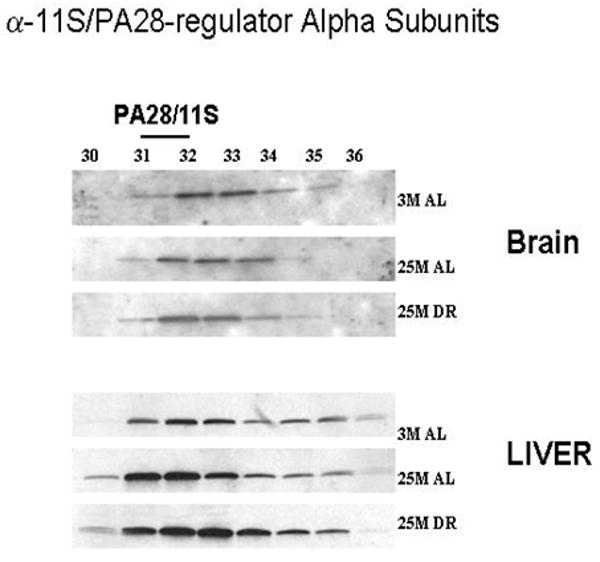
A 30 μl aliquot of each of the chromatography column fraction obtained after gel filtration of liver and brain lysates on Superose-6 HR10/300 column, were separated by SDS-PAGE and immunoblotted using 11S proteasome regulator antibodies against α- subunits. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent two sets of experiments done under similar conditions.
Figure 6. Increased expression of the proteasome biogenesis protein PAC1 with aging and caloric restriction.
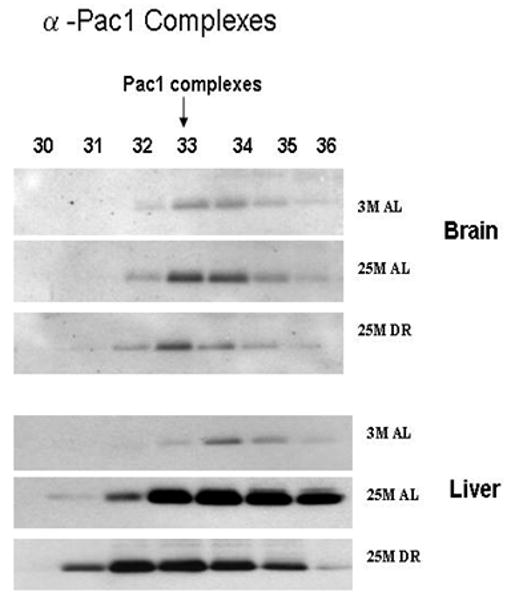
A 30 μl aliquot of each of the chromatography column fraction, separated on Superose-6 HR 10/300 column from each age group of the liver and brain lysates, were separated by SDS-PAGE and immunoblotted with proteasome assembly chaperone (PAC1) antibodies. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted. Results represent two sets of experiments done under similar conditions.
3.4. The brain and liver exhibit age-related increases in Hsp90-proteasome interactions
In the last set of experimentation we sought to elucidate whether we could detect Hsp90-proteasome interactions in both the brain and liver, and to elucidate whether aging and DR alter the level of such interactions. In the first step of this analysis we observed that in the brain there was no significant alteration in the level of Hsp90 within the crude lysate (Figure 7), while in the liver there was a significant decline in Hsp90 levels in the crude lysate that was significantly ameliorated by DR (Figure 7). In the analysis of Hsp90 proteasome interactions we observed that following immunoprecipitation with an antibody that recognizes multiple core subunits of the 20S complex (Gong et al., 2007), there was an enhanced level of Hsp90 immunoreactivity within the 20S proteasome immunoprecipitate of aged liver and aged brain fractions (Figure 7). Increased levels of Hsp90 interactions with the proteasome complex were sustained in the brain and liver of DR rodents (Figure 7), although there appeared to be a slight reduction in the levels of such interactions as compared to age-matched ad libitum animals.
Figure 7. The association of Hsp90 with proteasome complexes is altered by aging and dietary restriction.
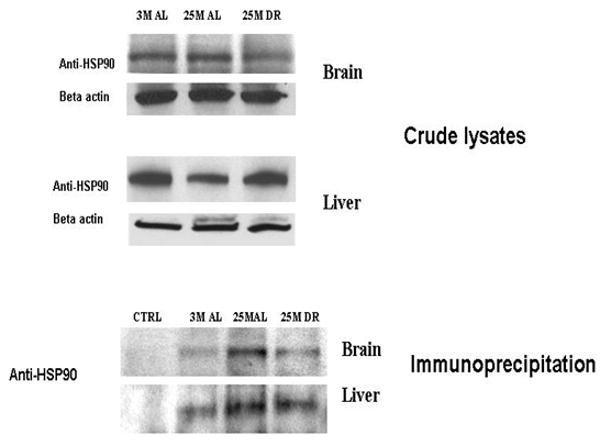
The levels of Hsp 90 in the crude lysates (A) of liver and brain were analyzed by immunoblotting with antibodies against Anti-Hsp90. Levels of Hsp 90 associated with 20S proteasomes in liver and brain lysates (B) were analyzed by immunoprecipitation with 20S proteasome antibodies against core subunits and immunoblotting with antibodies against Hsp90. Ctrl: control experiment where the lysates were incubated with beads alone without antibody for the background. 3 M: 3 month AL; 25 M: 25 month AL; DR, 25 month dietary restricted.
4. Discussion
Age-related declines in 20S and 26S proteasome activity occurred in both the brain and liver consistent with other studies that have demonstrated decreased levels of proteasome activity in these same tissues (Shibatini et al., 1996 a, b; Conconi et al., 1996; Keller et al., 2000 a, b). However in contrast to previous studies (Shibatini et al., 1996a, b; Conconi et al., 1996), in the current study we did not see significant effects of DR on the activity of the 26S proteasome in the liver. A significant effect of DR on 26S proteasome activity was observed in the brain consistent with previous studies (Keller et al., 2000 a, b). In neither the brain or liver did we see significant effects of DR on the 20S proteasome complex. While the basis for the differences between our studies and previous publications remains unclear, it is possible it is based on the fact many previous studies relied solely on analysis of proteasome activity in crude lysates as compared to purified proteasome complexes utilized in the present study. It is also important to note that previous publications using DR tissue have reported a lack of DR-induced preservation of proteasome function similar to our present findings (Shibatini et al., 1996 a, b; Conconi et al., 1996). Regardless, our studies are the first to study the levels of proteasome activity, proteasome plasticity, and proteasome biogenesis in either the liver or brain. Additionally, these are the first studies to simultaneously examine each of these aspects in the context of both DR and aging. Together, these data provide several important findings in regards to understanding how aging and DR differentially impact the proteasome complexes present in the liver and brain.
The brain was observed to have lower levels of proteasome activity as compared to the liver, consistent with our previous study (Dasuri et al., 2009), and to exhibit lower levels of proteasome plasticity. For example, age- and DR-related changes in alpha7/C8 19S/S6B, beta-5, and proteasome biogenesis were all more robust in the liver as compared to the brain. While the basis for these observations are not clearly known, one can reasonably speculate that higher mitotic capacity of the liver may be responsible for the observed findings. Alternatively, even though the brain and liver are highly metabolic tissues, it may be that the influence of aging and DR on the liver is more robust in terms of the effects of metabolic-proteasome interactions. Regardless, our studies strongly support the concept of the liver being able to exhibit significantly greater levels of proteasome plasticity as compared to the brain. Equally clear, our studies indicate that the 26S proteasome activity is preserved in the brain in response to DR, but not preserved in the liver in response to DR. Taken together, these data indicate that the level of proteasome activity may not be dictated by changes in proteasome plasticity or proteasome biogenesis per se. These studies suggest that the ability to suppress age-related changes in proteasome function may be mediated by events such as decreases in the oxidative modification to the proteasome complex (Louie et al., 2002; Viteri et al., 2004) as has been reported previously. The ability of DR to preferentially affect 26S as compared to the 20S proteasome complex is also in line with previous studies which suggested that the 26S is more vulnerable to oxidant-induced inactivation (Reinheckel et al., 1998; 2000). Currently it is not clear what the function of altered proteasome plasticity and proteasome biogenesis in response to aging and DR may serve. It may be possible for example that changes in composition arise as the result of proteasome subunits altering their expression in response to stressors. For example, it has been reported that cells tries to increase the levels of 11S during some oxidative stress conditions (Beedholm et al, 2004). Similarly, studies from our laboratory have demonstrated that oxidative stressors increase the expression of inducible proteasome subunits in neural cells (Ding et al., 2003). Regardless, our studies strongly caution against interpreting studies demonstrating proteasome plasticity and proteasome biogenesis, as being indicative of conditions where proteasome activity is preserved.
Previous studies indicate that the biogenesis of the 20S occurs in an ordered set of steps starting with the formation of alpha rings onto which beta subunits can assemble, which then results in the formation of precursor complexes involving several proteasome assembly chaperones (PACs) (Ramos et al., 2008). It was previously found in mammalian cells that low concentrations of proteasome inhibitor administration resulted in enhanced gene expression of proteasome subunits leading to increased de novo proteasome biogenesis (Meiners et al., 2003; Ding et al., 2004). Proteasome inhibition with MG132 in HEK293 resulted in accumulation of PAC1, PAC-2 in fractions with fully assembled proteasomes, indicating that PACs remain associated with assembly intermediates until they are degraded upon formation of mature proteasome (Hirano et al, 2005). Our immnunoblot analysis with FPLC separated fractions of liver and brain lysates indicates that there is an age-related increase in the level of PAC-1 protein complexes in fractions containing proteasome assembly intermediates. Western blot analysis of the fractions containing PAC-1 showed the presence of alpha subunits in those fractions, but did not show the presence of beta-5 subunits (results not shown). Our immunoblot analysis also did not show any signal for the presence of PAC-1 protein in fractions containing 20S proteasomes. These studies suggest that proteasome biogenesis in the liver and brain occurs in conditions where proteasome inhibition is present. These are the first data to demonstrate increased proteasome biogenesis in liver and brain in response to aging and DR. Based on our data and previous reports we believe such increases in biogenesis may be generated in an attempt to overcome the stresses of aging by increasing the capacity of proteasome mediated protein degradation, even though such increases in biogenesis appear ultimately unable to increase the amount of proteasome activity.
Increased levels of Hsp90 were observed in 20S proteasome fractions from 25 month-old AL and DR liver, as compared to 3-month liver and brain. Previous studies demonstrate that Hsp90 interactions with the proteasome are beneficial to the preservation of proteasome function (Conconi et al., 1996; 1998). While we are not certain of the cause for this increased recruitment of Hsp90 to the 20S proteasome during aging, we believe that there are at least two likely causes for this observation. Firstly, Hsp90 may be recruited to the proteasome as the result of the increase in oxidative modifications of the 20S proteasome during aging. Alternatively, the observed changes in proteasome composition with aging may also provide a basis for increased Hsp90 recruitment to the 20S proteasome during aging. For example, it is possible that Hsp90 association with 20S may be involved in rescuing only the trypsin like activity of 20S proteasome as reported previously in rat muscle tissue (Selsby et al., 2005), due to Hsp90 preferentially interacting with selected proteasome subunits. It is important to point out that the level of Hsp90 in the crude lysates is decreased in the aging liver relative to either young or DR rodents, which was not observed in the brain, and that even with such gross reductions in Hsp90 the levels of Hsp90 interactions with the 20S proteasome were observed to be increased with aging.
An increasingly studied area of aging research is focused on the topic of hormesis, whereby low levels of stress can stimulate cells to increase the levels of protective proteins and processes (Ali and Rattan, 2006; Rattan, 2006), which can subsequently protect cells from the toxicity associated with exposure to multiple stressors. Some studies have suggested that the proteasome may play an important role in hormesis by contributing to maintenance of protein turnover during periods of stress (Ali and Rattan, 2006). In previous studies from our laboratory we demonstrated that neural cells respond to low level oxidant exposure by increasing the expression of inducible proteasome subunits (Ding et al., 2003). Our present data indicate that as part of normal aging the liver and brain exhibit altered levels of individual proteasome subunits in the 26S and 20S complexes, and exhibit increased proteasome biogenesis. The current study suggest that such alterations may not rescue or preserve proteasome function, and highlight the complexity in understanding the relationship between proteasome expression/biogenesis and proteasome activity in tissues.
Acknowledgments
This work was generously supported by grants from the NIA (AG029885, AG025771) to J.N.K.. The authors thank Dr Rafael De Cabo at the National Institutes of Health for supplying some of the tissue for these studies.
References
- Aksensova MV, Aksensova MY, Carney JM, Butterfield DA. Protein oxidation and enzyme activity decline in old brown Norway rats are reduced by dietary restriction. Mech Ageing Dev. 1998;100:157–168. doi: 10.1016/s0047-6374(97)00133-4. [DOI] [PubMed] [Google Scholar]
- Ali RE, Rattan SI. Curcumin's biphasic hormetic response on proteasome activity and heat-shock protein synthesis in human keratinocytes. Ann N Y Acad Sci. 2006;1067:394–399. doi: 10.1196/annals.1354.056. [DOI] [PubMed] [Google Scholar]
- Amici M, Lupidi G, Angeletti M, Fioretti E, Eleuteri AM. Peroxynitrite-induced oxidation and its effects on isolated proteasomal systems. Free Radic Biol Med. 2003;34:987–996. doi: 10.1016/s0891-5849(02)01369-2. [DOI] [PubMed] [Google Scholar]
- Beedholm R, Clark BF, Rattan SI. Mild heat stress stimulates 20S proteasome and its 11S activator in human fibroblasts undergoing aging in vitro. Cell Stress Chaperones. 2004;9:49–57. doi: 10.1379/475.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Brooks P, Mason GG, Rivett AJ. Gamma-Interferon decreases the level of 26 S proteasomes and changes the pattern of phosphorylation. Biochem J. 2001;353:291–297. doi: 10.1042/0264-6021:3530291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, Hendil KB, Tanaka K, Dyson J, Rivett J. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J. 2000;346:155–161. [PMC free article] [PubMed] [Google Scholar]
- Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol. 2003;35:728–732. doi: 10.1016/s1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- Cecarini V, Gee J, Fioretti E, Amici M, Angeletti M, Eleuteri AM, Keller JN. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim Biophys Acta. 2007;773:93–104. doi: 10.1016/j.bbamcr.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J Cell Biochem Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Conconi M, Szweda LI, Levine RL, Stadtman ER, Friguet B. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch Biochem Biophys. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- Conconi M, Petropoulos I, Emod I, Turlin E, Biville F, Friguet B. Protection from oxidative inactivation of the 20S proteasome by heat-shock protein 90. Biochem J. 1998;333:407–415. doi: 10.1042/bj3330407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteosomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dahlmann B, Kuehn L, Reinauer H. Studies on the activation by ATP of the 26S proteasome complex from rat skeletal muscle. Biochem J. 1995;309:195–202. doi: 10.1042/bj3090195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasuri K, Nguyen A, Zhang L, Fernandez-Kim OS, Bruce-Keller AJ, Blalock BA, Cabo RD, Keller JN. Comparison of rat liver and brain proteasomes for oxidative stress-induced inactivation: Influence of ageing and dietary restriction. Free Radic Res. 2009;43:28–36. doi: 10.1080/10715760802534812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Ding Q, Reinacker K, Dimayuga E, Nukala V, Drake J, Butterfield DA, Dunn JC, Martin S, Bruce-Keller AJ, Keller JN. Role of the proteasome in protein oxidation and neural viability following low-level oxidative stress. FEBS Lett. 2003;546:228–232. doi: 10.1016/s0014-5793(03)00582-9. [DOI] [PubMed] [Google Scholar]
- Ding Q, Bruce-Keller AJ, Chen Q, Keller JN. Analysis of gene expression in neural cells subject to chronic proteasome inhibition. Free Radic Biol Med. 2004;36:445–455. doi: 10.1016/j.freeradbiomed.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Eleuteri AM, Cuccioloni M, Bellesi J, Lupidi G, Fioretti E, Angeletti M. Interaction of Hsp90 with 20S proteasome: thermodynamic and kinetic characterization. Proteins. 2002;48:169–177. doi: 10.1002/prot.10101. [DOI] [PubMed] [Google Scholar]
- Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- Friguet B, Szweda LI. Inhibition of the multicatalytic proteinase by 4-hydroxy-2-nonenal cross linked protein. FEBS lett. 1997;405:21–25. doi: 10.1016/s0014-5793(97)00148-8. [DOI] [PubMed] [Google Scholar]
- Gong X, Kole L, Iskander K, Jaiswal AK. NRH:quinone oxidoreductase 2 and NAD(P)H:quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67:5380–5388. doi: 10.1158/0008-5472.CAN-07-0323. [DOI] [PubMed] [Google Scholar]
- Goto S, Takahashi R, Radak Z, Sharma R. Beneficial biochemical outcomes of late-onset dietary restriction in rodents. Ann N Y Acad Sci. 2007;1100:431–441. doi: 10.1196/annals.1395.048. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J Biol Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata SA. Heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell. 2006;24:977–84. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Kapphahn RJ, Bigelow EJ, Ferrington DA. Age-dependent inhibition of proteasome chymotrypsin-like activity in the retina. Exp Eye Res. 2007;84:646–54. doi: 10.1016/j.exer.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech Ageing Dev. 2000a;113:61–70. doi: 10.1016/s0047-6374(99)00101-3. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000b;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Keller JN. Age-related neuropathology, cognitive decline, and Alzheimer's disease. Ageing Res Rev. 2006;5:1–13. doi: 10.1016/j.arr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guérois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–74. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang L, Craddock J, Bruce-Keller AJ, Dasuri K, Nguyen A, Keller JN. Aging and dietary restriction effects on ubiquitination, sumoylation, and the proteasome in the heart. Mech Ageing Dev. 2008;129:515–521. doi: 10.1016/j.mad.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie JL, Kapphahn RJ, Ferrington DA. Proteasome function and protein oxidation in the aged retina. Exp Eye Res. 2002;75:271–284. [PubMed] [Google Scholar]
- Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Krüger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19S and 20S subcomplexes of the 26S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- Petropoulos I, Friguet B. Maintenance of proteins and aging: the role of oxidized protein repair. Free Radic Res. 2006;40:1269–1276. doi: 10.1080/10715760600917144. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Radák Z, Takahashi R, Kumiyama A, Nakamoto H, Ohno H, Ookawara T, Goto S. Effect of aging and late onset dietary restriction on antioxidant enzymes and proteasome activities, and protein carbonylation of rat skeletal muscle and tendon. Exp Gerontol. 2002;37:1423–1430. doi: 10.1016/s0531-5565(02)00116-x. [DOI] [PubMed] [Google Scholar]
- Ramos PC, Höckendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- Ramos PC, Dohmen RJ. PACemakers of proteasome core particle assembly. Structure. 2008;16:1296–1304. doi: 10.1016/j.str.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Rattan SI. Hormetic modulation of aging and longevity by mild heat stress. Dose Response. 2006;3:533–546. doi: 10.2203/dose-response.003.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T, Ullrich O, Sitte N, Grune T. Differential impairment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- Selsby JT, Judge AR, Yimlamai T, Leeuwenburgh C, Dodd SL. Life long calorie restriction increases heat shock proteins and proteasome activity in soleus muscles of Fisher 344 rats. Exp Gerontol. 2005;40:37–42. doi: 10.1016/j.exger.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Shibatani T, Nazir M, Ward WF. Alteration of rat liver 20S proteasome activities by age and food restriction. J Gerontol A Biol Sci Med Sci. 1996;51:B316–322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- Shibatani T, Ward WF. Effect of age and food restriction on alkaline protease activity in rat liver. J Gerontol A Biol Sci Med Sci. 1996;51:B175–B178. doi: 10.1093/gerona/51a.2.b175. [DOI] [PubMed] [Google Scholar]
- Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Sullivan PG, Dragicevic NB, Deng JH, Bai Y, Dimayuga E, Ding Q, Chen Q, Bruce-Keller AJ, Keller JN. Proteasome inhibition alters neural mitochondrial homeostasis and mitochondria turnover. J Biol Chem. 2004;279:20699–20707. doi: 10.1074/jbc.M313579200. [DOI] [PubMed] [Google Scholar]
- Tsubuki S, Saito Y, Kawashima S. Purification and characterization of an endogenous inhibitor specific to the Z-Leu-Leu-Leu-MCA degrading activity in proteasome and its identification as heat-shock protein 90. FEBS Lett. 1994;344:229–233. doi: 10.1016/0014-5793(94)00388-2. [DOI] [PubMed] [Google Scholar]
- van Tijn P, Hol EM, van Leeuwen FW, Fischer DF. The neuronal ubiquitin-proteasome system: murine models and their neurological phenotype. Prog Neurobiol. 2008;85:176–193. doi: 10.1016/j.pneurobio.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Viteri G, Carrard G, Birlouez-Aragón I, Silva E, Friguet B. Age-dependent protein modifications and declining proteasome activity in the human lens. Arch Biochem Biophys. 2004;427:197–203. doi: 10.1016/j.abb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Wagner BJ, Margolis JW. Age-dependent association of isolated bovine lens multicatalytic proteinase complex (proteasome) with heat-shock protein 90, an endogenous inhibitor. Arch Biochem Biophys. 1995;323:455–462. doi: 10.1006/abbi.1995.0067. [DOI] [PubMed] [Google Scholar]
- Whittier JE, Xiong Y, Rechsteiner MC, Squier TC. Hsp90 enhances degradation of oxidized calmodulin by the 20 S proteasome. J Biol Chem. 2004;279:46135–46142. doi: 10.1074/jbc.M406048200. [DOI] [PubMed] [Google Scholar]
- Widmer R, Ziaja I, Grune T. Protein oxidation and degradation during aging: role in skin aging and neurodegeneration. Free Radic Res. 2006;40:1259–1268. doi: 10.1080/10715760600911154. [DOI] [PubMed] [Google Scholar]
- Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Krüger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7 (beta 5i) incorporation into 20 S proteasomes. J Mol Biol. 2004;301:1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- Yamano T, Mizukami S, Murata S, Chiba T, Tanaka Kl, Udono H. Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. J Biol Chem. 2008;283:28060–28065. doi: 10.1074/jbc.M803077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li F, Dimayuga E, Craddock J, Keller JN. Effects of aging and dietary restriction on ubiquitination, sumoylation, and the proteasome in the spleen. FEBS Lett. 2007;581:5543–5547. doi: 10.1016/j.febslet.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]


