Abstract
Meiosis in triploids results in four highly aneuploid gametes because six copies of each homolog must be segregated into four meiotic products. Using DNA microarrays and other physical approaches, we examined meiotic chromosome segregation in triploid strains of Saccharomyces cerevisiae. In most tetrads with four viable spores, two of the spores had two copies of a given homolog and two spores had only one copy. Chromosomes segregated randomly into viable spores without preferences for generating near haploid or near diploid spores. Using single-nucleotide polymorphisms, we showed that, in most tetrads, all three pairs of homologs recombined. Strains derived from some of the aneuploid spore colonies had very high frequencies of mitotic chromosome loss, resulting in genetically diverse populations of cells.
POLYPLOIDY is very common in plants; for example, the frequency of polyploidy in angiosperms is 30–80% (Hegarty and Hiscock 2008). Most polyploidy is thought to arise by whole-genome duplication of diploids, producing tetraploids. One mode of producing a triploid is by union of a haploid gamete produced by a “normal” diploid with a diploid gamete produced by a tetraploid. Triploids produced by this mechanism have been detected in animals as well as plants. In a region with both diploid and tetraploid Palearctic green toads, triploid male toads resulting from crosses of diploid females and tetraploid males were observed (Stock et al. 2009). Triploids can also arise from a cross of diploid individuals in which one individual has tetraploid germinal tissue (Bridges and Anderson 1925). Although the fertility of triploid Drosophila is reduced, viable offspring between diploids and triploids can be readily obtained. In humans, triploidy is responsible for 15–18% of spontaneous abortions, but only 1 in 1200 fetuses live after birth; all of the live-born individuals die within a few months (Iliopoulos et al. 2005).
Triploid yeast strains have been observed in the wild (Ezov et al. 2006) and have been generated in the lab by forced matings between haploids and diploids (Pomper et al. 1954). Since diploids of the MATa/MATα genotype do not mate, it is likely that the forced matings selected for rare diploids that had become homozygous for MATa or MATα as a consequence of a mitotic recombination at the mating-type locus on chromosome III. In most recent studies, triploids are generated by mating haploids to diploids that have been constructed to be homozygous at the mating-type locus (Parry and Cox 1970; Campbell et al. 1981; Campbell and Doolittle 1987).
The meiotic products derived from sporulating a triploid would be expected to be highly aneuploid, containing chromosome numbers varying between the haploid number of 16 and the diploid number of 32. Since even extra copies of single chromosomes often adversely affect cellular growth rates (Torres et al. 2007), it is unsurprising that the viability of spores derived from triploids is lower than those derived from diploids. There is, however, a wide range of spore viabilities reported in different labs varying between 15 and 18% (Parry and Cox 1970; Campbell et al. 1981) to about 75% (Pomper et al. 1954).
The first detailed analysis of chromosome segregation in viable spores derived from triploids was carried out by Parry and Cox (1970). The viable spores were crossed with haploids that had multiple auxotrophic markers and the resulting diploids were sporulated. Departures from 2:2 segregation of the markers were used to diagnose aneuploidy. Markers on 14 of the 16 yeast chromosomes were used. Of 34 segregants examined, 3 had one copy of all chromosomes tested, and 2 had two copies of all chromosomes tested. The other 29 segregants had between one and five disomic chromosomes. Parry and Cox (1970) noted that disomes of chromosomes V, VIII, IX, XII, and XIII were more frequently observed in the viable spores than disomes of the other chromosomes. In addition, they suggested that the ability of the triploid-derived spores to tolerate aneuploidy was limited, since most of the spores had less than five disomic chromosomes or were diploid.
A more extensive analysis of viable spores derived from triplods was done by Campbell et al. (1981). For chromosome III, they observed that about half of the spores had one chromosome and about half had two. They concluded that during meiotic segregation in the triploid, two spores received two copies of each homolog and two received one. For most chromosomes, the frequency of spores with two copies was significantly less than the frequency with one copy. They attributed this difference to loss of disomic chromosomes during mitotic growth of the spore cultures.
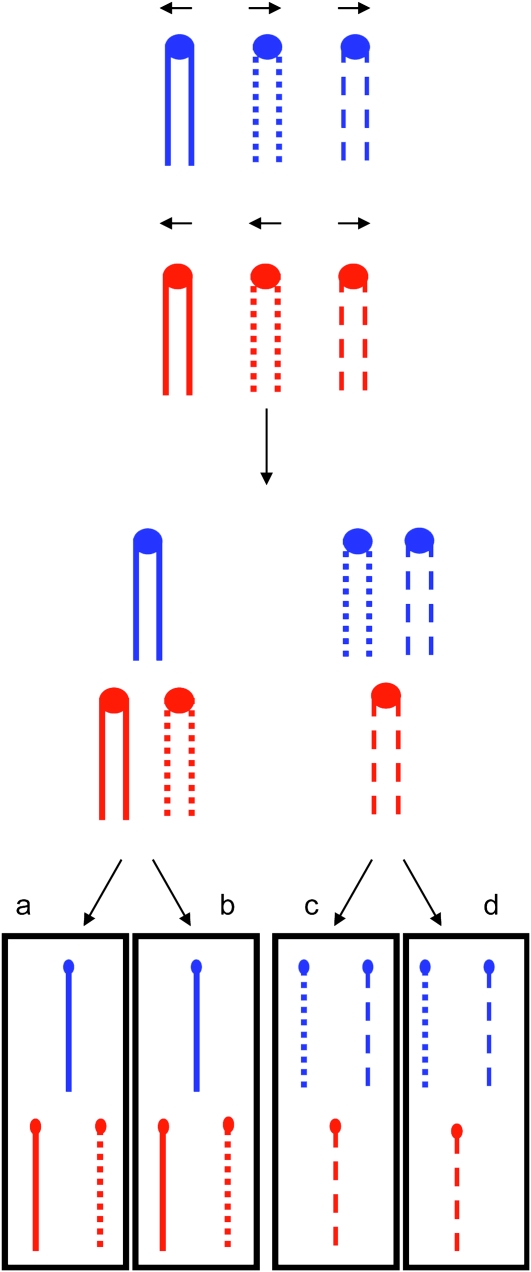
In a subsequent study, Campbell and Doolittle (1987) examined chromosome compositions in triploid-derived tetrads that had two viable spores. If these two viable spores are sister spores either both should have two copies of any given chromosome or both spores should have one copy (Figure 1); below, we use the term “monosomy” to describe spores that have only one copy of a homolog, although this term is also used to describe diploid cells that lack a single chromosome. These two classes of events should be equally frequent. The observed frequencies of tetrads in which both viable spores were disomic for the same chromosome varied between 0 (chromosome VII) and 0.5 (chromosome XIII) with an average of about 0.19 (Campbell and Doolittle 1987). The deviation between the expected (0.5) and observed frequencies of two disomic spores was explained as a consequence of mitotic chromosome loss.
Figure 1.—
Expected meiotic segregation pattern of chromosomes of a triploid. In this figure, we show the meiotic segregation of two homologs in blue and in red. The three copies of each homolog are shown as lines with long dashes, short dashes, or an uninterrupted straight line. Following the second meiotic division, one would expect to get two disomic spores and two monosomic spores for each chromosome. In this depiction, the sister spores “a” and “b” are disomic for the red homolog and monosomic for the blue homolog, whereas the sister spores “c” and “d” have the reciprocal pattern. With equal frequency, one would observe a pair of doubly disomic sister spores and a pair of spores that are monosomic for both chromosomes. In this figure, we have not shown recombination between the homologs, although such exchanges occur in most tetrads.
Because of the low spore viability, meiotic recombination in triploid yeast has not been examined previously. However, meiotic recombination in otherwise diploid cells that are trisomic or tetrasomic for a single chromosome has been studied by a number of researchers. Shaffer et al. (1971) suggested two possible mechanisms by which trisomic chromosomes can pair, recombine, and segregate during meiosis. According to the bivalent/univalent-pairing model, two homologs pair, recombine, and segregate to opposite poles while the third homolog randomly segregates to either pole. By the second model (trivalent pairing), all three homologs pair, possibly recombine, and randomly segregate to either pole. A distinguishing feature of the trivalent model is that two chromosomes that have recombined with each other are capable of segregating to the same pole during meiosis I. The majority of studies support the trivalent-pairing model (Shaffer et al. 1971; Culbertson and Henry 1973; Riley and Manney 1978; Koller et al. 1996). From the genetic data, it is impossible to determine whether all three homologs pair and recombine at the same time or whether there are multiple cycles of pairing and recombination although, by visualizing the synaptonemal complex in triploid strains, Loidl (1995) showed that simultaneous pairing of all three homologs was common.
Some of the inferences concerning meiotic segregation and recombination in past studies of triploids were indirect for two reasons. First, not all of the meiotic products from a single tetrad were examined. Second, mitotic chromosome loss occurred during the time-consuming assay of disomic chromosomes during growth of the spore cultures or in the diploid strains derived from mating the spore cultures. In the current study, we examine the meiotic segregation of chromosomes in triploid yeast in tetrads with four viable spores using physical methods (microarray analysis and a PCR-based examination of single-nucleotide polymorphisms) that are more rapid and sensitive than the methods used previously. We show that meiotic chromosome segregation in triploids is generally accurate, resulting in two spores with two copies and two spores with one copy of each homolog. The mean number of aneuploid chromosomes for each spore is initially close to 8, but we show directly that mitotic chromosome loss in spore cultures can be very rapid. In addition, we show that triploid chromosomes, like trisomic chromosomes, usually undergo trivalent pairing.
MATERIALS AND METHODS
Strain construction:
Our experiments were conducted using two triploid strains: MH10 and JSC2. The three copies of each homolog in MH10 are identical except for alterations introduced by transformation, whereas the three copies of each homolog in JSC2 are derived from three diverged haploid strains. The genotypes of all strains used in our study are given in supporting information, File S1 and Table S1. MH10 was generated by crossing MS71 (MATα strain) to JLMy133, an isogenic diploid (MATa/matα∷URA3). Thus, all three homologs of MH10 have the same DNA sequence except for the mating-type locus on chromosome III. The JSC2 triploid was constructed to contain three diverged homologs of each yeast chromosome. JSC2 was generated by mating the haploid PSL4 to the diploid JSC1. The haploid PSL4 (MATα mating type) was derived from the YJM789 genetic background (Wei et al. 2007; Lee et al. 2009). The JSC1 diploid is a MATa/matα∷NAT derivative of JAY306, a diploid generated by crossing FY834 (a haploid isogenic with S288c; Winston et al. 1995) with JAY291 (a recently sequenced haploid derived from an ethanol-producing strain; Argueso et al. 2009).
Genetic methods and media:
We used standard genetic and media protocols (Guthrie and Fink 1991). Strains were mated by mixing the parental strains on a plate containing rich growth medium (YPD) and incubating the mixture for 6 hr at 30°. We then picked zygotes and sporulated the resulting triploids. Following tetrad dissection of sporulated triploids, the dissection plates were incubated at 30° for up to 5 days. The entire spore colony of tetrads with four viable spores was transferred as a patch to a YPD plate and grown for 1–2 more days at 30°. Glycerol stocks were then prepared using the entire patch.
Comparative Genome Hybridization (CGH) microarrays:
DNA for CGH microarrays was isolated from 5 ml YPD cell cultures grown for 1–2 days at 30°. The isolation procedure was the same as that used for preparing DNA samples for contour-clamped homogeneous electric field (CHEF) gel analysis (Narayanan et al. 2006). DNA was isolated from the agarose plugs using a QIAgen gel extraction kit. After the plugs were dissolved with buffer QG from the QIAgen kit, the DNA was sonicated. After sonication, DNA was isolated using the standard QIAgen protocol. DNA from spore cultures was labeled with Cy5–dUTP and the control DNA was labeled with Cy3–dUTP (Lemoine et al. 2005). For the analysis of DNA from MH10-derived spore cultures, MS71 was used as the control DNA; for JSC2-derived spore samples, we used JSC2 DNA for the control. The experimental and control DNAs were competitively hybridized overnight at 65° to microarrays that contained both ORFs and intergenic regions from the yeast genome (Argueso et al. 2008).
Following hybridization, the slides were scanned using a GenePix 4000B scanner and GenePix Pro 6.0 software. Data files were analyzed using the University of North Carolina Microarray Database (http://genome.unc.edu). The log2 ratios (hybridization values of experimental and control DNA samples) for each element on the array were first normalized using the log2 median ratio for the whole data set. If the experimental strain has a mixture of disomic and monosomic chromosomes, following this normalization, the monosomic chromosomes are expected to have an average log2 ratio of less than zero. Consequently, we determined the average log2 ratio for all chromosomes with a log2 ratio less than zero and added this number to the average log2 ratio for each individual chromosome. If this sum was greater than 0.17, then the chromosome was considered disomic; if it was less than 0.17, then the chromosome was considered monosomic. A log2 ratio of 0.17 represents disomy of a chromosome in 12.5% of the population. The patterns of hybridization were depicted using CGH Miner (Wang et al. 2005).
Diagnostic PCR and restriction digests:
The primers listed in Table S2 were used to PCR amplify polymorphic regions near the centromere using genomic DNA from JSC2 spores. By treating the resulting fragments with various restriction enzymes (Table S2), we could determine the source of the homologs (S288c, YJM789, or JAY291) in each spore. The identity of chromosomes XIII and XIV were also diagnosed by amplification of centromeric regions that generated DNA fragments differing by 9–13 bp in size. These PCR products were resolved on a 2% agarose gel. Right and left arm identities were also diagnosed using this method with primers located near the telomeres of each chromosome arm. Primers and restriction enzymes used to diagnose these distal markers are listed in Table S3. We also used PCR methods to specifically amplify the centromere-linked JAY291 SNP on chromosome I and the YJM789-specific polymorphism on chromosome XIII. The names and sequences of the primers used for this analysis were DIST 1.1F (5′ CCACGCCAGGGAATCATCC) and DIST 1.1R (5′ TGCTACAGCATCTCGGCCC) for chromosome I and DIST 13.1F (5′ TTGTCCATAGCAGCAATCATACCAGCACCACC) and DIST 13.2R (5′ AGAAACGCGTCTGGCTTATCTACCGC) for chromosome XIII.
Statistical analysis:
The VassarStats Website for Statistical Computation (http://faculty.vassar.edu/lowry/VassarStats.html) was used for the chi-square goodness-of-fit tests and Fisher exact tests. We used the Benjamini and Hochberg (1995) test to control for the false discovery rate when 12 or more comparisons were made.
RESULTS
Description of experimental methods:
We examined meiotic chromosome segregation in two different types of triploids. In the triploid MH10, all three copies of each homolog were identical in DNA sequence, except for a very small number of changes introduced by transformation. In the triploid JSC2, the three copies of each homolog were derived from three different haploid yeast strains, each of which is diverged in DNA sequence from the other two by about 0.1%. As explained below, this sequence divergence allowed a sensitive assay for disomy in spores derived from JSC2 and also allowed detection of meiotic recombination events. Diploids derived from the diverged haploids had good spore viability, indicating that these strains do not have translocations or inversions (data not shown).
Both triploid strains were sporulated and dissected. Spore viability for both strains was about 50% (Table S4). This spore viability is considerably higher than that observed by Parry and Cox (1970) and Campbell et al. (1981) (15% and 17.9%, respectively) but less than that observed by Pomper et al. (1954) (69–83%). In addition, the distribution of the classes of tetrads (4 live:0 dead, 3 live:1 dead, 2 live:2 dead, 1 live:3 dead, 0 live:4 dead) deviated significantly from that expected by a binomial expansion (Fisher exact test, P < 0.001; Table S4). We do not know the reasons for the differences in spore viabilities in different studies. The differences may reflect technical issues (for example, the length of time that asci were treated with glusulase) or variation in the tolerance of aneuploidy in different genetic backgrounds.
About 16% of the tetrads derived from MH10 and JSC2 had four viable spores. All cells in each spore colony were transferred as patches to rich growth solid medium and frozen stocks were prepared. DNA was isolated for microarray analysis by growing large inocula derived from the frozen stocks. This protocol was designed to limit loss or duplication of chromosomes in the mitotic divisions following meiosis. We examined DNA isolated from all four spores for 10 tetrads derived from MH10 and 6 tetrads derived from JSC2. The chromosome compositions of all of these spore cultures were examined by CGH microarrays; we also determined that both of the starting triploids were euploid. In addition, we analyzed the chromosomes in JSC2 spore cultures by examining SNPs as described below.
Analysis of meiotic segregants of the triploid MH10:
Figure 2 shows a representative example of analysis of DNA samples from four spores of an MH10 tetrad by CGH microarrays. DNA from MH10 spores and an isogenic haploid reference strain (MS71) were labeled with different fluorescent nucleotides (Cy3 and Cy5) and competitively annealed to a microarray containing all yeast ORFs and intergenic regions. In the depiction of the microarray shown in Figure 2, each of the 16 chromosomes is represented by a line. Along each line are vertical lines whose length and position represents relative hybridization differences between the reference and sample genomic DNA at each element of the microarray. Red and green represent duplications or deletions of the experimental strain with respect to the control strain. For example, the spore MH10-32a (Figure 2A) was disomic for chromosomes X and XIII. In many of the microarrays, chromosomes had significantly elevated hybridization signals, although the hybridization level was not twofold higher than for the other chromosomes. It is likely that this effect is a consequence of loss of the disomic chromosomes in some fraction of the subcultured cells during vegetative growth.
Figure 2.—
CGH microarray analysis of DNA derived from four spores of a tetrad derived from the triploid MH10. (A) Analysis of the MH10-32 tetrad by CGH microarray. DNA was isolated from individual spore cultures and labeled with Cy5-tagged nucleotides. DNA from the isogenic haploid strain was labeled with Cy3-tagged nucleotides. The individual samples from spore cultures were mixed with the labeled control DNA and the mixture was hybridized to microarrays containing all ORFs and intergenic region. Each chromosome is depicted by a horizontal line whose length is proportional to the length of the chromosome. Red indicates gene amplification relative to the reference strain. The meiotic sister pairs of spores are MH10-32a/MH10-32c and MH10-32b/MH10-32d. Extensive loss of disomic chromosomes is evident in spore MH10-32b. (B) Photograph of MH10-32 spore colonies on the tetrad dissection plate.
The spore MH10-32c had the chromosome composition identical to that of MH10-32a (Figure 2A). The other two spores, MH10-32b and MH10-32d, were both disomic for chromosomes I, II, IX, XI, and XIV. MH10-32d was also disomic for chromosomes III, V-VIII, XII, XV, and XVI. The nonsister pairs of spores had nonoverlapping patterns of disomy, and all chromosomes were disomic in at the least one spore except chromosome IV, the largest chromosome. From the pattern of chromosome segregation shown in Figure 1, one would expect that two pairs of spores (the sister spores) should have reciprocal patterns of disomy, and, for each homolog, one pair of sisters would have two copies and one pair would have one copy. Although chromosomes I, II, IX, X, XI, XIII, and XVI met this expectation, the other chromosomes did not. It should be noted that the pairs of spores that had similar patterns of disomic chromosomes had growth properties similar to those of spore colonies on the dissection plates (Figure 2B).
One simple explanation of this discrepancy is that the disomic chromosomes are frequently lost during mitotic growth. For example, the sister spores MH10-32a and MH10-32c initially may have had two copies of chromosomes X and XIII and one copy of all of the other chromosomes. The sister spores MH10-32b and MH10-32d had one copy of chromosomes X and XIII and two copies of all of the other chromosomes. The strain MH10-32b subsequently lost many of the chromosomes originally present in two copies, whereas MH10-32d lost only chromosome IV. It is also possible that sister spores MH10-32a and MH10-32c were originally disomic for chromosome IV, but the extra chromosome was lost in both spore cultures. One alternative explanation of the results is that chromosome loss is very frequent during meiotic segregation in triploids at meiosis I (leading to loss of two copies) or meiosis II (leading to loss of one copy). Another possibility is that there is a high rate of chromosome loss in the triploid prior to meiosis. These issues are discussed in more detail below.
We examined nine other tetrads with four viable spores derived from MH10 by CGH arrays and these data are summarized in Table 1. For the most part, only meiotic sisters were disomic for the same chromosome in any given tetrad. Most tetrads consisted of colonies that were of two different sizes that corresponded with meiotic sisters as determined by disomy of the same chromosome (Table 1).
TABLE 1.
CGH analysis of disomy and monosomy in spores derived from the triploid MH10
| Chromosomes |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spore | Colony size | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI |
| 4a | Small | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 4b | Big | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2a | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| 4c | Small | 2 | 1 | 2 | 1 | 2 | 2a | 2 | 2a | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| 4d | Big | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| 22a | Small | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| 22b | Big | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 |
| 22c | Small | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| 22d | Big | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24a | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| 24b | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| 24c | Small | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| 24d | Small | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 |
| 25a | Small | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| 25b | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 |
| 25c | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 |
| 25d | Big | 2 | 2 | 1 | 2 | 2a | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 |
| 28a | Small | 1 | 2a | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 2 |
| 28b | Big | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| 28c | Small | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 28d | Big | 2 | 2a | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 |
| 29a | Average | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 29b | Average | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 29c | Average | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 29d | Average | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 30a | Big | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| 30b | Small | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| 30c | Big | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| 30d | Small | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| 32a | Small | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| 32b | Big | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 1 |
| 32c | Small | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 |
| 32d | Big | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 |
| 34a | Small | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| 34b | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 2 |
| 34c | Small | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| 34d | Big | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 37a | Average | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 |
| 37b | Average | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| 37c | Average | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| 37d | Average | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| Averageb | 5.7 | 5.7 | 5.3 | 4.7 | 5.7 | 4.9 | 5.2 | 5.7 | 5.4 | 5.5 | 5.7 | 5.1 | 5.1 | 5.5 | 4.9 | 5.2 | |
Based on CGH microarrays, we determined whether the spore cultures were disomic (indicated by 2 in the table) or monosomic (indicated by 1) for each homolog. Boldface indicates a tetrad in which one sister spore was monosomic, although the other sister was disomic, indicating a chromosome loss event in the monosomic spore.
In these spores, there was a possible chromosome gain. The extra chromosomes were not counted in the averages at the bottom of the table.
Average number of individual homologs per tetrad.
After we compiled the results of all 10 tetrads analyzed by CGH microarray, it was apparent that spores derived from the triploids are subject to extensive chromosome loss (Table 1). Assuming that there should be six chromosome copies of each homolog per tetrad (following meiotic DNA synthesis), there should be 96 chromosomes summed over the four spores of the tetrad. On average, however, each tetrad had about 85 chromosomes, representing a loss of about 11 chromosomes distributed over the four spores. Chromosomes IV, VI, and XV were lost at relatively high frequencies and chromosomes I, II, V, VIII, and XI at relatively low frequencies (Table 1).
Previous studies have already shown that spores derived from triploids have unstable karyotypes (Parry and Cox 1970; Campbell et al. 1981), although direct measurements of chromosome stability in the spores were not done. In addition, these studies did not determine whether meiotic chromosome segregation was fundamentally accurate in a triploid (Parry and Cox 1970; Campbell et al. 1981). To investigate this issue with a more sensitive assay for aneuploidy than CGH arrays, we analyzed sequence polymorphisms that distinguished the three copies of each homolog in the triploid JSC2.
Analysis of meiotic chromosome segregation in a triploid (JSC2):
We generated the triploid JSC2 with equal contributions of DNA sequences from three different sequenced backgrounds: S288c, YJM789 (Wei et al. 2007), and JAY291 (Argueso et al. 2009). The details of the construction of the triploid are in materials and methods. YJM789 and JAY291 have 6.1 and 5.4 single nucleotide polymorphisms (SNPs) per kilobase, respectively, relative to S288c (Wei et al. 2007; Argueso et al. 2009). Although the genome alignments of YJM789 to JAY291 have not been completed, in one 49-kb interval aligned in these strains, there were about 10 SNPs/kb (Argueso et al. 2009). Using a CGH microarray, we confirmed that JSC2 had three copies of every homolog prior to sporulation.
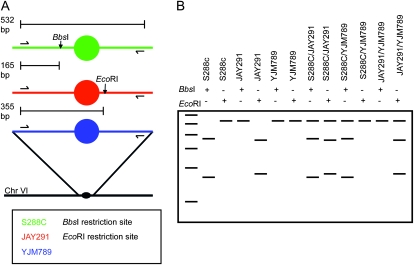
We were able to distinguish each of the three copies of each homolog for all homologs in JSC2 by PCR amplification of sequences within 5 kb of the centromeres, followed by treatment of the PCR fragment with restriction enzymes that distinguish the amplified regions of each strain. For this analysis, we utilized SNPs that altered restriction enzyme recognition sites. In Figure 3, we illustrate how the three copies of chromosome VI were distinguished by this approach. As shown in Figure 3A, chromosome VI of S288c has a BbsI site near CEN6 that is not present in JAY291 or YJM789, and JAY291 has a CEN6-linked EcoRI site that is not present in either S288c or YJM789. Using PCR with primers that flank these heterozygous sites, we generated a DNA fragment of about 530 bp. By examining the sizes of the fragments produced following treatment of the 530-bp fragment with EcoRI and BbsI, we can distinguish the three homologs individually (lanes 2–7; Figure 3B) and in pairs (lanes 8–13, Figure 3B). Table S2 lists all the primers and restriction enzymes used to diagnose each chromosome. We refer to this method of analysis as SPA (single-nucleotide-polymorphism PCR analysis).
Figure 3.—
Use of SNPs to determine the number and identity of homologous chromosome homologs within a JSC2-derived spore. For this analysis, we designed primers that could be used to amplify SNP-containing regions located very near (within 10 kb) the centromere of each of the 16 chromosomes (Table S2). By digestion of the resulting PCR products with the relevant restriction enzyme(s), we could diagnose the source of the various copies of each homolog. (A) Depiction of the differential restriction sites present in each strain in the amplified region of chromosome VI. The PCR-amplified regions of S288c, JAY291, and YJM789 are depicted in green, red, and blue, respectively. Positions of PCR primers are indicated by horizontal arrows, and the centromere is indicated by circles. There is a BbsI restriction site on the S288c PCR fragment (shown as a vertical arrow) that is not present in JAY291 or YJM789 and an EcoRI restriction site on the JAY291 PCR fragment that is not present in the other two strains. The BbsI and EcoRI restriction sites are located at 165 and 355 bp, respectively, from the left end of the PCR fragment. (B) Demonstration of how differential restriction sites can be used to diagnose the source(s) of homologs in spore colonies. Digestion of each possible monosome and heterozygous disome with both BbsI and EcoRI produces a unique restriction pattern shown as a schematic gel. The leftmost lane is a 100-bp DNA standard ranging from 100 to 600 bp. The horizontal rows above the gel indicate which lane corresponds to which digest with plus signs indicating treatment with that enzyme.
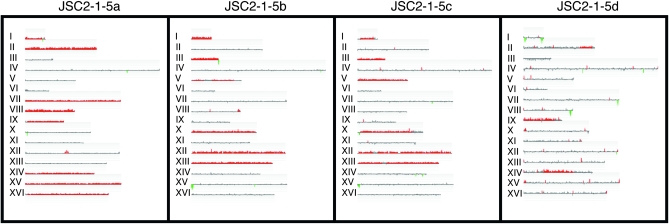
We analyzed 10 JSC2 tetrads that had four viable spores by SPA, and these data are summarized in Table 2. Additionally, six JSC2 tetrads were also analyzed by CGH microarrays (Table S5). Figure 4 shows a CGH analysis of tetrad 5 of JSC2; the SPA data for this same tetrad are in the first four rows of data of Table 2. By both methods, it is clear that the two pairs of meiotic sister spores are JSC2-1-5a and -5d, and JSC2-1-5b and -5c. The two methods of analysis are completely concordant for spores JSC2-1-5b and -5c, both methods showing that these strains are disomic for chromosomes I, III, V, X, XII, and XIII.
TABLE 2.
Analysis of disomy and monosomy in spores derived from the triploid JSC2-1
| Chromosomes |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spore | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI |
| 5a | Ja | J Y | Y | Yb | Y | S Y | S Y | J Y | S J | J | S J | Y | S | J Y | S Y | S J |
| 5b | S Y | S | S J | S | S J | J | J | S | Y | S Y | Y | S J | J Y | S | J | Y |
| 5c | S Y | S | S J | S | S J | J | J | S | Y | S Y | Y | S J | J Y | S | J | Y |
| 5d | J | J Y | Y | J Y | Y | S Y | S Y | J Y | S J | J | S J | Y | S | J Y | S Y | S J |
| 6a | J | J | Y | S | S | Ja | J | S | J Y | J | J Y | Y | Y | S Y | S | S |
| 6b | S Y | S Y | S J | J Y | J Y | S Y | S Y | J Y | S | S Y | S | S J | S J | J | J Y | J Y |
| 6c | J | J | Y | S | S | J | J | S | J Y | J | J Y | Y | Y | S Y | S | S |
| 6d | S Y | S Y | S J | J Y | J Y | S Y | S Y | J Y | S | S Y | S | S J | S J | J | J Y | J Y |
| 8a | S | S J | J Y | J Y | J | J | S | S J | Y | S Y | J | Ya | S J | S Y | S | S J |
| 8b | J Y | Y | S | S | S Y | S Y | J Y | Y | S J | J | S Y | S J | Y | J | J Y | Y |
| 8c | J Y | Y | S | S | S Y | S Y | J Y | Y | S J | J | S Y | S J | Y | J | J Y | Y |
| 8d | S | S J | J Y | J Y | J | J | S | S J | Y | S Y | J | Jc | S J | S Y | S | S J |
| 9a | Y | S J | J | S | S | S Y | S J | J | S Y | Y | S J | S J | S | S J | S Y | J Y |
| 9b | S J | Y | S Y | J Y | J Y | J | Y | S Y | J | S J | Y | Y | J Y | Y | J | S |
| 9c | S J | Y | S Y | J Y | J Y | J | Y | S Y | J | S J | Y | Y | J Y | Y | J | S |
| 9d | Y | S J | J | S | S | S Y | Sb | J | S Y | Y | S J | S J | S | S J | S Y | J Y |
| 10a | J Y | J Y | J Y | J | J Y | S | S Y | Y | J | S Y | J | S J | S | S J | J | J Y |
| 10b | JdY | Yb | Jb | J | J Y | S | S Y | Y | J | S Y | J | S J | S | Jb | J | Yb |
| 10c | S | S | S | S Y | S | J Y | J | S J | S Y | J | S Y | Ya | J Y | Y | S Y | S |
| 10d | S | S | S | Yb | S | J Y | J | S J | S Y | J | S Y | Y | Yb | Y | S Y | S |
| 11a | S Y | Y | Y | J | S Y | S | S J | Y | J | S Y | Y | Y | Y | S Y | S | J Y |
| 11b | J | S J | S J | S Y | J | J Y | Y | S J | S Y | J | S J | S J | S J | J | J Y | S |
| 11c | J | S J | S J | S Y | J | J Y | Y | S J | S Y | J | S J | S J | S J | J | J Y | S |
| 11d | S Y | Y | Y | J | S Y | S | S J | Y | J | S Y | Y | Y | Y | S Y | S | J Y |
| 12a | Y | S J | J | J | J | S Y | Y | S Y | S Y | S | S J | S | J | S Y | S Y | S J |
| 12b | Y | S J | J | J | J | S Y | Y | Sab | S Y | S | S J | S | J | S Y | S Y | S J |
| 12c | S J | Y | S Y | S Y | S Y | J | Sb | J | J | J Y | Y | J Y | S Sc | J | J | Y |
| 12d | S J | Y | S Y | S Y | S Y | J | S J | J | J | J Y | Y | J Y | S Sc | J | J | Y |
| 13a | J | S J | S | S J | S Y | Y | J Y | S J | J | S | J | S Y | S | J | S Y | S |
| 13b | S Y | Y | J Y | Y | J | S J | S | Y | S Y | J Y | S Y | J | J Y | S Y | J | J Y |
| 13c | S Y | Y | J Y | Y | J | S J | S | Y | S Y | J Y | S Y | Ja | J Y | S Y | J | J Y |
| 13d | J | S J | S | S Je | S Y | Y | J Y | S J | J | S | J | S Y | S | J | Ybe | S |
| 14a | J | Y | S J | S | Y | J | S | Y | J | J | Y | S J | S | S | S Y | S |
| 14b | J | Y | S J | S | Y | J | S | Y | J | J | Y | S J | S | S | S Y | S |
| 14c | S Y | S J | Y | J Y | S J | S Y | J Y | S J | S Y | S Y | S J | Y | J Y | J Y | J | J Y |
| 14d | S Y | S J | Y | J Y | S J | S Y | J Y | S J | S Y | S Y | S J | Y | J Y | J Y | J | J Y |
| 15a | S Y | S J | S J | Y | J | S J | S J | S | J | J | J | J Y | J Y | S Y | Y | J Y |
| 15b | J | Y | Y | S J | S Y | Y | Y | J Y | S Y | S Y | S Y | S | S | J | S J | S |
| 15c | J | Y | Y | S J | S Y | Y | Y | J Y | S Y | S Y | S Y | S | S | J | S J | S |
| 15d | S Y | S J | S J | Y | J | S J | S J | S | J | J | J | J Y | J Y | S Y | Y | J Y |
| Averagef | 6 | 5.9 | 5.9 | 5.8 | 6 | 6 | 5.8 | 5.9 | 6 | 6 | 6 | 6 | 5.9 | 5.9 | 5.9 | 5.9 |
Using SPA (described in text), we determined whether spores were monosomic or disomic, and the source of each homolog (S, S288c; J, JAY291; Y, YJM789).
Extra chromosome homolog detected by CGH microarrays. These extra chromosomes were not counted in the averages shown at the bottom of this table.
Chromosome loss.
Putative gene conversion event.
Boldface indicates that homologs were detected using PCR and primers that exclusively amplified only one of the strain-specific SNPs.
Translocation between chromosome IV and XV detected by CGH microarray and gel analysis.
Average number of individual homologs per tetrad.
Figure 4.—
Analysis of a JSC2 tetrad by CGH microarrays. DNA was isolated from spore colonies of a triploid-derived tetrad of JSC2 and examined by the same method described in Figure 2. The meiotic sister pairs are JSC2-1-5a/JSC2-1-5d and JSC2-1-5b/JSC2-1-5c. Extensive loss of disomes from spore JSC2-1-5d is evident.
The spores JSC2-1-5a and -5d are substantially different when examined by CGH and, in addition, show a number of discrepancies when analyzed by CGH vs. SPA. By CGH, JSC2-1-5a is disomic for chromosomes I, II, VII-IX, and XIV-XVI, whereas JSC2-1-5d is only disomic for chromosomes II, IX, and XIV. One obvious explanation for this discrepancy is that many of the disomic chromosomes originally present in the JSC2-1-5d spore were rapidly lost during the mitotic divisions required to produce the cultures for the CGH analysis. In support of this explanation, when we examined DNA isolated from JSC2-1-5d by SPA, we observed disomy for chromosomes II, IV, VI–IX, XI, and XIV–XVI. This increased level of disomy detected by SPA is the result of two factors. First, it is easier to detect disomy present in a small fraction (less than 12.5%) by SPA than by CGH analysis. Second, we found that some of the spore cultures had two types of cells in approximately equal amounts, one population that had one variant for a particular homolog and one that had a different variant. An example of such a spore culture is shown in Figure 5. We performed SPA on 10 colonies derived from the JSC2-1-8d spore culture; eight had the SNP characteristic of the S288c-derived chromosome II, one had the SNP characteristic of the JAY2981 homolog, and one had both types of homolog, although the S288c-derived homolog predominated. Thus, in this spore colony, although most of cells have only a single copy of chromosome II, the original spore must have had two copies.
Figure 5.—
Analysis of SNPs in colonies derived from a single meiotic spore culture (JSC2-1-8d). We isolated DNA from single colonies derived from individual cells of the spore culture JSC2-1-8d and DNA from the original spore culture. By CGH microarrays, this spore had one copy of chromosome II. The region near CEN2 was amplified in all DNA samples and treated with the restriction enzymes DraI and BbsI, and the resulting fragments were separated by gel electrophoresis. By SPA, the spore colony was disomic for chromosome II with copies derived from S288C and JAY291. In this figure, the DraI digests of the PCR fragment are shown. The digest of DNA isolated from the original spore colony (third lane from the right) indicates that more cells in the culture have the S288c chromosome (uncut band) than the JAY291 chromosome (cut band). This conclusion is confirmed by examining single-colony isolates (labeled 1–10).
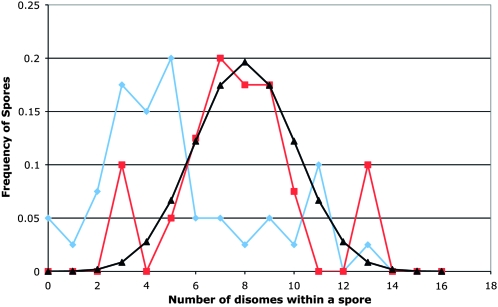
As shown in Table 2, in most of the tetrads, most of the homologs segregated to produce two disomic spores and two monosomic spores. This result demonstrates that meiotic chromosome segregation in triploids is accurate, and most of the chromosome loss events detected by CGH arrays represent mitotic chromosome loss. Analyzing 10 MH10 tetrads with CGH arrays, we found 107 chromosome loss events. Analyzing 10 JSC2 tetrads using SPA, we found only 15 chromosome loss events, reflecting the greater sensitivity of SPA in detecting disomy. At least some of these 15 loss events also represent mitotic loss (as discussed further below), presumably at a very early stage in the growth of the spore colony. Figure 6 shows a comparison of the number of recovered disomes for each method. A line depicts the expected binomial distribution of disomes, assuming that the probabilities of a chromosome being disomic or monosomic are both equal to 0.5. The peaks for the JSC2 spores are shifted to the right relative to the MH10 spores, reflecting the more sensitive nature of SPA compared to CGH. Additionally, the line representing JSC2 spores is symmetrical around eight disomes. Since we examined only complete tetrads, this symmetry is expected.
Figure 6.—
Comparison of the detection of disomes in spores derived from the triploids MH10 and JSC2. The graph shows the number of disomic chromosomes per spore as a function of the frequency of spores with that chromosome composition. The blue line with diamonds represents the data from the MH10 triploid (based on CGH), and the red line with squares shows the data from the JSC2 triploid (based on SPA). The black line with triangles is the curve predicted by the binomial expansion with the assumption that there is the same probability (0.5) that a spore will be monosomic or disomic.
There were two other observations of interest concerning the JSC2 spore analysis. First, in comparing the CGH and SPA data, we found six examples of apparent chromosome duplication. For example, by CGH, the spores JSC2-1-5a, -5b, and -5c are disomic for chromosome I. By SPA, the spores JSC2-1-5b and -5c both have the S288c-derived and the YJM789-derived copies of chromosome I, but the spore JSC2-1-5a has only the JAY291 homolog. We interpret this result as indicating that a single copy of chromosome I originally present in spore JSC2-1-5a underwent a chromosome gain, either by nondisjunction or by rereplication.
The last unusual class of tetrad likely reflects meiotic gene conversion, the nonreciprocal transfer of information from one chromosome to the other. One example of conversion is JSC2-1 tetrad 8. In this tetrad, for chromosome XII, there were two chromosomes with the centromere-linked SNP characteristic of S288c, three chromosomes with the SNP characteristic of the JAY291, and only one chromosome with the SNP characteristic of YJM789. This pattern could be explained if there was a gene conversion that substituted the chromosome XII region on one of the YJM789-derived homologs with information derived from the JAY291-derived homologs. Since the amount of DNA transferred in meiotic gene conversion is usually limited to about 1–3 kb (Petes et al. 1991; Mancera et al. 2008), in the spore representing the putative conversion (JSC2-1-8d), we examined (by SPA) a SNP located on chromosome XII on the opposite side of the centromere located about 4 kb from the original SNP (primers described in Table S2). As expected, this polymorphism had the SNP characteristic of the YJM789 chromosome. We also observed one tetrad (JSC2-1-12) in which two chromosomes appeared to have undergone gene conversion: four copies of the S288c-associated SNP, no copies of the YJM789-associated SNP, and two copies of the JAY291-associated SNP. Although this tetrad may have undergone a double meiotic event (two conversions of the YJM789 SNP to the S288c SNP), on the basis of the rarity of the single conversion events, it is more likely that the observed pattern reflects a mitotic gene conversion.
Mitotic chromosome loss:
Our analysis of JSC2 demonstrates that most tetrads have two spores with two copies and two spores with one copy of each homolog (Table 2). However, in some tetrads, one or more chromosomes was lost. As discussed above, such losses have three possible sources: (1) loss of one or more chromosomes in the triploid prior to meiosis, (2) loss of chromosomes during the first or second meiotic divisions, and (3) loss of chromosomes during the mitotic growth of the spores. Several arguments strongly support the third mechanism as the principle factor.
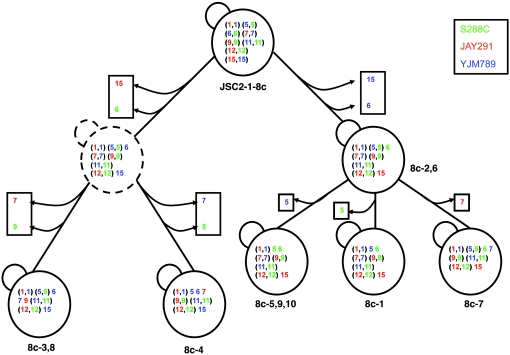
First, we can directly detect chromosome instability in some of the aneuploid spores. We examined the distribution of disomic chromosomes in 10 individual colonies derived from JSC2-1-8c. Our SPA analysis of the chromosome distribution of the original spore colony showed that this spore was disomic for eight chromosomes (I, V–VII, IX, XI, XII, and XV). As shown in Figure 7, none of the 10 colonies (labeled 8c-1 to 8c-10) examined retained disomy for all eight chromosomes. All disomic chromosomes, except I, XI, and XII, were lost in at least 1 of the 10 colonies. In Figure 7, we show a pathway by which the chromosomes may have been lost; although other pathways could be depicted, the one shown requires the fewest steps.
Figure 7.—
Mitotic loss of disomic chromosomes from spore JSC2-1-8c. The spore JSC2-1-8c was originally disomic for eight chromosomes (I, V-VII, IX, XI, XII, and XV). We examined 10 single-colony isolates of JSC2-1-8c (labeled 8c-1-10) by SPA, and found six genotypes (shown in solid-line depictions of yeast cells). In this figure, we show a pathway of chromosome loss consistent with these genotypes. The silhouette of a yeast cell with dashed lines is a hypothetical intermediate that was not detected. The numbers within each circle are the chromosome numbers, and the colors indicate the haploid from which the chromosome was derived.
Although it is difficult to calculate a rate of chromosome loss from this analysis, we estimated the rate of loss of chromosome V in JSC2-8c by another method. The initial spore isolate had two copies of chromosome V, one with the wild-type URA3 allele and one with the mutant allele. Thus, the spore was initially Ura+. Since the URA3 gene is centromere linked, the rate of sectored Ura+/Ura− colonies observed when JSC2-8c colonies cultured on rich growth medium are replica plated to medium lacking uracil is an estimate of the rate of loss of the chromosome with the wild-type allele. Of 8200 colonies examined, 223 were sectored, a rate of about 0.027. This rate of loss is much higher than the rate of chromosome V loss in a wild-type diploid (10−5; Hartwell and Smith 1985). The frequency of chromosome loss was somewhat spore specific, and lower rates of loss were observed in JSC2-1-5b (3.5 × 10−3) and JSC2-1-6d (no sectored colonies observed).
Since the triploid JSC2 had one copy of chromosome V with a wild-type URA3 gene and two copies of chromosome V with mutant ura3 genes, we also examined sectored colonies in this strain. No sectored Ura+/Ura− colonies were observed in 24,000 examined, suggesting a low rate of chromosome loss (<4 × 10−5). This result is consistent with the measurement of chromosome loss rate of 1.4 × 10−5 obtained by Mayer and Aguilera (1990). Thus, mitotic loss of chromosomes in the JSC2 triploid prior to sporulation is unlikely to be a substantial contributor to the observed loss in the spores derived from JSC2.
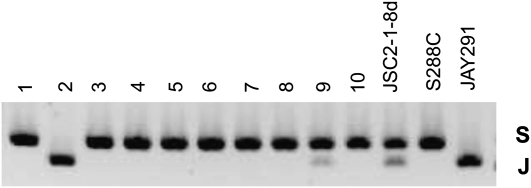
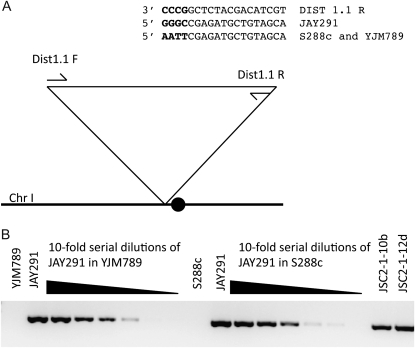
A second argument for a high rate of chromosome loss during mitotic growth of spores of the triploid is that we find a lower rate of chromosome loss when we examine the spores with a method of detection that is more sensitive (SPA vs. CGH). For two JSC2 spores, we extended our analysis using a method that was even more sensitive than SPA. By our initial SPA analysis, spore cultures of JSC2-1-10b and JSC2-1-12d were monosomic for chromosome I, although their meiotic sisters were disomic. By SPA, JSC2-1-10b had only the “Y” homolog whereas the sister spore had both the Y and J homologs; for JSC2-1-12d, only the S form was detectable by SPA, although the sister spore had both the S and J homologs. To determine whether cells with the J homolog of chromosome I were present in very low amounts in spore cultures of JSC2-1-10b and JSC2-1-12d, we designed a pair of primers that would specifically amplify a centromere-linked sequence from the J homolog (primer sequence in materials and methods). As shown in Figure 8, by PCR, we found that the J homolog was present in both spore cultures. A similar assay was used to detect missing chromosome XIII in spores JSC2-1-9c and JSC2-1-14c (data not shown). In summary, taking into account these results, there are maximally 11 meiotic chromosome loss events in JSC2 of a total of 960 total chromosomes, establishing an upper limit for the frequency of meiotic chromosome loss events in triploids as 0.01.
Figure 8.—
PCR method of detecting very small amounts of JAY291-derived chromosome I in strains with S288c- or YJM789-derived chromosomes. Although the sister spores of JSC2-1-10b and JSC2-1-12d were disomic for chromosome I and contained the JAY291-derived homolog, we could not detect this homolog using SPA. Consequently, to detect very small amounts of this homolog, we designed primers to specifically amplify DNA with this polymorphism. (A) Primer combinations DIST1.1F/DIST1.1R amplify a region ∼12 kb from the centromere of chromosome I. The 3′-end of DIST1.1R anneals to a 4-bp mismatch region that is present only in the JAY291 strain. (B) 1% agarose gel of a PCR products generated with the DIST1.1F/DIST1.1R primer pair. Genomic DNA from JAY291 was diluted into YJM789 genomic DNA in 10-fold serial dilutions from 10−1 to 10−6 in lanes 3–8. The same analysis is shown with JAY291 genomic DNA diluted into S288c in lanes 11–16. Lanes 17 and 18 contain the products of the same PCR reaction with genomic DNA from spores JSC2-1-10b and JSC2-1-12d, showing that these samples have the JAY291-specific sequence.
Chromosome pairing during meiosis:
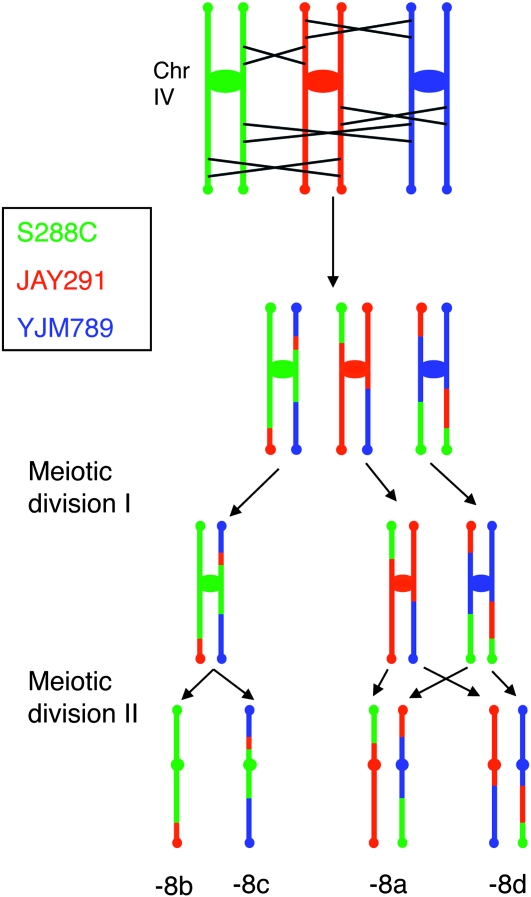
As discussed in the introduction, in near-diploid strains with one trisomic chromosome, all three copies of the trisomic chromosome recombine (Shaffer et al. 1971). To determine whether the three copies of each homolog in triploid also engage in trivalent pairing, we examined meiotic recombination for four chromosomes (IV, V, IX, and XIII) using the same centromere-linked SNPs as in our previous analysis and SNPs located near (within 50 kb) of the left and right telomeres of each chromosome. Table S3 lists the primers and restriction enzymes used for the telomere-linked SNPs, and Table S6 summarizes the results. The evidence for trivalent pairing is that at least one spore contains a chromosome in which there has been a crossover between the centromere-linked markers for the S, J, and Y SNPs and the telomere-linked SNPs. One example of this analysis (for chromosome IV of JSC2-1-8) is shown in Table S6 and Figure 9. In spores with a single copy of chromosome IV (spores -8b and -8c), the coupling of the chromosome IV markers is unambiguous. For spores that are disomic for IV, the coupling relationships of the markers are unclear. For some spores of this type, we screened multiple colonies derived from the original spore colony to identify derivatives that had lost one of the two disomic chromosomes. Analysis of the SNPs in these derivatives clarified the coupling relationships. In this tetrad, there is redundant information demonstrating trivalent pairing. For example, the existence of a single copy of chromosome IV in JSC2-1-8a-2 (a colony derived from the -8a spore) with the centromere-linked Y marker, the left telomere J marker, and the right telomere S marker (Table S6) is sufficient to argue for trivalent pairing in this meiosis.
Figure 9.—
Chromosome pairing and recombination during meiosis in a triploid. As discussed in the text, using SNPs located near the centromere and telomeres, we mapped crossovers on four chromosomes in 10 tetrads (data in Table S6). This figure shows patterns of crossovers consistent with the recombination events detected in tetrad JSC2-1-8 on chromosome IV. The centromeres are shown as circles and ovals at the telomeres represent SNPs; the left and right telomeres are at the top and bottom of the chromosome. The individual spore numbers corresponding to those in Table S6 are shown at the bottom of the figure. In this tetrad, every chromatid engaged in at least one exchange. The pattern of exchanges is shown in this figure is not the only possible pathway for generating the recombinant chromosomes at the bottom of the picture.
Chromosomes IV and V had trivalent pairing in all 10 tetrads. Six of 10 tetrads had trivalent pairing for chromosome IX. Four tetrads (JSC2-1-5, -6, -12, and -13) could not be completely diagnosed because we were unable to isolate derivatives of disomic spores that had only copy of IX; in such spores, we could not determine the coupling arrangements of the markers. Chromosome XIII underwent trivalent pairing in 9 of 10 tetrads, but, in 1 tetrad (JSC2-1-5), the coupling arrangements of the markers on chromosome XIII also could not be determined. In summary, of 35 events in which we could unambiguously determine the recombination patterns, the chromosomes showed trivalent pairing in all cases.
DISCUSSION
In this study, we showed that meiotic segregation of chromosomes in triploids is usually accurate with two spores receiving one copy and two spores receiving two copies of each homolog. As expected from previous studies, spore viability was low (about 50%) compared to the viability of spores derived from diploids. No particular combinations of two or three disomic chromosomes were favored in the viable spores. The aneuploid strains derived from the spores often had very high rates of mitotic chromosome loss. In most meioses, all three copies of each homolog pair and recombine.
Spore viability:
We studied meiotic chromosome segregation in two very different triploids. MH10 has three copies of a haploid set derived from MS71 (Strand et al. 1995) and JSC2 is a triploid derived from three different haploid strains with diverged sequences: S288c, YJM789, and JAY291. Despite this difference, both triploids had similar spore viabilities which were greater than those observed in most past triploid studies (Parry and Cox 1970; Campbell et al. 1981; Campbell and Doolittle 1987; Loidl 1995). The similar spore viability between our strains indicates that the 0.4–1% sequence divergence between homologs in JSC2 does not affect spore viability in triploids.
We examined patterns of chromosome segregation to determine whether certain combination of disomic chromosomes led to spore inviability. All 120 two-chromosome disomy combinations and all 560 three-chromosome disomy combinations within the MH10 and/or JSC2 tetrads were observed in viable spores (Tables 1 and Tables 2). No combination of two or three coincident disomies were significantly overrepresented or underrepresented in the viable spores (analysis described in File S1), consistent with previous studies (Campbell et al. 1981).
Parry and Cox (1970) observed that most triploid-derived spores were aneuploid for five chromosomes or less, with an average of 2.6 disomes per spore. They suggested that either triploid segregation produced two near-haploid and two near-diploid spores or that aneuploid chromosomes were rapidly eliminated during vegetative growth of the spore cultures. Our results demonstrate that the second explanation is correct, although the strains in our study seem more tolerant of high levels of disomy (Figure 6) than those examined by Parry and Cox (1970).
Patterns of chromosome loss:
Complete or partial loss of disomic chromosomes during the vegetative growth of spore cultures was very common in our study. Two types of observations demonstrate a high rate of chromosome loss. First, the loss events (chromosomes with less than six copies in the tetrad) were much more common when we analyzed the spore cultures by CGH than when we analyzed them by the more sensitive SPA procedure; for six tetrads of JSC2, both methods were used on the same DNA samples. Second, we directly showed that subcultures of some of the disomic spores had unstable karyotypes (Figure 5 and 7).
In MH10-derived spores, we frequently observed that one sister spore was disomic for a particular chromosome and the other was not, suggesting a chromosome loss event. There were 53 such loss events (indicated in boldface in Table 1). These loss events were nonrandomly distributed among the spores; 70% of the losses were in 5 of the 40 spores. In addition, the two sister spores often had very different loss rates. One extreme example is the sister spores MH10-4a and MH10-4c. The MH10-4a spore lost 11 chromosomes (becoming a euploid haploid strain), whereas the MH10-4c spore did not lose any chromosomes.
Two different statistical tests were applied to these data (Table S7). First, for each of the 40 spores, we determined whether there was a significantly elevated rate of chromosome loss compared to the expected rate of loss per disomic chromosome for all 40 spores (0.2 loss events/disomic chromosome). By the chi-square goodness-of-fit test, spores MH10-4a, -22d, -25a, -28b, -32b, and -37c had significantly elevated rates. Second, we used the Fisher exact test to compare chromosome loss events in pairs of sister spores. By this test, five pairs of sister spores had significantly different numbers of loss events (Table S7). Thus, multiple pairs of sister spores in MH10, which presumably initially had identical karyotypes and genotypes, had different rates of chromosome loss. We suggest two explanations of this interesting observation. One possibility is that certain spores receive a partially defective spindle pole body and this defect persists for a number of generations following spore germination, leading to an elevated rate of chromosome loss. An alternative possibility is that a stochastic loss of one of the disomic chromosomes from a spore results in a much elevated rate of loss of other chromosomes.
Several more points concerning these observations should be made. First, in analyzing the MH10 data, we did not consider chromosome loss events in which all four spores were monosomic, since we could not determine which of the four spores had lost chromosomes. Second, the observation that certain spores have very high rates of chromosome loss in MH10 was considerably less obvious in the JSC2 data (Table S8). Of the 24 JSC2 spore cultures examined by microarrays, we detected 33 examples of loss in one sister but not the other (total of 160 events). Five spores had lost five or more chromosomes, whereas the other 19 spores had lost two or fewer chromosomes. Despite this asymmetry, none of the spores had a significantly elevated rate of chromosome loss compared to the average loss rate for the six tetrads (0.21 events/disomic chromosome). Since the sister spores of JSC2 are not isogenic (unlike those of MH10), it is possible that genotype-dependent mechanisms of chromosome loss prevent the detection of genotype-independent mechanisms of loss.
Another important point to emphasize is that we do not know the mechanism of chromosome loss. If the loss of the disomic chromosome is a consequence of nondisjunction, then we should observe trisomic derivatives at the same frequency as monosomic derivatives. Trisomic strains were not observed, although it is possible that such strains would have a severe growth disadvantage. Although we found a few examples of monosomic chromosomes becoming disomic consistent with chromosome nondisjunction events (Tables 1 and Tables 2), such events were much less frequent than chromosome losses.
An alternative mechanism for chromosome loss is failure to replicate one or both of the disomic chromosomes before cell division. Although we have no direct evidence in favor of this mechanism, the pattern of chromosome loss diagrammed in Figure 7 has several branch points in which symmetrical chromosome loss (consistent with a defect in chromosome replication) is postulated. For example, we show a symmetric loss of chromosomes VI and XV at the first branch point. This pattern is consistent with failure to replicate VI and XV and segregation of the unreplicated homologs to the two daughter cells. To prove this mechanism, however, one would need to perform pedigree analysis on the aneuploid strains.
Torres et al. (2007) constructed 13 yeast strains, each disomic for a different yeast chromosome. All disomic strains had proliferative defects, increased glucose uptake, and were sensitive to cycloheximide, a protein synthesis inhibitor. Strains disomic for chromosomes IV and XV were among those with the most profound proliferative defect, and these chromosomes were among those most frequently lost from the MH10 tetrads. Torres et al. (2007) were unable to isolate a strain that was disomic only for chromosome VI and suggested that such strains could not be recovered because of the imbalance between levels of β-tubulin (located on chromosome VI) and α-tubulin (located on chromosome XIII). In support of this suggestion, they were able to obtain strains disomic for both VI and XIII. Similarly, in our experiments, chromosome VI was frequently lost from MH10 tetrads (Table 1), although we were able to recover strains that were disomic for VI and monosomic for XIII.
The rate of chromosome loss that we observed in the spore cultures is clearly much greater than that of wild-type diploid cells. The rate of chromosome V loss in a wild-type diploid is about 10−5/cell division (Hartwell and Smith 1985). Since many of the disomic chromosomes were lost in less than 50 cell divisions, the lost rate is likely to be considerably greater than 10−2/division. This high loss rate probably reflects defective structures involved in segregation or replication of the chromosomes resulting from nonstochiometric amounts of the proteins required to form these structures. In addition, since most disomic yeast strains grow more slowly than wild type (Torres et al. 2007), those strains that lose chromosomes have a selective advantage. In our study, two of the haploid strains derived by sporulating MH10 (MH10-4a and MH10-22d) lost all disomic chromosomes to revert to haploidy (Table 1).
There is also evidence that chromosome segregation may be inherently less accurate in triploids and tetraploids compared to diploids. Gerstein et al. (2008) observed that subcultured triploid and tetraploid strains underwent a reduction in chromosome number to diploidy. Since this reduction occurred quickly, they suggested that there was a mechanism of concerted chromosome loss. Mayer and Aguilera (1990) showed that triploids had chromosome loss rates elevated about 30-fold compared to a diploid (Mayer and Aguilera 1990; Andalis et al. 2004). In addition, Mayer and Aguilera found that loss of one chromosome from the triploid or tetraploid increased the likelihood of loss of a second chromosome. Storchova et al. (2006) showed that tetraploids have an elevated level of monopolar kinetochore attachments and suggested that tetraploids had partially defective spindle pole bodies, spindles, and/or kinetochores as a consequence of defective “scaling” of complex structures in yeast cells with high ploidies.
Chromosome pairing and recombination:
We first examined whether there was any evidence of preferential segregation of homologs derived from the three strains used to construct JSC2. More specifically, using the centromere-linked polymorphism data (Table 2), we tested each of the sixteen homologs for whether the disomic spores had a significant excess of the chromosomes derived from any of the three possible pairs of S288c, YJM789, or JAY291. After correcting for the rate of false discoveries with the Benjamini and Hochberg (1995) test, no significant strain-specific selection for any pairing combination was observed (detailed discussion of the statistical analysis in File S1).
Although meiotic recombination in triploids has not been examined in any detail, several studies have shown that all three copies of a trisomic chromosome in a near-diploid strain frequently engage in recombination in a single meiosis (Shaffer et al. 1971; Culbertson and Henry 1973; Riley and Manney 1978; Koller et al. 1996). In our study, we examined meiotic recombination on chromosomes IV, V, IX, and XIII. As discussed above, in all of the 35 events in which we could unambiguously diagnose meiotic exchanges, all three copies of these homologs recombined. Our results are consistent with the cytological studies of Loidl (1995) in which he observed that all three homologs in triploids were involved in synaptonemal complex formation.
Acknowledgments
We thank all members of the Petes and Jinks-Robertson labs for their discussions and technical help, and we thank John McCusker, Jennifer McCulley, Hengshan Zhang, Joseph Heitman, and Sue Jinks-Robertson for comments on the manuscript. We thank Lucas Argueso and Piotr Mieczkowski for generating the microarrays, and Einat Hazkani-Covo for help with the statistical analyses. This research was supported by grants from the National Institutes of Health (GM24110, GM52319, and 1RC1ES018091-01).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121533/DC1.
References
- Andalis, A. A., Z. Storchova, C. Styles, T. Galitski, D. Pellman et al., 2004. Defects arising from whole-genome duplication in Saccharomyces cerevisiae. Genetics 167 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, J. L., M. F. Carazzolle, P. A. Mieczkowski, F. M. Duarte, O. V. Netto et al., 2009. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res. 19 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso, J. L., J. Westmoreland, P. A. Mieczkowski, M. Gawel, T. D. Petes et al., 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57 289–300. [Google Scholar]
- Bridges, C. B., and E. A. Anderson, 1925. Crossing over in the X chromosome of triploid females of Drosophila melanogaster. Genetics 10 418–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, D., J. S. Doctor, J. H. Feuersanger and M. M. Doolittle, 1981. Differential mitotic stability of yeast disomes derived from triploid meiosis. Genetics 98 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, D. A., and M. M. Doolittle, 1987. Coincident chromosomal disomy in meiotic dyads from triploid yeast. Curr. Genet. 12 569–576. [DOI] [PubMed] [Google Scholar]
- Culbertson, M. R., and S. A. Henry, 1973. Genetic analysis of hybrid strains trisomic for the chromosome containing a fatty acid synthetase gene complex (fas1) in yeast. Genetics 75 441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezov, T. K., E. Boger-Nadjar, Z. Frenkel, I. Katsperovski, S. Kemeny et al., 2006. Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from “Evolution Canyon”: microsatellite polymorphism, ploidy and controversial sexual status. Genetics 174 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, A. C., R. M. McBride and S. P. Otto, 2008. Ploidy reduction in Saccharomyces cerevisiae. Biol. Lett. 4 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego.
- Hartwell, L. H., and D. Smith, 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of Saccharomyces cerevisiae. Genetics 110 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty, M. J., and S. J. Hiscock, 2008. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18 R435–R444. [DOI] [PubMed] [Google Scholar]
- Iliopoulos, D., G. Vassiliou, E. Sekerli, V. Sidiropoulou, A. Tsiga et al., 2005. Long survival in a 69, XXX triploid infant in Greece. Genet. Mol. Res. 4 755–759. [PubMed] [Google Scholar]
- Koller, A., J. Heitman and M. N. Hall, 1996. Regional bivalent-univalent pairing versus trivalent pairing of a trisomic chromosome in Saccharomyces cerevisiae. Genetics 144 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P. S., P. W. Greenwell, M. Dominska, M. Gawel, M. Hamilton et al., 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5 e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, K. Lobachev and T. D. Petes, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase α: a model for chromome fragile sites. Cell 120 587–598. [DOI] [PubMed] [Google Scholar]
- Loidl, J., 1995. Meiotic chromosome pairing in triploid and tetraploid Saccharomyces cerevisiae. Genetics 139 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera, E., R. Bourgon, A. Brozzi, W. Huber and L. M. Steinmetz, 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, V. W., and A. Aguilera, 1990. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mut. Res. 231 177–186. [DOI] [PubMed] [Google Scholar]
- Narayanan, V., P. A. Mieczkowski, H. M. Kim, T. D. Petes and K. S. Lobachev, 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125 1283–1296. [DOI] [PubMed] [Google Scholar]
- Parry, E. M., and B. S. Cox, 1970. The tolerance of aneuploidy in yeast. Genet. Res. 16 333–340. [DOI] [PubMed] [Google Scholar]
- Petes, T. D., R. E. Malone and L. S. Symington, 1991. Recombination in yeast, pp. 407–521 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. Broach, E. W. Jones and J. R. Pringle. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pomper, S., K. M. Daniels and D. W. McKee, 1954. Genetic analysis of polyploid yeast. Genetics 39 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, M. I., and T. R. Manney, 1978. Tetraploid strains of Saccharomyces cerevisiae that are trisomic for chromosome III. Genetics 89 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, B., I. Brearley, R. Littlewood and G. R. Fink, 1971. A stable aneuploid of Saccharomyces cerevisiae. Genetics 67 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock, M., J. Ustinova, D. K. Lamatsch, M. Schartl, N. Perrin et al., 2009. A vertebrate reproductive system involving three ploidy levels: hybrid origin of triploids in a contact zone of diploid and tetraploid Palearctic green toads (Bufo viridis subgroup). Evolution 64 944–959. [DOI] [PubMed] [Google Scholar]
- Storchova, Z., A. Breneman, J. Cande, J. Dunn, K. Burbank et al., 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443 541–547. [DOI] [PubMed] [Google Scholar]
- Strand, M., M. C. Earley, G. F. Crouse and T. D. Petes, 1995. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92 10418–10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli et al., 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317 916–924. [DOI] [PubMed] [Google Scholar]
- Wang, P., Y. Kim, J. Pollack, B. Narasimhan and R. Tibshirani, 2005. A method for calling gains and losses in array CGH data. Biostatistics 6 45–58. [DOI] [PubMed] [Google Scholar]
- Wei, W., J. H. McCusker, R. W. Hyman, T. Jones, Y. Ning et al., 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., C. Dollard and S. L. Ricupero-Hovasse, 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11 53–55. [DOI] [PubMed] [Google Scholar]