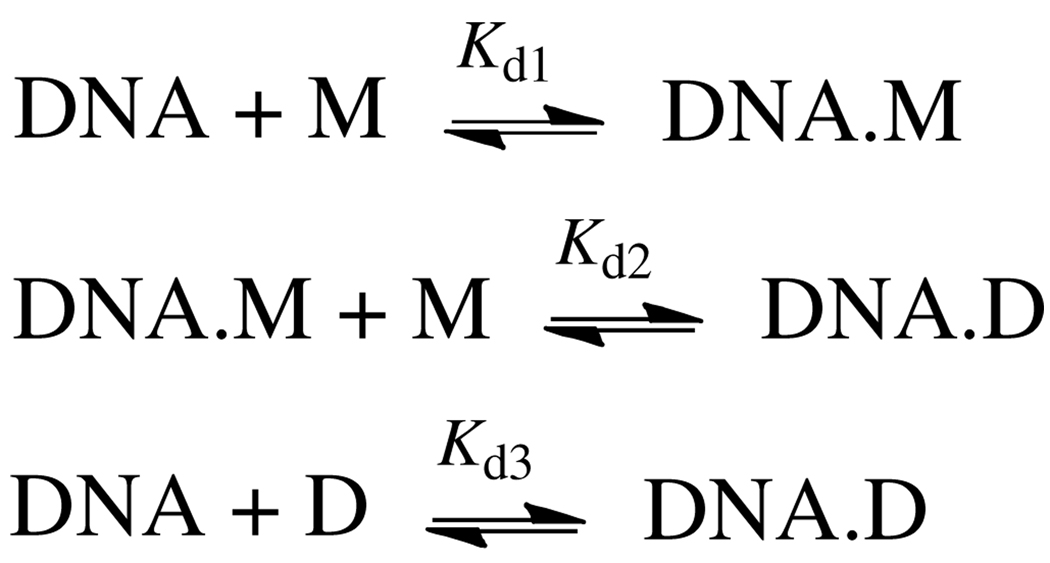Abstract
The expression of the gene products in many methicillin-resistant Staphylococcus aureus (MRSA) strains is regulated by the gene repressor BlaI. Here we show that BlaI is a mixture of monomer and dimer at in vivo concentrations, binds to the operator regions preferentially as a monomeric protein, and the measured dissociation constants and in vivo concentrations account for the basal level transcription of the resistance genes. These observations for the first time provide a quantitative picture for the processes that take place in the cytoplasm that lead to the induction of antibiotic resistance factors to counter the challenge by β-lactams.
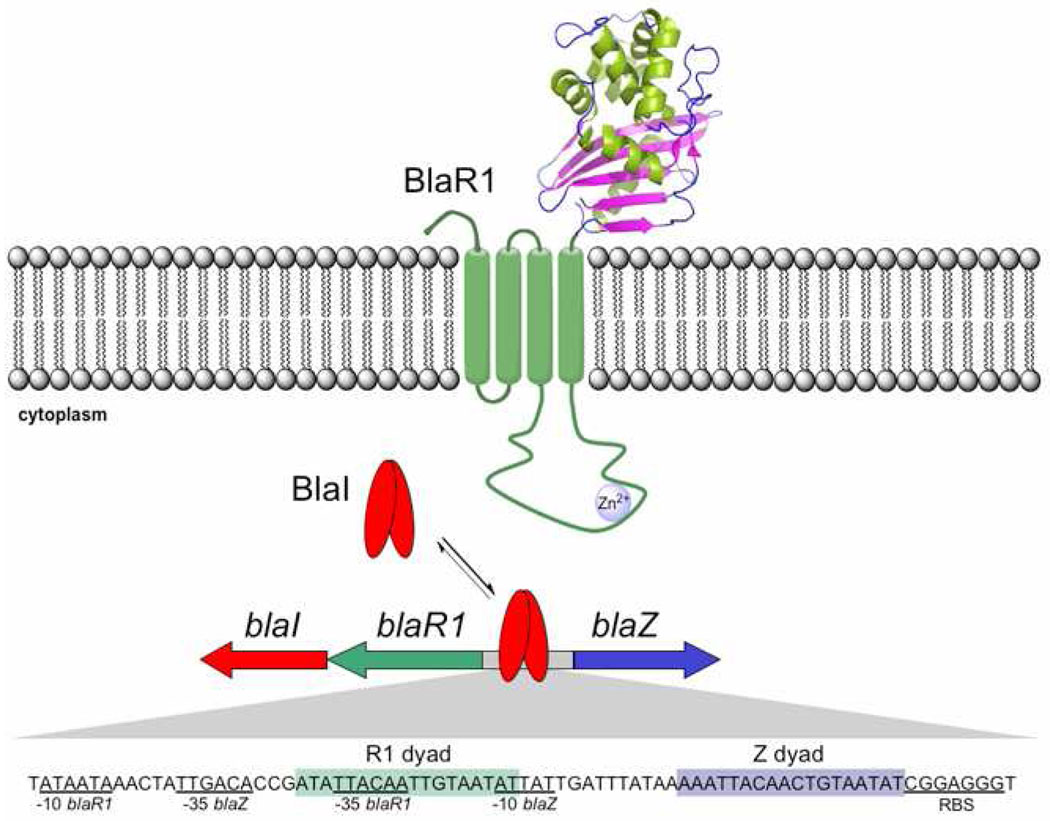
A problematic bacterial strain was identified in 1961 shortly after the introduction of the second-generation penicillins to the clinic, which has since come to be known as methicillin-resistant Staphylococcus aureus (MRSA) (1–3). MRSA remains to the present day as a global scourge (4–8). A collection of acquired genes in MRSA confers resistance to virtually all known β-lactam antibiotics. An inducible and highly regulated system in MRSA, known as the bla operon, manifests its antibiotic resistance mechanisms by expression of the PC1 β-lactamase (the product of the blaZ gene) and/or a unique penicillin-binding protein, PBP2a (the product of the mecA gene). Expression of the PC1 β-lactamase is governed by a membrane-bound β-lactam sensor/signal transducer protein, BlaR1, which senses the presence of the antibiotic in the milieu and regulates transcription of the antibiotic resistance genes, including its own (i.e., blaR1) (9–14). This regulation is via derepression of the requisite genes by the proteolytic activity of the cytoplasmic domain of BlaR1 (15). It degrades the gene repressor BlaI (blaI gene product), which leads to transcription of the genes blaI, blaR1 and blaZ (Figure 1). The influence of BlaI on gene transcription is central in manifestation of the deleterious MRSA phenotype, a process that is the subject of investigation in this report.
Figure 1.
The sensor and repressor proteins of the bla operon are involved in the onset of β-lactam resistance in MRSA.
We amplified by PCR the blaI gene of plasmid pI258 from S. aureus NRS128 (NCTC 8325) and cloned it in plasmid pET-24a(+). The protein BlaI was expressed and purified to homogeneity (Supporting Information). The bla operator consists of two distinct binding sites for the repressor, referred to as the R1 dyad (which is upstream of the blaR1 gene) and the Z dyad (located upstream of the blaZ gene; Figure 1).
The R1 dyad contains an 18-bp palindrome, while the Z dyad contains an 18-bp imperfect palindrome (16). A 13-bp linker separates the two dyads. We synthesized three pieces of double-stranded DNA, corresponding to the full-length bla operator and the individual R1 and Z dyads. One strand in each case was tagged by fluorescein at the 5’-end to make it suitable for fluorescence anisotropy measurements on binding to BlaI. The single-stranded synthetic DNA pieces were purified by HPLC, before annealing with their partner strands for use in anisotropy experiments. With these reagents in hand, we were poised to evaluate interactions of the gene repressor with the regulatory regions of the bla operator.
Sedimentation equilibrium experiments showed that BlaI is present in solution as both monomeric and dimeric species, with a dissociation constant, Kd, of 1.61 ±0.02 µM for the monomer-dimer equilibrium (Supporting Information). Subsequently, we evaluated the concentrations of BlaI in living MRSA strains S. aureus NRS70 and NRS128. The results were similar for both strains, with BlaI concentrations of 2.3–6.4 µM (NRS128) and 1.3–3.6 µM (NRS70) in the cells at the exponential phase of growth and of 0.98–2.7 µM (NRS128) and 0.75–2.1 mM (NRS70) in the stationary phase (Supporting Information). The approximately three-fold range for these in vivo determinations is a factor of the measured radii for S. aureus cells, which vary as much as 1.4-fold (17; 18). When this difference in radii is taken to the power of three in calculation of the cell volume, the difference amplifies to approximately three-fold. Based on the dimerization dissociation constant and the total concentration of BlaI in S. aureus, 45–70% of BlaI in the exponential growth phase and 59–78% in the stationary phase exist in the monomeric form. In the case of MRSA strains exposed to the penicillin CBAP, a high resistance inducer (19; 20). we could not detect by Western blotting the presence of any intact BlaI, nor any accumulated fragments thereof. This finding indicates that BlaI is turned over effectively by the protease action of the cytoplasmic domain of BlaR1 on exposure of the organism to the antibiotic.
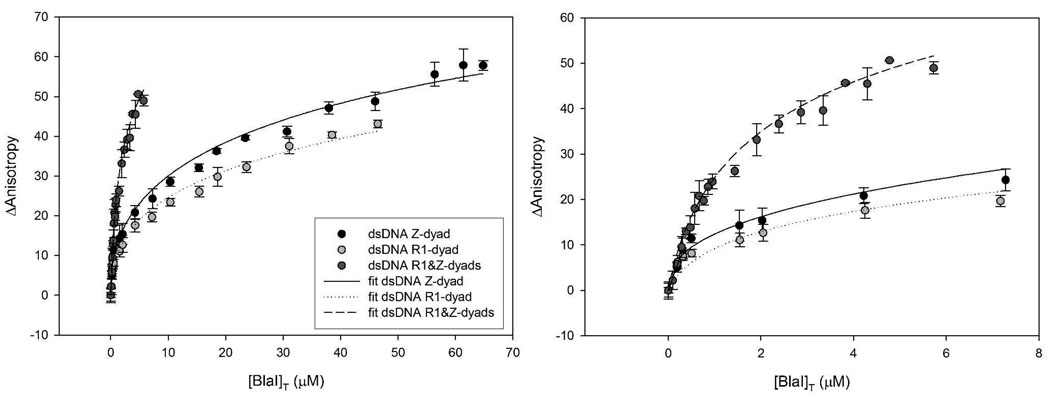
Next, we evaluated binding of the purified BlaI to the operator region of the bla operon by fluorescence anisotropy (Figure 2). The anisotropy data were fit using equations derived for different models (Supporting Information), taking into account the possibility of binding of BlaI as a monomer and/or dimer. The best fits were achieved for binding of BlaI to the given DNA stretch as a monomer (Kd1), as a second monomer binding to the complex of DNA-BlaI monomer (Kd2), and as a BlaI dimer binding to DNA (Kd3)(Table 1). From the consideration of the in vivo concentrations of BlaI that we reported above and from the results shown in Table 1, it is obvious that the BlaI monomer binding to any of these stretches of DNA is favorable. Furthermore, binding of the dimer to the Z dyad (that controls transcription of the blaZ gene) alone or to the full-length operator appears to be very favorable. The in vivo BlaI concentrations would appear to disfavor largely the possibility of the second monomer binding (the Kd2 event), although in the case of the full-length operator this might play a limited role in vivo (Table 1). Regardless, the in vitro binding studies of Fig. 2, which were not limited by the in vivo concentrations of BlaI, indicate an influence on the change in DNA anisotropy for the full-length operon, as revealed by a more favorable (among the three dsDNA pieces) Kd2 component. It is important to note that the data did not fit to models of simultaneous binding of two BlaI monomers to any of the dsDNA or of simultaneous binding of two dimers to the two sites of the full-length operator.
Figure 2.
Fluorescence anisotropy measurement of BlaI binding to DNA. The lines show the corresponding fits to the model shown in Scheme 1, where the equilibrium between BlaI monomer and dimer is characterized by Kd. The equations used are described in Supporting Information. The panel on the right is the same as the image on the left panel, except that the plot is given for a more narrow concentration range.
Table 1.
Binding of BlaI to dsDNA. Dissociation constants obtained from the fit of the anisotropy data to the model presented in Scheme 1. The anisotropy data were fit using equation E1, shown in Supporting Information.
| Kd1 (µM) | Kd2 (µM) | Kd3 (µM) | |
|---|---|---|---|
| R1 dyad | 0.8 ± 0.1 | 20 ± 1 | 10 ± 2 |
| Z dyad | 0.05 ± 0.04 | 10.7 ± 0.3 | 0.3 ± 0.3 |
| R1-Z dyads | 0.45 ± 0.07 | 2.6 ± 0.1 | 0.72 ± 0.07 |
In S. aureus NRS128, the bla operon is present in the low-copy plasmid pI258 (~5 copies per cell) (21), for which we estimate the in vivo concentration at 5–15 nM, based on the aforementioned variability of the cell radii. This in vivo determination, in conjunction with the dissociation constants of BlaI and dsDNA and the in vivo concentrations of BlaI that we disclosed above, indicates that the system has evolved to be exquisitely responsive to small fluctuations of the BlaI concentration, which is modulated by the proteolytic activity of BlaR1, once exposed to the antibiotic in the milieu. That is to say, proteolysis of a small fraction of BlaI within the cytoplasm would liberate a sufficient amount of the DNA from repression to allow manifestation of antibiotic resistance. Our simulations of these parameters based on the three binding equilibria that are operative simultaneously and the in vivo BlaI concentrations revealed that as much as 10% of the DNA should be uncomplexed by BlaI within MRSA in the absence of antibiotic. The implication of this observation is that these genes should be “leaky”, indicating that they would undergo a basal level of transcription, in the absence of antibiotic. This is exactly what is seen (16; 22).
A different situation was observed for binding of BlaI from Bacillus licheniformis to the three operator sequences in the bla operon of the same organism (23). In this case, no BlaI monomer.DNA complex was observed in band shift assays. Furthermore, the dissociation constants that were evaluated were in the low nanomolar range, hence tighter binding. The total BlaI concentration in B. licheniformis is 1.9 µM (23), similar to the concentrations that we measure for BlaI in S. aureus, but the dimer/monomer dissociation constant in B. licheniformis was evaluated at 25 µM, much larger than that of S. aureus, which indicates that BlaI is present mainly as monomer in the cytoplasm of B. licheniformis. The absence of BlaI monomer.DNA complex indicates that in B. licheniformis two favorable sequential events of monomer binding to DNA have to occur with a high degree of cooperativity (23), which was observed. Based on our disclosure here, the situation in S. aureus is different, which reveals the evolution of distinct regulatory systems in these two bacterial genera.
Whereas the structure of BlaI bound to the bla operator DNA is not known to date, the X-ray structure of the BlaI dimer in complex with a 32-bp dsDNA corresponding to the mec palindromic operator has been solved (24). One difficulty in elucidating the structure of the monomer complex with DNA is that the concentrations of BlaI and DNA needed for crystallization are well above the dissociation constants for formation of the dimer.DNA complex (Kd2 and Kd3), and hence the prevailing complex under the crystallization conditions would be the dimer.DNA complex.
As stated previously, the parameters here described for binding of BlaI to the two sequences in the operator region indicate a basal level of transcription of the bla operon in S. aureus which is critically important for the formation of sufficient BlaI to maintain transcriptional regulation and of sufficient BlaR1 and minimal β-lactamase as vanguards against the initial exposure to the antibiotic. Once exposure to the antibiotic takes place, the protease domain of BlaR1 is unleashed by its activation—through an unknown mechanism—for degradation of BlaI, which derepresses the genes to allow full-blown production of BlaR1 and the β-lactamase to meet the challenge of the antibiotic. To put it succinctly, BlaI in MRSA has evolved for its relatively poor ability to bind to the bla operon (nanomolar to micromolar)—in contrast to the case of the B. licheniformis—positioning the system just on the threshold of derepression of the genes that it controls.
Supplementary Material
Scheme 1.
Model for BlaI binding to DNA. Designations M and D denote BlaI monomer and dimer, respectively.
ACKNOWLEDGMENT
This work was supported by the National Institute of Health. Leticia I. Llarrull, Ph.D., is a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts. The opinions expressed are those of the authors and do not necessarily reflect the views of The Pew Charitable Trusts.
Footnotes
Supporting Information available: Procedures for blaI cloning and BlaI purification; BlaI sedimentation equilibrium experiments; BlaI quantification in S. aureus cells; binding of BlaI to operon regions by fluorescence anisotropy; estimation of the concentration of DNA bound species in vivo. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Jevons MP. British Medical Journal. 1961:124–125. [Google Scholar]
- 2.Knox R. British Medical Journal. 1961:124–125. [Google Scholar]
- 3.Rolinson GN. British Medical Journal 1961. 1961:125–126. [Google Scholar]
- 4.Boucher HW, Corey GR. Clin. Infect. Dis. 2008;46 Suppl 5:S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 5.Loffler CA, Macdougall C. Expert. Rev. Anti. Infect. Ther. 2007;5:961–981. doi: 10.1586/14787210.5.6.961. [DOI] [PubMed] [Google Scholar]
- 6.Llarrull LI, Fisher JF, Mobashery S. Antimicrob. Agents Chemother. 2009;53:4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durai R, Ng PC, Hoque H. AORN J. 2010;91:599–606. doi: 10.1016/j.aorn.2009.11.065. quiz 607–609. [DOI] [PubMed] [Google Scholar]
- 8.Gastmeier P. Int. J. Med. Microbiol. 2010 doi: 10.1016/j.ijmm.2010.04.007. doi:10.1016/j.ijmm.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Lewis RA, Curnock SP, Dyke KG. FEMS Microbiol. Lett. 1999;178:271–275. doi: 10.1111/j.1574-6968.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 10.Gregory PD, Lewis RA, Curnock SP, Dyke KG. Mol. Microbiol. 1997;24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 11.Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. J. Am. Chem. Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 13.Cha J, Mobashery S. J. Am. Chem. Soc. 2007;129:3834–3835. doi: 10.1021/ja070472e. [DOI] [PubMed] [Google Scholar]
- 14.Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. J. Biol. Chem. 2003;278:18419–18425. doi: 10.1074/jbc.M300611200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 16.Clarke SR, Dyke KG. J. Antimicrob. Chemother. 2001;47:377–389. doi: 10.1093/jac/47.4.377. [DOI] [PubMed] [Google Scholar]
- 17.Wyatt PJ. Nature. 1970;226:277–279. doi: 10.1038/226277a0. [DOI] [PubMed] [Google Scholar]
- 18.Beltramini AM, Mukhopadhyay CD, Pancholi V. Infect. Immun. 2009;77:1406–1416. doi: 10.1128/IAI.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leggate J, Holms WH. J. Bacteriol. 1968;96:2110–2117. doi: 10.1128/jb.96.6.2110-2117.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettinger GE, Lampen JO. J. Bacteriol. 1970;104:283–288. doi: 10.1128/jb.104.1.283-288.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Appl. Environ. Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke SR, Dyke KG. Microbiology. 2001;147:803–810. doi: 10.1099/00221287-147-4-803. [DOI] [PubMed] [Google Scholar]
- 23.Filee P, Vreuls C, Herman R, Thamm I, Aerts T, De Deyn PP, Frere JM, Joris B. J. Biol. Chem. 2003;278:16482–16487. doi: 10.1074/jbc.M210887200. [DOI] [PubMed] [Google Scholar]
- 24.Safo MK, Zhao Q, Ko TP, Musayev FN, Robinson H, Scarsdale N, Wang AH, Archer GL. J. Bacteriol. 2005;187:1833–1844. doi: 10.1128/JB.187.5.1833-1844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.