Abstract
A wide variety of biological experiments rely on the ability to express an exogenous gene in a transgenic animal at a defined level and in a spatially and temporally controlled pattern. We describe major improvements of the methods available for achieving this objective in Drosophila melanogaster. We have systematically varied core promoters, UTRs, operator sequences, and transcriptional activating domains used to direct gene expression with the GAL4, LexA, and Split GAL4 transcription factors and the GAL80 transcriptional repressor. The use of site-specific integration allowed us to make quantitative comparisons between different constructs inserted at the same genomic location. We also characterized a set of PhiC31 integration sites for their ability to support transgene expression of both drivers and responders in the nervous system. The increased strength and reliability of these optimized reagents overcome many of the previous limitations of these methods and will facilitate genetic manipulations of greater complexity and sophistication.
THE ability to express a gene of interest in a spatially restricted manner in a transgenic animal has greatly contributed to the use of Drosophila in a wide variety of biological studies. In conjunction with RNA interference and proteins that have been engineered to alter or report cell function in specific ways, directed gene expression enables the precise manipulation of the function of single cells or cell types, as well as their visualization. This ability is essential in cases such as the nervous system, where understanding function requires probing the structure and activity of individual, identified cells.
We previously described an approach for identifying a large set of enhancers that can each reproducibly drive expression of a reporter gene in a distinct, small subset of cells in the adult central nervous system (Pfeiffer et al. 2008). In the course of that work, we became aware of many limitations of current methods that employ the yeast GAL4 (Brand and Perrimon 1993; Duffy 2002) and bacterial LexA (Lai and Lee 2006) transcription factors to drive gene expression in Drosophila. In this article, we report our efforts to systematically improve these widely used methods.
The GAL4 transcriptional activator from Saccharomyces cerevisiae functions in Drosophila (Fischer et al. 1988), and the GAL4/UAS binary expression system (UAS denotes a GAL4 binding site) has become a powerful and widely used tool for directed gene expression (Brand and Perrimon 1993; Duffy 2002). In such two component systems, one transgenic construct drives the expression of a site-specific transcriptional activator and a second construct contains its binding sites positioned upstream of a responder gene. We evaluated many factors that affect the pattern and strength of GAL4-driven expression that had not been well characterized previously, including codon usage, transcriptional terminator, and activation domain. In addition, we show how varying the number of UAS sites or including introns or post-transcriptional regulatory elements in the UTRs influences the expression level of the target gene and the perdurance of its product. These manipulations allow the GAL4/UAS system to be tuned to adjust expression to desired levels.
The repressor LexA is a regulator of the SOS response to DNA damage in Escherichia coli (Walker 1984). LexA is a 202-amino-acid protein consisting of a DNA-binding domain and a dimerization domain and binds as a dimer with varying affinities to single or multiple copies of gene-specific LexA DNA-binding motifs (LexAop) found upstream of its target genes (Little and Mount 1982; Butala et al. 2008). Fusing a C-terminal activation domain derived from GAL4 or VP16 (Sadowski et al. 1988) to LexA allows it to drive in vivo transcription of reporter transgenes in Drosophila whose promoters contain LexAop motifs (Szuts and Bienz 2000; Lai and Lee 2006). Use of LexA/LexAop as a complementary binary system in conjunction with GAL4/UAS has proven useful in a variety of applications such as mosaic analysis (Lai and Lee 2006), GFP reconstitution across synaptic partners (GRASP) (Feinberg et al. 2008; Gordon and Scott 2009), and intersectional strategies for refining gene expression patterns (Shang et al. 2008). However, we found that the published LexA drivers tend to be less effective than GAL4 as transcriptional activators. Moreover, even in the absence of the LexA protein, there is detectable expression from the colE1-derived LexA binding sites used in the LexAop from Lai and Lee (2006). In this report we describe LexA drivers containing either the more potent extended GAL4 or human p65 (Schmitz and Baeuerle 1991) activation domains and a new reporter of LexA activity that uses LexA-binding sites from sulA (Cole 1983), another LexA target gene. Unlike a reporter containing colE1-derived LexA-binding sites, one derived from sulA has no apparent leak in the adult or larval CNS when assayed by histochemical methods.
The DNA-binding and transcription-activating functions of GAL4 are accomplished through different functional domains of the protein that can be separated into distinct polypeptides (Brent and Ptashne 1985; Keegan et al. 1986). When association of the separated domains is promoted by protein–protein interactions, the activity of GAL4 as a transcription factor is restored; this is the basis of the two-hybrid method for screening for protein–protein interactions (Fields and Song 1989). Luan et al. (2006) demonstrated that it is possible to drive GAL4's DNA-binding and activation domains separately, under different enhancers, thereby restricting reporter expression to the overlap of the two patterns, where the complete GAL4 is reconstituted by means of a leucine zipper attached to each domain (Split GAL4). However, the expression levels obtained with Split GAL4 were greatly reduced from those obtained with the same enhancers driving intact GAL4, even when the strong VP16 activation domain was used. We improved the efficacy of this method, primarily by the use of the p65 activation domain.
Another strategy for refining expression patterns in GAL4 lines is targeted suppression of GAL4 activity in a subset of cells comprising the pattern by expression of GAL80 (Lee and Luo 1999); GAL80 binds to GAL4 and neutralizes its transcription-promoting activity (Yun et al. 1991; Traven et al. 2006). The successful implementation of this strategy requires that GAL80 be expressed at a sufficiently high level (Johnston and Hopper 1982; Salmeron et al. 1989). We wanted to be able to use GAL80 to suppress GAL4-driven transcription when GAL4 and GAL80 are driven by enhancers of similar strength. To this end, we generated and tested vectors expressing a version of a GAL80 optimized for Drosophila codon usage and varied both the copy number and the presence of two post-transcriptional regulatory elements: an intron and the woodchuck post-transcriptional regulatory element (WPRE). The WPRE has been shown to increase protein expression from viral vectors in mammalian cells (Zufferey et al. 1999). We also tested mutant versions of GAL80 that were reported to provide greater suppression of GAL4 activity in yeast.
The ability to repeatedly target the same genomic sites using PhiC31 integrase (Groth et al. 2004) is a major advance in that it enables the experimenter to control for the influence of local genomic environment on transgene expression (Lewis 1950; Wilson et al. 1990). However, the many locations where the attP docking site has been inserted vary themselves in their local environments, and it is necessary to test each site for each desired property. We assayed 16 genomic attP docking sites to identify sites that showed minimal expression in the adult nervous system when a basal enhancer trap vector was inserted, allowed an exogenous enhancer to drive strong expression, and allowed a UAS construct to respond strongly to the presence of GAL4. Further, by using site-specific integration of transgenes (Groth et al. 2004), we were able to directly compare constructs without the variation that results from insertion at different genomic locations (Spradling and Rubin 1983).
Taken together, the efforts described in this article have generated and characterized a new set of vectors and methods that allow unprecedented control over the expression of exogenous genes in Drosophila. These vectors are modular to facilitate further improvements and allow the addition of enhancer fragments in a reproducible orientation by site-specific recombination using Gateway technology (Invitrogen, Carlsbad, CA). When integrated into well-characterized genomic locations, this approach maximizes the consistency and predictability of the resultant expression patterns.
MATERIALS AND METHODS
Codon optimization and gene synthesis:
Codon optimization was performed using Gene Designer (Villalobos et al. 2006; DNA 2.0). DNA 2.0 (Menlo Park, CA) synthesized the DNA.
Construction of GAL4, LexA, Split GAL4, and GAL80 vectors:
Standard molecular biology methods were used and constructs were sequence verified prior to injection into flies. Restriction enzymes and mung bean nuclease were from New England Biolabs (Ipswich, MA). PCR amplifications were performed with PfuUltra High-Fidelity DNA polymerase (Stratagene, La Jolla, CA). Vectors, maps, and sequences are available from Addgene (http://www.addgene.org/pgvec1).
Construction of GAL4 vectors:
Synthesized Drosophila codon-optimized GAL4 was cloned into pBPGUw (Pfeiffer et al. 2008) either as a 5′-KpnI to 3′-HindIII or a HindIII fragment to include the indicated UTR and terminator. The yeast transcriptional terminator was derived from pBPGUw. The SV40 transcriptional terminator was PCR amplified from Janelia Farm Reporter Construct (JFRC) pJFRC-MUH (see below) and cloned 5′-HindIII to 3′-SpeI, replacing the hsp70 terminator. GAL4 deletion variant II-9, which includes the published GAL4d (Ma and Ptashne 1987a) and the yeast transcriptional terminator, was excised from plasmid G610 (gift of Gary Struhl, Columbia University, New York) as a 5′-KpnI to 3′-HindIII fragment. A multistep cloning process was used to construct the codon-optimized GAL4d variant. First, the codon-optimized GAL4 DNA-binding domain (DBD) (amino acids 1–147) and the activation domain II (ADII) (amino acids 768–881) were PCR amplified from a codon-optimized GAL4 gene. Second, the PCR amplicons were cloned as a triple ligation into pBDP (Pfeiffer et al. 2008) 5′-EcoRI to 3′-NotI. The resultant vector, pBDP236, includes a BamHI linker between the GAL4 DBD and ADII protein domains. Finally, the codon-optimized GAL4d was excised from pBDP236 as a 5′-KpnI to 3′-HindIII fragment and cloned into pBPGUw. Codon-optimized GAL4∷p65 and GAL4∷VP16 were made by first liberating GAL4 DBD from pBDP236, as a 5′-KpnI to 3′-BamHI fragment, and then cloning it as a fusion to codon-optimized p65 (amino acids 283–551) or VP16 (amino acids 413–490) activation domains. GAL4∷p65 was cloned 5′-KpnI to 3′-PmeI into pBPLexA∷p65Uw from which LexA∷p65 had been removed. GAL4∷VP16 was cloned by excising GAL4∷p65 from pBPGAL4.2∷p65Uw as a HindIII fragment and replacing it with GAL4∷VP16.
Construction of LexA vectors:
LexA∷VP16 was PCR amplified from pBS_LexA∷VP16_SV40 (Lai and Lee 2006) and cloned 5′-KpnI to 3′-HindIII into pBPGUw to create pBPLexA∷VP16Uw. Codon-optimized LexA transgenes were cloned in pBPGUw as described above. LexA vectors pBPnlsLexA∷GADflUw and pBPnlsLexA∷p65Uw contain an N-terminal nuclear localization signal consisting of 11 amino acids (residues 125–135) derived from SV40 large T antigen (Smith et al. 1985).
Construction of Split GAL4 vectors:
Design of the Split GAL4 vectors has been described (Luan et al. 2006). However, we included codon-optimized GAL4 DNA-binding domain and p65 activation domain and corrected the Y → N mutation at amino acid 36 of the leucine zipper domain in the VP16AD and GAL4AD versions of Luan et al. (2006) to create pBPZpGAL4DBDUw and pBPp65ADZpUw.
Construction of GAL80 vectors:
Codon-optimized GAL80 was cloned 5′-KpnI to 3′-HindIII into pBPGUw and pBPGAL4.2Uw-2 to create pBPGAL80Uw-1 and pBPGAL80Uw-5. The Drosophila Myosin heavy chain intron 16 was PCR amplified from pJFRC2 (see below) and cloned 5′-KpnI to 3′-NheI in pBPGAL80Uw-1, making pBPGAL80Uw-3. The WPRE was PCR amplified from FLEX vector DNA (Atasoy et al. 2008) with flanking HindIII sites and cloned into pBPGAL80Uw-1 and pBPGAL80Uw-3 to generate pBPGAL80Uw-2 and pBPGAL80Uw-4, respectively. pBPGAL80Uw-6 was generated in the same fashion as pBPGAL80Uw-4, but using pBPGAL80Uw-5 instead of pBPGAL80Uw-1 as the starting vector.
Construction of pJFRC reporter vectors:
pJFRC-MUH was created in a multistep cloning process. First, pBDP (Pfeiffer et al. 2008) was cut with BglII, treated with DNA polymerase, and religated. Next, two copies of 5XUAS sites were PCR amplified from pUASBP (Horne-Badovinac and Bilder 2008) as 5′-NotI/HindIII to 3′-AvrII and 5′-AvrII to 3′-NheI/BamHI and cloned, as a triple ligation, 5′-NotI to 3′-BamHI into pBDP. The resultant plasmid, pBDP-10XUAS, was digested 5′-NheI to 3′-BamHI and ligated with a PCR-amplified hsp70basal promoter that included flanking 5′-AatII and 3′-BglII sites from pUASBP, generating pBDP-MUH. Mung bean nuclease was used to destroy the NotI site in pBDP-MUH. Finally, the SV40 terminator and multiple cloning site (MCS) were extracted from pUASBP as a BglII to FseI fragment and cloned into pBDP-MUH, yielding pJFRC-MUH.
Construction of vectors pJRC1–pJFRC8:
mCD8∷GFP was removed from pUAST (Lee and Luo 1999) as a XhoI to XbaI fragment and then cloned into pJFRC-MUH to make pJFRC1. To generate pJFRC2, the Drosophila Myosin heavy chain intron 16 was PCR amplified to include splice site consensus sequences using Phusion High-Fidelity polymerase (FINNZYMES, Espoo, Finland) from y; cn bw sp DNA (Adams et al. 2000) and cloned as a 5′-BglII to 3′-NotI fragment into pJFRC1. Promoters containing an hsp70 basal promoter and one or three UAS sites were cloned into pJFRC2 5′-HindIII to 3′-BglII to yield pJFRC3 and pJFRC4, respectively. Digestion of pJFRC2 with AvrII and NheI and subsequent ligation with the hsp70 promoter and intron removed 5 copies of the UAS site, generating pJFRC5. A modular cassette containing 10 additional UAS copies was cloned in pJFRC2 as a 5′-AatII to 3′-NheI fragment, yielding pJFRC7, followed by a subsequent digest with NheI and SpeI to yield pJFRC6. pJFRC8 was made by cutting pJFRC7 with HindIII, blunting with DNA polymerase, digesting with ZraI, and finally cloning as a blunt end 20XUAS fragment into ZraI-cut pJFRC7.
Construction of pJRC9–pJFRC11 tandem vectors:
To construct pJFRC9, pJFRC2 was first cut with HindIII and made blunt using DNA polymerase, followed by PmeI digestion and gel extraction. The resultant fragment was cloned into pJFRC2, which was cut with FseI and treated with DNA polymerase; products were screened for orientation. pJFRC10 and pJFRC11 were created in a multistep process. First, pJFRC2 was cut with FseI, followed by ligation with a synthesized gypsy-insulated spacer of 2.8 kb, generating pJFRC2-INS. pJFRC2-INS was cut with PmeI and ligated with a HindIII (blunt) to PmeI-cut pJFRC2 fragment (see above). Orientation was determined via PCR and restriction digest.
Construction of pJFRC12–pJFRC14:
DNA encoding the first 85 amino acids of Drosophila Src oncogene at 64B (Src64B) was PCR amplified from P{UAS-myr-mRFP} (gift of Henry Chang, Purdue University, West Lafayette, IN) as a 5′-XhoI to 3′-BamHI fragment and cloned into pJFRC2, replacing the mCD8 membrane tag. Next, a Drosophila codon-optimized GFP utilizing the same F64L and S65T mutations as in the published version (Lee and Luo 1999) was cloned 5′-BamHI to 3′-XbaI, creating pJFRC12. Codon-optimized GFP from pJFRC12 was generated by PCR amplification and used to replace the myr∷GFP transgene in pJFRC12 as a 5′-XhoI to 3′-XbaI fragment, making pJFRC13. Next, the WPRE was PCR amplified from FLEX vector DNA and cloned as a XbaI fragment into pJFRC13, yielding pJFRC14.
Construction of pJRC15–pJFRC18:
A modular cassette, containing eight LexA-binding sites from the sulA operator and an hsp70 basal promoter, was cloned in pJFRC1 as a 5′-HindIII to 3′-BglII fragment yielding pJFRC18. Five LexA-binding sites were PCR amplified from pJFRC18-8XLexAop2-mCD8∷GFP with the inclusion of a 3′-SpeI restriction site. Next, the PCR amplicon was cloned 5′-NheI to 3′-AatII into pJFRC18 to generate pJFRC15. pJFRC16 was made as follows: pJFRC18 vector was digested with HindIII and ZraI to liberate a fragment containing eight LexA-binding sites; the fragment was gel extracted and the HindIII sticky ends were filled in; the HindIII (blunt) to ZraI fragment was cloned into an AvrII-cut 8XLexAop2-mCD8∷GFP that was made blunt with DNA polymerase, creating pJFRC16. pJFRC17 was generated in a similar fashion, but using pJFRC15 as template DNA for all steps.
Adult brain dissection and histochemistry:
Flies for imaging GFP expression were bred from homozygous driver females crossed to homozygous reporter males. Brains of adult female flies were dissected in cold PBS and fixed in 0.8% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) in PBS at 4° overnight. After four 30-min washes in 0.5% BSA and 0.5% Triton X-100 in PBS (PAT), samples were blocked with 3% normal goat serum (NGS) in PAT for 2 hr at room temperature. Samples were then incubated overnight at 4° in a primary antibody solution containing mouse anti-nc82 (1:50; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti-GFP IgG (1:1000; no. A11122, Invitrogen), and 3% NGS in PAT. After four 30-min washes in PAT, samples were incubated overnight at 4° in a secondary antibody solution containing Alexa Fluor 568 goat anti-mouse IgG (1:1000, Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (1:1000, Invitrogen), and 3% NGS in PAT. After at least four 30-min washes in PAT, samples were rinsed in PBS and mounted in Vectashield mounting medium (Vector Labs, Burlingame, CA) within multiwell silicone adhesive spacers (Grace Bio-labs, Bend, OR). Samples were covered with a no. 1.5 glass coverslip before imaging.
Immunolabeled adult brains were imaged with a Model 510 laser scanning confocal microscope (Zeiss, Thornwood, NY) under 20× magnification. Twelve-bit z-stack images were scanned at 1-μm section intervals with a resolution of 1024 × 1024 pixels. Z-projection images were converted from 12-bit to 8-bit, inverted, and rotated using macros written for Fiji (http://pacific.mpi-cbg.de/wiki/index.php/Main_Page). Image processing macros are available upon request.
Larval dissection and histochemistry:
Larval tissues were dissected in PBS and fixed for 1–2 hr in 4% paraformaldehyde at room temperature. After multiple rinses in PBS with 1% Triton X-100 (PBS-TX), tissues were preblocked in 2% NGS in PBS-TX for 30 min and then incubated overnight at 4° with mouse anti-neuroglian (1:50 BP-104; Developmental Studies Hybridoma Bank) and rabbit anti-GFP IgG (1:1000) in PBS-TX. After multiple rinses in PBS-TX, tissues were incubated overnight in the cocktail of fluorescent secondary antibodies described above. Nervous systems were then washed two to three times in PBS-TX, mounted on polylysine (Sigma, St. Louis)-coated coverslips, dehydrated through a graded ethanol series, cleared in xylene, and mounted in DPX Mountant (Sigma).
Immunolabeled larval nervous systems were imaged under 40× magnification as a 2 × 3 array of tiled stacks. Each stack was scanned as an 8-bit image with a resolution of 512 × 512 and a z-step interval of 2 μm. For a given series, all images were taken at the same gain settings.
Quantitative reverse transcriptase PCR:
Flies were anesthetized with CO2 and then frozen on dry ice. Heads were removed with a scalpel and immediately placed in Eppendorf tubes containing TRIzol reagent (Invitrogen) on ice. Heads were homogenized in TRIzol using a pellet pestle (Kontes, Vineland, NJ) and RNA was extracted according to Invitrogen instructions. RNA concentration was read using a NanoDrop ND-1000 spectrophotometer (Nano Drop Technologies, Wilmington, DE). RNA integrity was confirmed on a nondenaturing agarose gel. Reverse transcription was performed on 1 μg of RNA treated with gDNA Wipeout Buffer using the Quantitect Reverse Transcription kit (QIAGEN, Valencia, CA) in a final volume of 20 μl. PCR was carried out using 2.5 μl of the reverse transcription reaction and 15 μl of Brilliant SYBR Green QPCR Master Mix (Stratagene) in a total volume of 30 μl. Cycling conditions were as follows: 95° for 10 min followed by 40 cycles at 95° for 30 sec, 55° for 1 min, and 72° for 30 sec. QRT–PCR reactions were run in the Stratagene Mx3005P and analyzed with the accompanying MxPro software. Experiments included no-reverse-transcriptase controls for each template and no-template controls for each pair of primers. Percentage of error was determined by dividing the upper and lower relative quantity (RQ) value limits by the reported value and multiplying by 100. The given error is the largest resultant percentage of error from all samples. Primers used were as follows: GFP, CM29F 5′-ATTGGCGATGGCCCTGTCCT-3′ and CM30R 5′-GTTCATCCATGCCATGTGTAATCC-3′; and Drosophila melanogaster beta-Tubulin at 56D (NM_079071) (normalizer), CM57F 5′-ATCCCGCCCCGTGGTCTG-3′ and CM59R 5′-AAAGCCTTGCGCCTGAACATAGC-3′.
Drosophila stocks:
Flies were reared at 25° and raised on standard cornmeal/molasses food. In addition to the materials presented in this study we used three published transgenic flies: y w; UAS-mCD8∷GFP; + and y w; Pin/CyO; UAS-mCD8∷GFP (Lee and Luo 1999) and w; Sp/Cyo; LexAop-rCD2∷GFP (Lai and Lee 2006).
RESULTS
Optimization of vectors for GAL4 expression:
We examined the effects of altering the sequence of the GAL4 gene and the UTR sequences that flank it in commonly used vectors. Figure 1 diagrams the structure of the vector we used to test these variants and Table 1 lists the resultant constructs. Codon-optimizing genes for expression in heterologous hosts can improve levels of protein expression (Gustafsson et al. 2004). We found that using a variant of the yeast GAL4 gene that had been optimized for both Drosophila codon-usage and translation-initiation sequence increased levels of GAL4-driven GFP by ∼50% (compare Figure 2B with 2C).
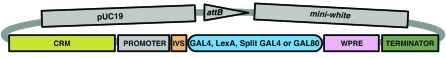
Figure 1.—
Diagram of pBP cloning vectors for GAL4, LexA, GAL80, and Split GAL4. All constructs contain the pUC19-derived bacterial origin of replication and ampicillin resistance gene, the PhiC31 attB site, the mini-white marker for identification of transformants in Drosophila, and the DSCP basal promoter (Pfeiffer et al. 2008). The vector backbone is modular to allow for many possible combinations: gray shading indicates components that were held constant, while the colored elements were varied between the constructs we describe in this report. Abbreviations: CRM, conserved regulatory module (generally a 2- to 3-kb enhancer-containing fragment of Drosophila DNA); IVS, intervening sequence within the 5′-UTR; WPRE, a woodchuck hepatitis virus post-transcriptional regulatory element; and TERMINATOR, the transcriptional terminator.
TABLE 1.
New GAL4, LexA, Split GAL4, and GAL80 vector backbones used in this study
| Name | 5′-UTR IVS | Transgene | 3′-UTR WPRE | Terminator |
|---|---|---|---|---|
| pBPGAL4.1Uw | GAL4a | hsp70 | ||
| pBPGAL4.2Uw | GAL4a | hsp70 | ||
| pBPGAL4.2Uw-2 | GAL4a | SV40 | ||
| pBPGAL4dUw | GAL4 deletion variant II-9 (GADd) | hsp70 | ||
| pBPGAL4d.2Uw | GAL4 deletion variant II-9a (GADd) | hsp70 | ||
| pBPGAL4.2∷VP16Uw | GAL4∷VP16a | hsp70 | ||
| pBPGAL4.2∷p65Uw | GAL4∷p65a | hsp70 | ||
| pBPLexA∷GADdUw | LexA∷GADda | hsp70 | ||
| pBPLexA∷GADflUw | LexA∷GADfla | hsp70 | ||
| pBPnlsLexA∷GADflUw | nlsLexA∷GADfla | hsp70 | ||
| pBPLexA∷VP16Uw | LexA∷VP16 | hsp70 | ||
| pBPLexA∷p65Uw | LexA∷p65a | hsp70 | ||
| pBPnlsLexA∷p65Uw | nlsLexA∷p65a | hsp70 | ||
| pBPZpGAL4DBDUw | Zip-GAL4DBDa | hsp70 | ||
| pBPp65ADZpUw | p65AD-Zipa | hsp70 | ||
| pBPGAL80Uw-1 | GAL80a | hsp70 | ||
| pBPGAL80Uw-2 | GAL80a | + | hsp70 | |
| pBPGAL80Uw-3 | + | GAL80a | hsp70 | |
| pBPGAL80Uw-4 | + | GAL80a | + | hsp70 |
| pBPGAL80Uw-5 | GAL80a | SV40 | ||
| pBPGAL80Uw-6 | + | GAL80a | + | SV40 |
BP plasmid vector backbones are derived from pBPGUw (Pfeiffer et al. 2008) and contain the pUC19-derived bacterial origin of replication and ampicillin resistance gene, the PhiC31 attB site, the mini-white marker for identification of transformants in Drosophila, and the DSCP basal promoter. For more details see Figure 1 and Addgene plasmid 17575. Abbreviations: U, DSCP basal promoter; w, mini-white marker; nls, nuclear localization signal; IVS, intervening sequence within the 5′-UTR; WPRE, a woodchuck hepatitis virus post-transcriptional regulatory element within the 3′-UTR; TERMINATOR, the transcriptional terminator; and pBP, plasmid BP backbone.
Drosophila codon-optimized transgene.
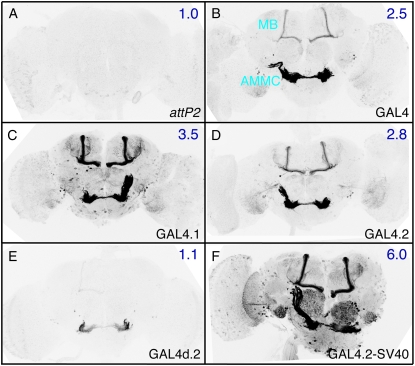
Figure 2.—
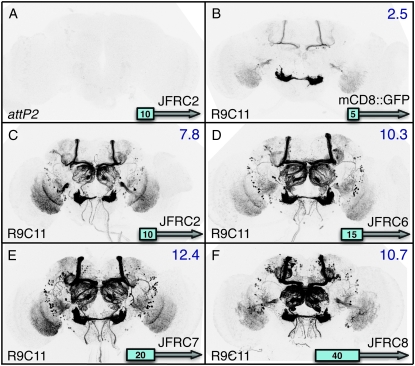
Effects of codon optimization and terminators on GAL4-driven GFP transgene expression. Adult brains are shown after immunostaining to reveal GFP expression. (A) As a control for transcription of the UAS-mCD8∷GFP reporter construct (Lee and Luo 1999) in the absence of a GAL4 driver, as well as for the background of the immunohistochemistry procedure, UAS-mCD8∷GFP was crossed to the attP2 site with no integrated construct. (B–F) The CRM R9C11 fragment (Pfeiffer et al. 2008) was used to drive GAL4 expression in constructs that are integrated into the attP2 site and crossed to the UAS-mCD8∷GFP reporter. (B) The GAL4 gene from Brand et al. (1994), which contains 45 bp of the hsp70 5′-UTR and transcriptional terminators from both the GAL4 gene and the hsp70 gene. CRM R9C11 drives expression prominently in the antennal mechanosensory and motor center (AMMC) and mushroom body (MB). (C) GAL4.1, the same construct as in B, but with a GAL4 coding sequence optimized for Drosophila codon usage. (D) GAL4.2, the same construct as in C, but with the 45-bp hsp70 5′-UTR and yeast transcriptional terminator removed. (E) Same as D, but with GAL4 deletion variant II-9 (pMA236; Ma and Ptashne 1987a) replacing the full-length GAL4 gene. (F) Same as D, but with the SV40 terminator replacing the hsp70 terminator. Relative quantities of GFP mRNA expression levels as measured by QRT–PCR in homogenates of heads of each genotype relative to the control (calibrator) in A, which was arbitrarily set at 1.0, are indicated in the top right of each panel. Assays were done in triplicate except for F, which was done in duplicate; error was within 9% of reported values.
The GAL4 gene from pGaTB and pGawB (Brand and Perrimon 1993), present in most enhancer trap vectors, contains two additional DNA segments whose effects on the transcription of the GAL4 gene have not been systematically investigated: 45 bp of hsp70 5′-UTR and the yeast 3′-GAL4 transcriptional terminator (Brand et al. 1994). It has been suggested that sequences within the cloned hsp70 5′-UTR of GAL4 might direct the larval salivary gland background expression common to most P-element-generated GAL4 insertions (Gerlitz et al. 2002; Hrdlicka et al. 2002). We found that removal of one or both of these elements modestly decreased the level of GAL4-driven transgene expression (compare Figure 2C with 2D, and data not shown). Moreover, our collection of GAL4 drivers contains these sequences, but lacks the salivary gland background (Pfeiffer et al. 2008). If indeed the P-element vector carries a cryptic salivary gland enhancer, it seems to have been lost in our constructs.
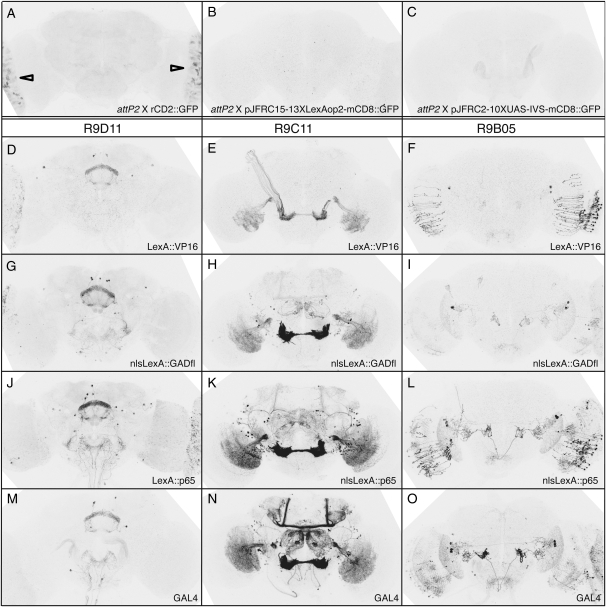
Most GAL4 constructs carry the hsp70 3′-UTR, which contains degradation sequences that cause rapid turnover in non-heat-shock conditions (Petersen and Lindquist 1989). Replacing the hsp70 transcriptional terminator used in our GAL4 vectors with the SV40 UTR is expected to result in greater mRNA stability, consistent with the increased expression levels we observed (Figure 2F). It may also result in perdurance of expression from earlier developmental stages, one explanation for the extra cells observed when using CRM R9C11-GAL4-SV40 (Figure 2F). Similar results were obtained using enhancers R9C11, R9B05, and R9D11 (supporting information, Figure S1, and Pfeiffer et al. 2008).
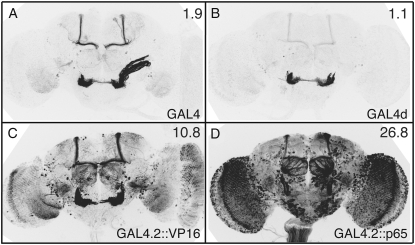
Choice of activation domain can strongly affect levels of transcription (Ptashne 1988; Melcher 2000). We compared the GFP expression patterns driven by GAL4 constructs with four different activation domains: (1) GAL4; (2) GAL4 deletion variant II-9 (GAL4d, GAL4 amino acid residues 1–147 fused by a small linker with 768–881; Ma and Ptashne 1987a), which has been used in LexA (Lai and Lee 2006) and Split GAL4 vectors (Luan et al. 2006); (3) herpes simplex virus protein VP16 (Sadowski et al. 1988); and (4) human p65 (Schmitz and Baeuerle 1991), which has been successfully used to drive transcription of reporter transgenes in Drosophila as part of GeneSwitch (Osterwalder et al. 2001; Roman et al. 2001). Figure 3 shows expression driven by these GAL4 variants. R9C11-GAL4d (Figure 3B) drove GFP at a greatly reduced level, relative to intact GAL4 (Figure 3A). Conversely, replacing the GAL4 activation domain with either VP16 (Figure 3C) or p65 (Figure 3D) resulted in severalfold increases in GFP mRNA; the observed expression patterns were also broader, presumably because weakly expressing cells became visible. Although GAL4∷p65 yielded the highest transcription levels, it reduced transformant viability. Cytotoxicity has been previously reported when GAL4 is expressed at very high levels (Kramer and Staveley 2003).
Figure 3.—
Activation domain choice has a large effect on GAL4-driven levels. Drosophila adult brains were immunostained for GFP after crossing drivers to the UAS-mCD8∷GFP reporter, as in Figure 2. All GAL4 constructs are directed by CRM R9C11, integrated into attP2, and contain an hsp70 terminator. (A) GAL4 from pGawB (Brand and Perrimon 1993). (B) GAL4d, containing the GAL4 deletion variant II-9 (pMA236; Ma and Ptashne 1987a). The 45-bp segment of hsp70 5′-UTR has been removed, but the yeast transcriptional terminator from the GAL4 gene is present. (C) GAL4.2∷VP16, containing a fusion of the GAL4 DNA-binding domain to the VP16 activation domain. The entire coding region has been optimized for Drosophila codon usage and the hsp70 5′-UTR and yeast transcriptional terminator have been removed. (D) GAL4.2∷p65, as in C, but with the activation domain from p65. Relative GFP QRT–PCR values, standardized to attP2 crossed to UAS-mCD8∷GFP (set at 1.0), are indicated in the top right of each panel. Assays were done in triplicate; error was within 5% of reported values.
Improved UAS reporter constructs:
Using a modular backbone (Figure 4), we generated a series of UAS-mCD8∷GFP reporter constructs (Table 2) and used them to determine the effects on GFP expression of (1) the choice of basal promoter, (2) the presence of an intron in the 5′-UTR, (3) the number of UAS sites, (4) the copy number of the reporter construct, (5) the nature of the protein localization signals fused to the GFP protein, and (6) the presence of the WPRE regulatory element in the 3′-UTR.
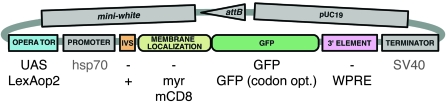
Figure 4.—
Diagram of pJFRC reporter constructs. All constructs contain the pUC19-derived bacterial origin of replication and ampicillin resistance gene, the PhiC31 attB site, the mini-white marker for identification of transformants in Drosophila, an hsp70 basal promoter, and an SV40 transcriptional terminator. The vector backbone is modular to allow for many possible combinations: gray shading indicates components that were held constant, while the colored elements were varied between the constructs we describe in this report. Examples of some of these alternatives are listed below colored elements; see text for more details.
TABLE 2.
New reporter transgenes used in this study
| Name | Tandem construct | Operator | 5′-UTR IVS | Reporter | 3′-UTR WPRE |
|---|---|---|---|---|---|
| pJFRC-MUH | 10XUAS | ||||
| pJFRC1 | 10XUAS | mCD8∷GFP | |||
| pJFRC2 | 10XUAS | + | mCD8∷GFP | ||
| pJFRC3 | 1XUAS | + | mCD8∷GFP | ||
| pJFRC4 | 3XUAS | + | mCD8∷GFP | ||
| pJFRC5 | 5XUAS | + | mCD8∷GFP | ||
| pJFRC6 | 15XUAS | + | mCD8∷GFP | ||
| pJFRC7 | 20XUAS | + | mCD8∷GFP | ||
| pJFRC8 | 40XUAS | + | mCD8∷GFP | ||
| pJFRC9 | TH1 | 10XUAS | + | mCD8∷GFP | |
| 10XUAS | + | mCD8∷GFP | |||
| pJFRC10 | THS2 | 10XUAS | + | mCD8∷GFP | |
| 10XUAS | + | mCD8∷GFP | |||
| pJFRC11 | TTS3 | 10XUAS | + | mCD8∷GFP | |
| 10XUAS | + | mCD8∷GFP | |||
| pJFRC12 | 10XUAS | + | myr∷GFPa | ||
| pJFRC13 | 10XUAS | + | GFPa | ||
| pJFRC14 | 10XUAS | + | GFPa | + | |
| pJFRC15 | 13XLexAop2 | mCD8∷GFP | |||
| pJFRC16 | 16XLexAop2 | mCD8∷GFP | |||
| pJFRC17 | 26XLexAop2 | mCD8∷GFP | |||
| pJFRC18 | 8XLexAop2 | mCD8∷GFP |
Janelia Farm Reporter Construct (JFRC) backbones are derived from pBDP (Pfeiffer et al. 2008) and contain the pUC19-derived bacterial origin of replication and ampicillin resistance gene, the PhiC31 attB site, and the mini-white marker for identification of transformants in Drosophila. For more details see Figure 4 and Addgene plasmid 17566. In addition, all vectors also contain a basal promoter derived from hsp70 and an SV40 transcriptional terminator (Brand and Perrimon 1993). Abbreviations: IVS, intervening sequence within the 5′-UTR; and WPRE, a woodchuck hepatitis virus post-transcriptional regulatory element within the 3′-UTR.
Drosophila codon-optimized GFP.
We tested two basal promoters, the Drosophila synthetic core promoter (DSCP) (Pfeiffer et al. 2008) and the hsp70 basal promoter (Brand and Perrimon 1993) to determine the levels of expression they gave when paired with UAS sites in the presence or absence of GAL4. The DSCP contains a large set of core promoter elements to promote robust expression with a broad range of enhancers that bind different activator proteins, which themselves might have specific preferences for one or more of these promoter motifs (Pfeiffer et al. 2008). While the DSCP promoter works well with a variety of enhancers, we found that in the specific case of GAL4-driven UAS expression, the hsp70 basal promoter yielded twofold higher expression levels than the same construct built with the DSCP, while still displaying nearly undetectable leak in the absence of GAL4 (data not shown). We therefore used the hsp70 basal promoter in subsequent UAS constructs.
Intervening sequences (IVS, introns) placed in the 5′-UTR of genes can boost expression by promoting steady-state export of spliced mRNAs to the cytoplasm (Huang and Gorman 1990; Duncker et al. 1997; Zieler and Huynh 2002). We added a 67-bp intron from Drosophila Myosin heavy chain (Mhc IVS16) to pJFRC1, our 10XUAS-mCD8∷GFP vector to make pJFRC2. GAL4-driven GFP mRNA levels from pJFRC2 were elevated 20% compared to the same construct without the intron (data not shown). We also tested four other introns: synthetic intron IVS8 (Invitrogen), white intron 2 (Lee and Carthew 2003), ftz intron (Ni et al. 2009), and a hybrid intron (Choi et al. 1991). As none were substantially better than Mhc IVS16 in terms of expression levels and background (data not shown), we chose to use Mhc IVS16 in subsequent constructs.
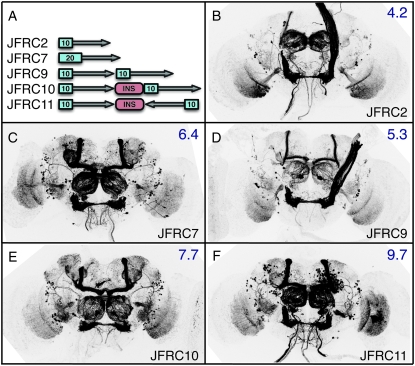
pJFRC2 contains 10 copies of an optimized GAL4 DNA-binding site, the “ScaI 17-mer” (Webster et al. 1988), and drives GFP expression more than twofold higher than the UAS-mCD8∷GFP construct described by Lee and Luo (1999) that contains 5 sites (compare Figure 5B with 5C). We varied the number of UAS sites from 5 to 40 and assayed levels of GFP expression driven from R9C11-GAL4 (Figure 5). Maximal GFP expression was obtained with 20 sites (pJFRC7; Figure 5E). Similar results were obtained by Ni et al. (2008) with RNAi Notch hairpin-induced phenotypes.
Figure 5.—
Increasing the number of GAL4 DNA-binding sites boosts GFP levels. Drosophila adult brains were immunostained for GFP after crossing R9C11-GAL4 to mCD8∷GFP reporters with 5 to 40 UAS sites. With the exception of the UAS-mCD8∷GFP construct of Lee and Luo (1999), which is a P-element insertion on the second chromosome, all constructs are integrated into attP2. (A) attP2 (no GAL4 driver) crossed to pJFRC2 (negative control). (B–F) R9C11-GAL4 crossed to reporters containing (B) 5, (C) 10, (D) 15, (E) 20, and (F) 40 UAS sites. We also tested a 5XUAS construct in the pJFRC backbone (pJFRC5; see Table 2), which gave similar results to UAS-mCD8∷GFP (data not shown). Relative GFP QRT–PCR values, normalized to a level of 1.0 for attP2 × UAS-mCD8∷GFP (see Figure 2A), are indicated in the top right of each panel. Assays were done in triplicate; error was within 7% of reported values.
Finally, while inclusion of gypsy insulators has been shown to boost transgene expression (Markstein et al. 2008), we did not observe notable increases from the inclusion of these elements in pJFRC2 or pJFRC12 when integrated in attP2 (data not shown).
We also tested expression driven from tandem constructs, containing two copies of pJFRC2 in a single insert (Figure 6; Table 2), as the ability to drive multiple UAS constructs without complicated genetics can be helpful. A tandem construct with two copies of the internal components of pJFRC2 in a tail-to-head orientation, pJFRC9 (Figure 6D), gave more expression than pJFRC2 (Figure 6B), but less than pJFR7 (Figure 6C), which has the same number of UAS sites upstream of a single GFP gene. The addition of a 2.8-kb gypsy insulated spacer between the tandem copies of these same components in pJFRC10 (Figure 6E) increased expression to above that of pJFRC7. The relative orientation of the tandem genes was significant; a tail-to-tail construct with a spacer, pJFRC11 (Figure 6F), gave the highest level of GFP expression. Both the spacer and a tail-to-tail orientation appear to be required to obtain the expected twofold increase over pJFRC2. While helpful in increasing expression levels, such tandem constructs will likely have their greatest utility when used to direct the expression of two different responder genes under the control of the same GAL4 driver.
Figure 6.—
Tandem reporters can be used to increase output. (A) Diagram of reporter constructs. Note variation in UAS copy number (indicated in the blue box), tandem orientation (indicated by the arrows), and presence of a gypsy-insulated spacer of 2.8 kb (INS). The synthesized spacer was designed by independently randomizing a minimum of five times (Shuffle program GCG Version 11.1; Accelrys, San Diego) a sequence derived from kanamycin CDS (base pairs 1–810) and a sequence derived from the E. coli lacZ CDS (base pairs 799–2000) and then fusing these randomized sequences. The spacer was then flanked on either end with 424 bp from the 5′-UTR of gypsy (base pairs 647–1074) that contains 12 binding sites for the su(Hw) protein (Marlor et al. 1986; Spana et al. 1988). (B–F) Drosophila adult brains immunostained for GFP after crossing R9C11-GAL4 driver to indicated reporter. All constructs integrated into attP2. (B and C) Controls showing driver pattern with (B) 10 UAS copies (pJFRC2) or (C) 20 copies (pJFRC7). (D) A tandem 10XUAS-mCD8∷GFP reporter in a tail-to-head orientation (pJFRC9). (E) As in D, but with inclusion of a gypsy-insulated 2.8-kb spacer between inserts (pJFRC10). (F) As in E, but reporters are inserted tail to tail (pJFRC11). Relative GFP QRT–PCR values, standardized to attP2 × UAS-mCD8∷GFP, are indicated in the top right corners. Assays were done in duplicate; error was within 13% of reported values.
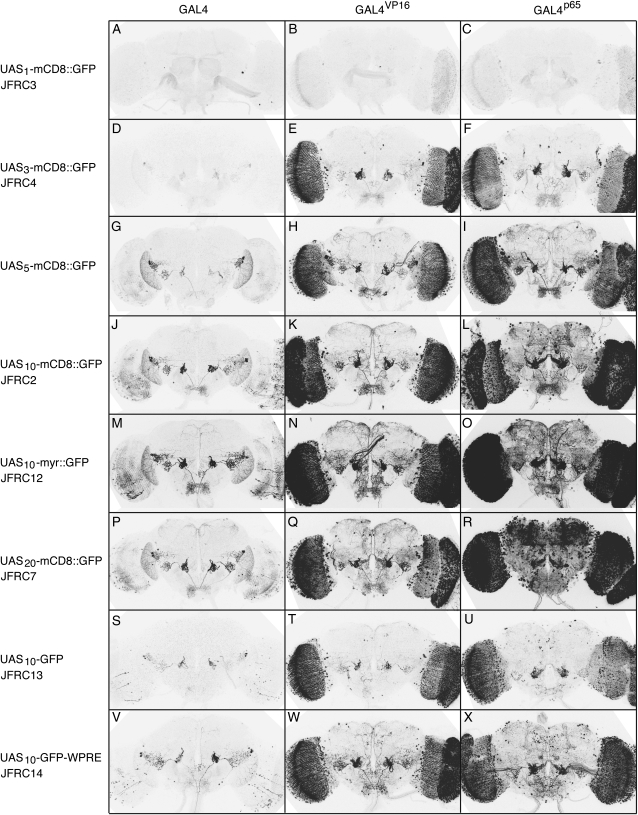
For many experiments, it is desirable to generate the same level of reporter expression while utilizing enhancers of very different strengths. For example, decreasing the number of UAS sites when using a strong enhancer-GAL4 driver will result in lower levels of reporter expression. We sought to evaluate our ability to modulate expression levels by varying the strength of the activation domain of the GAL4 driver and the number of UAS sites carried by the reporter construct (Figure 7). mCD8∷GFP driven from a single UAS site was almost undetectable, even with our strongest GAL4 driver, GAL4∷p65 (Figure 7C). The expression levels obtained increased with UAS number from 3 to 20 and with activation domain strength (Figure 7, D–R). Thus, using a single enhancer, we can obtain a range of expression levels from undetectable to undesirably high and cytotoxic, by simply modulating the strength of the activation domain and UAS responder. The toxicity apparent with the GAL4∷p65 driver and reporter constructs with ≥10 UAS sites (Figure 7, L, O, and R) appears to result largely from reporter protein expression levels, as this same driver with a 3X or a 5XUAS reporter results in labeled cells of apparently normal morphology (Figure 7, F and I; also see Figure S2).
Figure 7.—
Tuning expression levels by varying the strength of the activation domain and UAS responder. Drosophila adult brains were immunostained for GFP. With the exception of the 5× UAS-mCD8∷GFP (Lee and Luo 1999) construct all transgenes are integrated into attP2. CRM R9B05 was used to drive three GAL4 variants: standard GAL4 (as used in the constructs described by Pfeiffer et al. 2008), GAL4.2∷VP16, or GAL4.2∷p65. These three GAL4 drivers were crossed to different responders as indicated, which vary in number of UAS sites, localization tag, and inclusion of a WPRE: (A–C) The 1XUAS-mCD8∷GFP (pJFRC3). (D–F) The 3XUAS-mCD8∷GFP (pJFRC4). (G–I) The 5XUAS-mCD8∷GFP of Lee and Luo (1999). (J–L) The 10XUAS-mCD8∷GFP (pJFRC2). (M–O) The 10XUAS-myr∷GFP (pJFRC12): myristoylated, codon-optimized GFP. (P–R) The 20XUAS-mCD8∷GFP (pJFRC7). (S–U) The 10XUAS-GFP (pJFRC13): untagged (cytoplasmic), codon-optimized GFP. (V–X) As in S–U, but containing a WPRE in the 3′-UTR. Note that strength of the activation domain and UAS number can drive undesirable levels of GFP and cause cytotoxicity. (P and R) Weakly expressing cells in the optic lobe become prominent, while those that expressed robust levels are gone (see Figure S2).
Depending on the experiment, it may be desirable to target the reporter protein to either membrane or cytoplasmic sites. pJFRC2 employs the first 222 amino acids of the mouse CD8 protein to target GFP to the plasma membrane as first described by Lee and Luo (1999). We also evaluated a second method of targeting proteins to the membrane, N-myristoylation (Resh 1999); pJFRC12 employs the first 85 amino acids encoded by the Drosophila Src oncogene at 64B, Src64B, which has been used previously in Drosophila to target proteins to the membrane (Mauss et al. 2009). Myristoylation improved signal strength; with 10XUAS, the N-myristoylated GFP reporter (pJFRC12; Figure 7, M–O) expresses GFP at levels similar to our 20× UAS-mCD8∷GFP reporter (pJFRC7; Figure 7, P–R).
Signal strength observed with our cytoplasmic GFP reporter (pJFRC13; Figure 7, S–U) was weak compared to that obtained with an equivalent membrane-targeted GFP construct (pJFRC2; Figure 7, J–L). However, inclusion of the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) (Zufferey et al. 1999) in the 3′-UTR (pJFRC14; Figure 7, V–X) substantially increased GFP levels, making them comparable to those seen with pJFRC2. However, the addition of the WPRE did not cause a notable increase in GFP expression from either the myristoylated or the mCD8 membrane-targeted reporters (data not shown).
Improved LexA drivers and operators:
Lai and Lee (2006) showed that a C-terminal fusion with either the GAL4 activation domain II (GADd, residues 768–881 of GAL4) or the more potent VP16 activation domain from herpes simplex virus allows LexA-driven transcription of reporter transgenes in a GAL80-sensitive or -insensitive manner, respectively. We sought to improve the potency of these published LexA drivers by fusing LexA with two alternative activation domains. First, we used an extended version of the GAL4 activation domain, GADfl (residues 148–881 of GAL4), which includes activation domains I and II (Ma and Ptashne 1987a). Second, we replaced VP16 with the human p65 activation domain. Also, because LexA is a prokaryotic protein and is not specifically targeted to the nucleus in eukaryotic cells (Brent and Ptashne 1984), we reasoned that a nuclear localization signal (NLS) might increase its ability to promote transcription.
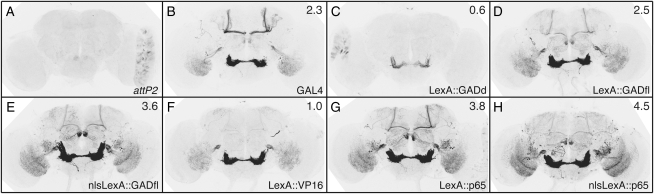
We constructed R9C11 enhancer-directed LexA drivers with a codon-optimized GADd, VP16 (codon optimization of VP16 did not significantly improve LexA-driven transgene expression; data not shown), codon-optimized GADfl, or codon-optimized p65, with or without an NLS (Table 1). We compared the patterns and strength of expression observed when these constructs drove LexAop-rCD2∷GFP (Lai and Lee 2006). LexA drivers containing GADfl (Figure 8D) or p65 (Figure 8G) activation domains drove GFP expression at higher levels than GADd (Figure 8C) or VP16 (Figure 8F). In all cases, we observed the expected expression pattern, based on R9C11-GAL4 driving mCD8∷GFP (Figure 8B). Surprisingly, expression levels of GFP driven from LexA∷GADfl were only slightly lower than those obtained with LexA∷p65. Although a rigorous quantitative comparison cannot be made, as the GFP reporter constructs used differ in structure and genomic location, the level of LexAop-rCD2∷GFP expression driven from LexA∷GADfl seemed comparable to that seen with R9C11-GAL4 driving the mCD8∷GFP reporter of Lee and Luo (1999). Further, as expected, tub-GAL80 (Lee and Luo 1999) was able to suppress GFP expression from LexA∷GADfl (data not shown). Addition of a nuclear localization signal to the LexA∷GADfl (Figure 8E) or the LexA∷p65 (Figure 8H) proteins modestly increased their efficacy.
Figure 8.—
Improved LexA drivers with GADfl and p65 activation domains. Drosophila adult brains were immunostained for GFP after crossing LexA drivers to a published LexAop-rCD2∷GFP (Lai and Lee 2006). All GAL4 and LexA constructs are directed by CRM R9C11 and integrated into attP2. (A) attP2 (no LexA driver) crossed to published LexAop-rCD2∷GFP (negative control; note “leak” expression in the lamina). (B) R9C11-GAL4 with UAS-mCD8∷GFP. (C and D) R9C11-LexA drivers containing GAL4 activation domain variants GADd (C) or GADfl (D), crossed to rCD2∷GFP. (E) As in D, but with a nuclear localization signal (nls). (F and G) R9C11-LexA drivers with GAL80-insensitive activation domains VP16 (F) or p65 (G), crossed to rCD2∷GFP. (H) As in G, but with an nls. Relative GFP mRNA levels as measured by QRT–PCR, standardized to attP2 × UAS-mCD8∷GFP (set as 1.0), are indicated in the top right of each panel. Assays were done in duplicate; error was within 14% of reported values.
LexA protein binds, with various affinities, to a 20-bp motif found in the promoters of >20 E. coli genes (Schnarr et al. 1991; Wade et al. 2005). Drosophila LexA reporters (Lai and Lee 2006) have used the colE1-binding motif because it was found to have one of the highest affinities for LexA (Ebina et al. 1983). Although the colE1 sequence permits robust transgene expression in Drosophila, these LexA reporters allow low levels of expression, or leak, in the absence of LexA, presumably due to affinity of endogenous transcription factors for the colE1-derived LexA-binding sites (Figure 9A and data not shown). In an attempt to identify other LexA-binding sites that would support strong expression in the presence of LexA, but not show any expression in its absence, we compared leak and levels of GFP induction from reporter transgenes containing sites from colE1 (Ebina et al. 1983), sulA (Cole 1983), umuDC (Kitagawa et al. 1985; Perry et al. 1985), or a synthetic 22-bp lexA operator (Brent and Ptashne 1984). The latter two operator sequences did not perform well in our assays and were dropped from further studies (data not shown). Moreover, the published colE1 LexA-binding sites showed strong leak in both larval and adult brains when used to drive mCD8-GFP under the DSCP promoter in attP2 (data not shown). Thus, we focused on sulA-derived LexA-binding sites for subsequent constructs. We also varied the number of binding sites, which we expected would influence levels of transgene expression on the basis of studies with the GAL4/UAS system (this study; Ni et al. 2008). We built mCD8∷GFP reporters containing 8, 13, 16, or 26 LexA DNA-binding sites from the sulA operator and found the 13× version to give optimal expression with minimal leak (Figure 9 and data not shown). This reporter (pJFRC15) reproduced the expected expression pattern for three different enhancer–LexA constructs (Figure 9).
Figure 9.—
A LexA operator containing 13 binding sites from sulA provides robust and nonleaky expression. Drosophila adult brains were immunostained for GFP. With the exception of LexAop-rCD2∷GFP (Lai and Lee 2006) all constructs are integrated into attP2. (A) attP2 (no LexA driver) crossed to LexAop-rCD2∷GFP (negative control; note “leak” expression in the lamina indicated by arrowheads). (B) attP2 (no LexA driver) crossed to pJFRC15-13XLexAop2-mCD8∷GFP (pJFRC15; negative control, no detectable leak), a LexAop reporter containing 13 LexA-binding sites derived from sulA. (C) attP2 (no GAL4 driver) crossed to pJFRC2 (negative control). (D–L) Enhancers driving LexA variants (indicated in the bottom right of each panel) crossed to the LexA reporter pJFRC15: (D, G, and J) R9D11-LexA; (E, H, and K) R9C11-LexA; (F, I, and L) R9B05-LexA. (M–O) GAL4 driven by the same three enhancers crossed to pJFRC2: (M) R9D11-GAL4, (N) R9C11-GAL4, and (O) R9B05-GAL4.
Refinement of expression patterns using Split GAL4:
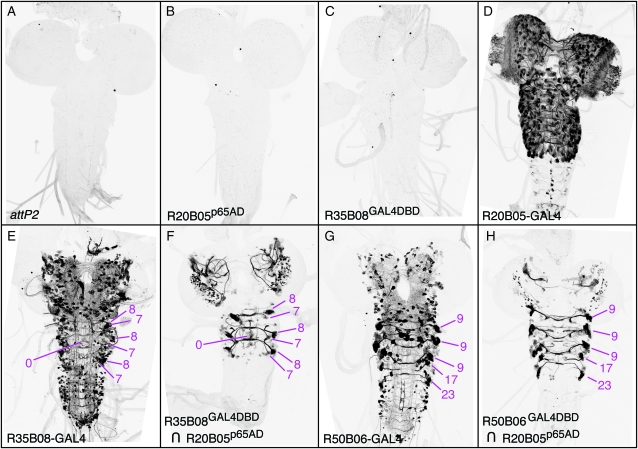
Luan et al. (2006) used GADd and VP16 as activation domains in their implementation of the Split GAL4 system. However, the low levels of reconstituted GAL4 expression obtained limit the utility of this intersectional approach. We tested whether the GADfl and p65 activation domains, which we have shown drive much higher expression levels than the GADd and VP16 domains when fused to the LexA DNA-binding domain, could similarly improve expression in the Split GAL4 approach. We used enhancer R20B05 to drive either GADfl or p65 Split GAL4 activation domains. These were combined with the GAL4 DBD driven under either R35B08 or R50B06 and their ability to drive pJFRC2 was assayed. The p65 domain performed well. Both R35B08Gal4DBD ∩ R20B05p65AD and R50B06Gal4DBD ∩ R20B05p65AD resulted in robust and specific GFP expression in the predicted cells, where the expression patterns of the two parent enhancers overlapped; expression outside the overlap was not detected (Figure 10). In contrast, the GADfl domain performed poorly; neither R35B08Gal4DBD ∩ R20B05GADfl nor R50B06Gal4DBD ∩ R20B05GADfl yielded detectable GFP expression (data not shown).
Figure 10.—
Refinement of GFP expression using Split GAL4. Drosophila third instar central nervous systems were immunostained for GFP. All constructs are integrated into attP2. (A) attP2 (no GAL4 driver) crossed to pJFRC2-10XUAS-IVS-mCD8∷GFP (negative control). (B and C) R20B05p65AD or R35B08GAL4DBD with pJFRC2 (negative control: no detectable leak). (D) R20B05-GAL4 crossed to pJFRC2 drives expression in immature neurons of the optic lobes and in all of the lineages of secondary neurons in the central brain and ventral nerve cord but none of the primary neurons that are born during embryogenesis. (E) R35B08 crossed to pJFRC2 drives expression in primary neurons of the central brain and ventral nerve cord, as well as eight secondary lineages in the central brain and secondary lineages 0, 8, and 7 in the thoracic neuromeres of the ventral nerve cord (for a description of lineages see Truman et al. 2004). (F) Crossing Split GAL4 DNA-binding domain driven by R35B08 (R35B08GAL4DBD; enhancer fragment R35B08 cloned in vector pBPZpGAL4DBDUw) with a stock of R20B05p65AD (enhancer fragment R20B05 cloned in vector pBPp65ADZpUw); pJFRC2 (attP40; attP2) yields GFP expression restricted to the overlap between the two parent patterns. (G) R50B06 crossed to pJFRC2 drives expression in primary neurons of the brain and central nervous system, secondary lineage BAmv3 (for a description of brain lineages, see Pereanu and Hartenstein 2006) in the central brain, and secondary lineages 9, 17, and 23 of the ventral nerve cord. (H) Crossing Split GAL4 DNA-binding domain driven by R50B06 (R50B06GAL4DBD) with a stock of R20B05p65AD; pJFRC2 (attP40; attP2) yields GFP expression restricted to the overlap between the two parent patterns. In both F and H, expression in primary neurons is eliminated, as predicted.
Luan et al. (2006) reported instances where Split GAL4 intersections showed expression in cells not observed in either parent pattern and we also observed this phenomenon, to varying extents, in many of the Split GAL4 intersections we examined in the adult brain (A. Nern, B. D. Pfeiffer and G. M. Rubin, unpublished results). One possible explanation is that stronger activation domains were used in the Split GAL4 constructs than in the parent lines; we have shown that both the VP16 and the p65 activation domains drive broader transcription than the GAL4 activation domain (Figure 7). We therefore made additional attempts to develop Split GAL4 reagents, employing the GADfl domain. Since the GADfl peptide is approximately seven times larger than GADd, GADfl may introduce steric hindrance or other deleterious protein–protein interactions. However, our attempts to address this problem by increasing the length of the polyglycine linker connecting the leucine zipper to the GADfl domain from 20, 40, or 80 residues were unsuccessful (data not shown). In summary, when using the p65 activation domain, the Split GAL4 intersectional method performs well in a majority of cases. However, it is important to verify the resultant expression pattern by direct assays.
Subtraction of GAL4 expression by GAL80:
One of the features of the modular system of enhancers and vectors we are generating (Pfeiffer et al. 2008 and this study) is that, once the pattern of expression produced by an enhancer has been established, we expect to be able to use that enhancer to drive another protein in the same pattern. This assumption is particularly important for experiments using GAL80 to refine the patterns of GAL4-driven expression, as it has not been feasible to assay GAL80 expression directly. For the results to be predictable, the vectors used to express the two proteins must differ as little as possible in sequences that affect expression patterns. Moreover, GAL80 must be expressed at levels similar to or higher than GAL4, as it acts by directly binding to GAL4.
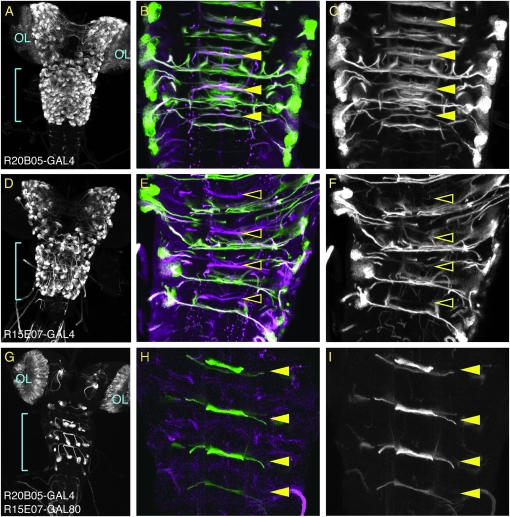
We first tested two enhancers that drive GAL4 in overlapping patterns and then moved one of the enhancers to a GAL80 vector. When these GAL4 and GAL80 constructs were present in the same fly, we obtained the expected pattern in which GAL4 activity is detected only in those cells that do not overlap between the patterns (Figure 11).
Figure 11.—
Targeted refinement of a GAL4 pattern using GAL80. Dissected third instar central nervous systems were immunostained for GFP (green) and neurotactin (magenta). All GAL4 and GAL80 constructs are integrated in attP2; the reporter is a P-element insertion of UAS-mCD8∷GFP on the second chromosome (Lee and Luo 1999). (A) R20B05 drives expression in the immature neurons of the optic lobes (OL) and in all of the lineages of secondary neurons in the central brain and ventral nerve cord in third instar larvae. (B) A single optical section of the region indicated by the blue line in A at the level of the intermediate commissures. R20B05 drives GFP expression in both anterior and posterior commissures of thoracic segments T1–T3 and abdominal segment A1: anterior commissures are indicated by solid yellow arrowheads. (C) As in B, but only the green channel (anti-GFP) is shown. (D) R15E07 drives expression in most of the secondary lineages in the central brain and VNC, with the notable exception of the neuroblast lineages whose axon bundles comprise the anterior intermediate commissure in the thoracic segments. It does not drive optic lobe expression. (E) A single optical section of the region indicated by the blue line in D. Note that R15E07 drives GFP expression in only the posterior commissures; the anterior commissures lacking GFP expression are indicated by open yellow arrowheads. (F) As in E, but only the green channel (anti-GFP) is shown. (G) R15E07-GAL80-SV40 (attP40) crossed to w; UAS-mCD8∷GFP; R20B05-GAL4. As predicted, GFP expression is now restricted to the optic lobes (OL), a few lineages in the central brain, and the thoracic and abdominal lineages that form the anterior intermediate commissure. (H) A single optical section of the region indicated by the blue line in G showing GFP expression restricted to the lineages of the anterior commissure: anterior commissures are indicated by solid yellow arrowheads. (I) As in H, but only the green channel (anti-GFP) is shown.
In the experiment described above the GAL80 vector used a 3′-UTR from Simian virus 40 (SV40), which had been used in previous GAL80 constructs (Lee and Luo 1999; Stoleru et al. 2004; Suster et al. 2004). However, after observing the strong effect on expression of including the SV40 UTR in GAL4 vectors (Figure 2 and also see Figure S1), we realized that it would be important to use the same 3′-UTR in both our GAL4 and GAL80 vectors.
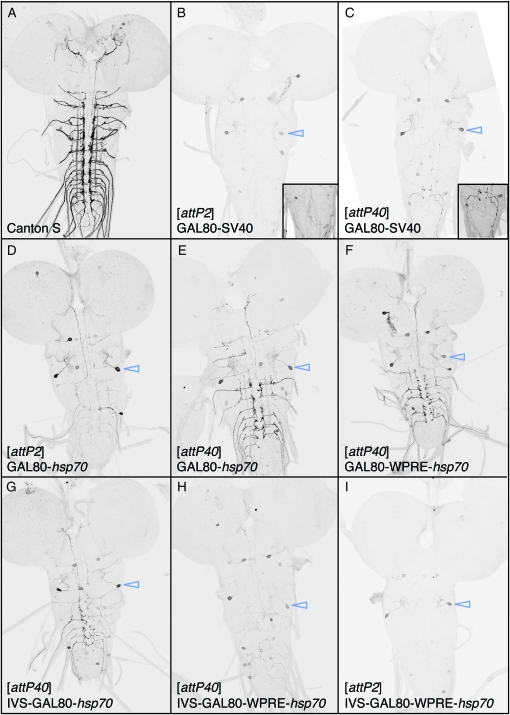
Thus, we generated a series of vectors for the expression of GAL80 that contain either the SV40 or the hsp70 3′-UTR (Table 1) and tested them with the enhancer R11F05 (Figure 12). R11F05 directs reporter expression to ∼100 sensory neurons that project into the ventral nerve cord of the third instar larva. The constructs were integrated into two genomic docking sites: attP2, on the third chromosome, which shows robust expression with GAL4 drivers, and attP40, on the second chromosome, which supports weaker expression (see below). A single copy of R11F05-GAL80-SV40 in attP2, but not attP40, reduced GFP expression generated by R11F05-GAL4 (in attP2) and UAS-mCD8∷GFP (Lee and Luo 1999) to undetectable levels (Figure 12, B and C). The increased suppression seen when the GAL80 construct is inserted in attP2 is presumably a simple reflection of the increased levels of transcription seen from constructs inserted at this genomic docking site compared with attP40. We also generated a tandem construct with two copies of R11F05-GAL80-SV40; when integrated in either attP40 or attP2, this construct was able to suppress GFP expression to undetectable levels (data not shown).
Figure 12.—
GAL80 suppression of GAL4 is improved with post-transcriptional regulatory elements (intron and WPRE). Drosophila third instar central nervous systems were immunostained for GFP. Enhancer R11F05 was used to drive different GAL80 constructs. These constructs were then tested against their parent enhancer using a recombinant reporter line of R11F05-GAL4 (attP2) and a P-element insertion of UAS-mCD8∷GFP on the third chromosome. Both GAL4 and GAL80 constructs use an hsp70 terminator, unless otherwise noted. GFP expression was assayed after crossing R11F05-GAL4 (attP2) UAS-mCD8∷GFP to (A) Canton S (positive control), showing GFP expression in a subset of sensory neurons that project into the ventral nerve cord, and (B) R11F05-GAL80-SV40 (no post-transcriptional regulatory elements), integrated in attP2. The inset at the bottom right shows a portion of the VNC at higher gain. (C) As in B, but integrated in attP40. The inset at the bottom right shows a portion of the VNC at higher gain. (D) R11F05-GAL80, integrated into attP2. A small number of neurons in the pattern escape suppression (express GFP). (E) As in D, but integrated into the weaker site attP40. Note increased incidence of neurons that escape suppression. (F–I) Inclusion of post-transcriptional regulatory elements to R11F05-GAL80 increases the level of GFP suppression: (F) R11F05-GAL80 with a WPRE, in attP40; (G) R11F05-GAL80 with an IVS, in attP40; (H) R11F05-GAL80 with both IVS and WPRE, in attP40; (I) R11F05-GAL80 with both IVS and WPRE, in attP2. Weak background GFP expression from GAL4-independent expression from the UAS-mCD8∷GFP reporter was present in a small group of cells in all preparations (examples indicated by arrowheads).
When we substituted the hsp70 UTR for the SV40 UTR in the GAL80 vector, suppression levels decreased (Figure 12, D and E). This decrease allowed us to test other methods for raising levels of GAL80 expression. We explored the use of two post-transcriptional regulatory elements thought to act by increasing RNA transport from the nucleus to the cytoplasm, rather than mRNA stability: intron 16 (IVS) from Drosophila Myosin heavy chain, and the WPRE (Zufferey et al. 1999). One or both elements were added to R11F05-GAL80-hsp70 (Table 1) and then assayed as above (Figure 12, F–I). Addition of either IVS or WPRE to R11F05-GAL80-hsp70 increased the extent of GFP suppression. When both were added, a single copy of GAL80 was able to reduce GAL4-driven GFP expression to almost undetectable levels when inserted in attP40 and suppressed it completely when inserted in attP2. Thus, R11F05-IVS-GAL80-WPRE-hsp70 suppresses GAL4 at least as well as the SV40 version. We expect the addition of IVS and WPRE would further enhance GAL4 suppression when added to a GAL80-SV40 construct, as might be required when using a weak enhancer to drive GAL80 expression; for this reason we constructed pBPGAL80Uw-6 (Table 1).
Nogi and Fukasawa (1984) cloned and sequenced mutants of GAL80 (GAL80s) that suppress GAL4 in both inducing and noninducing conditions in S. cerevisiae. One of these mutants, GAL80s-1, contains a glycine-to-arginine substitution at amino acid 323, which is thought to give it a higher affinity for GAL4 (Salmeron et al. 1990). We thought this mutation might also increase its GAL4 suppression in Drosophila, so we generated lines in which the R11F05 enhancer drives GAL80s-1or a triple-mutant GAL80 containing the S-0, S-1, and S-2 mutations, with or without the addition of IVS and WPRE. These GAL80s constructs varied in their ability to suppress GAL4-driven GFP expression, but none was substantially better than 11F05-IVS-GAL80-WPRE-hsp70 (data not shown).
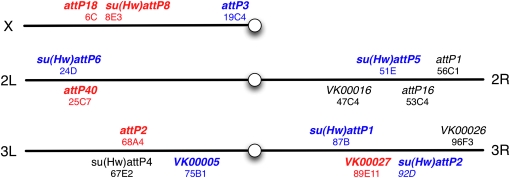
Assaying position effects at genomic attP docking sites:
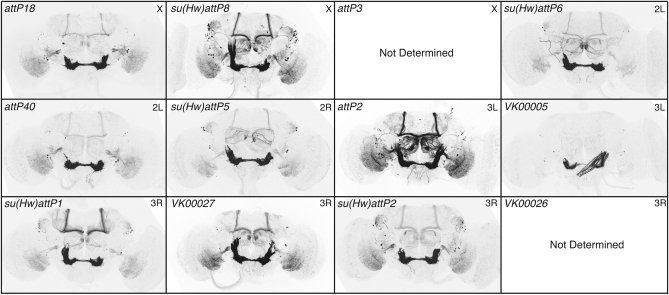
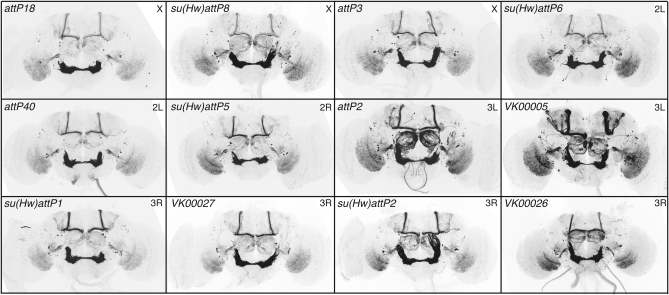
To generate complex genotypes it will be necessary to have several attP sites with similar and favorable properties. To identify such a set, we began with 16 PhiC31 genomic attP docking sites (Figure 13) and assayed their ability to support (1) expression in the adult nervous system when an enhancer trap vector was inserted, (2) expression driven by an exogenous enhancer, and (3) expression from a UAS construct responding to a GAL4 driver. We tested for criterion 1 by constructing an enhancer detector (O'Kane and Gehring 1987; Bellen et al. 1990), pBDPGAL4Uw, and integrating it into each of these sites. We crossed lines containing pBDPGAL4Uw with UAS-mCD8∷GFP (Lee and Luo 1999) and assayed for GFP in the adult brain (data not shown). In four cases, strong position effects were observed, while the remaining 12 candidate attP sites showed minimal or no detectable expression of GFP in the adult brain. We confirmed these results using the stronger GFP reporter pJFRC2 (see Figure S3).
Figure 13.—
Map of evaluated genomic attP sites. The indicated 16 PhiC31 genomic attP integration sites were assayed for four properties: (1) expression in the adult nervous system when an enhancer trap vector was inserted, (2) expression from an exogenous enhancer, (3) expression from a UAS construct responding to a GAL4 driver, and (4) transgene integration rate. Sites shown in red meet all four criteria, while those shown in blue performed well with UAS reporter constructs, but not with enhancer-GAL4 drivers. Sites shown in black were rejected. See text for details. Genomic attP site references: attP1, attP2, attP3, attP18, and attP40 (Groth et al. 2004; Markstein et al. 2008); attP16 (Markstein et al. 2008); VK00005, VK00016, VK00026, and VK00027 (Venken et al. 2006); and su(Hw)attP1, su(Hw)attP2, su(Hw)attP4, su(Hw)attP5, su(Hw)attP6, and su(Hw)attP8 (Ni et al. 2009; this study).
We then tested for the ability of these 12 selected attP sites to allow robust expression of GAL4 driven by an enhancer and a strong core promoter, the DSCP promoter (Pfeiffer et al. 2008). Of the 12 sites, 2 gave integrants at low efficiency rates and were not analyzed further (Figure 14). We integrated three GAL4 drivers, R9C11, R9B05, and R9D11 (Pfeiffer et al. 2008), which have different expression patterns, into the remaining 10 sites and crossed them to pJFRC2 (in attP2) to measure reproducibility and fidelity of expression between sites (Figure 14; also see Figure S4 and Figure S5). Five sites (labeled in red in Figure 13) were identified as superior on the basis of their displaying little modification of the expression patterns observed when the three test drivers were inserted in attP2, a site known to support robust expression (Markstein et al. 2008; Pfeiffer et al. 2008).
Figure 14.—
Chromatin effects on R9C11-GAL4. Drosophila adult brains were immunostained for GFP. R9C11-GAL4 was integrated into 10 attP docking sites and crossed to pJFRC2 in attP2. The genomic docking site is shown in the top left corner of each panel and the chromosome arm of the insertion site in the top right. We were unable to get transformants in the attP3 and VK00026 sites. Four sites showed reproducible and robust expression, comparable to our standard attP2 insertion: attP18, su(Hw)attP8, attP40, and VK00027. Genomic docking sites flanked with gypsy insulators [indicated by su(Hw) in the name] share a common background leak in a half-dozen cells in the lateral horn (see Figure S3 and text).
Chromatin environment can influence transgene expression unpredictably, and thus the same genomic landing sites might not work well for both enhancer-driven GAL4 expression and UAS reporters. We therefore also evaluated the 12 sites for their ability to support expression from an integrated UAS construct in response to GAL4. We crossed pJFRC2 integrated into each of the 12 different attP genomic sites to three GAL4 drivers (all in attP2), see above, to measure pattern fidelity and reliability of expression relative to the same construct inserted in attP2. Reproducibility and fidelity of pJFRC2 expression patterns were consistent across 11 of the 12 insertion sites (Figure 15; also see Figure S6 and Figure S7). Unlike enhancer–GAL4 constructs, the UAS transgenes seem more refractory to chromatin influence and thus offer a larger choice for genomic integration sites, an observation also reported by Bischof et al. (2007). As summarized in Figure 13, by screening 16 different attP docking sites we were able to identify 5 sites (labeled in red) that meet all three of our criteria as well as 6 additional sites (labeled in blue) that appear suitable for UAS–effector constructs but not for enhancer–GAL4 constructs.
Figure 15.—
Chromatin effects on pJFRC2, assayed by R9C11-GAL4. Drosophila adult brains were immunostained for GFP. pJFRC2 was integrated in 12 docking sites and crossed to R9C11-GAL4 in attP2. The genomic docking site is shown in the top left corner of each panel and the chromosome arm of the insertion site in the top right. With the exception of VK00026, all docking sites showed strong, reproducible expression (also see Figure S6 and Figure S7). UAS transgenes seem to be less susceptible to chromatin influence than enhancer-GAL4 constructs; for example, VK00005 works well for UAS, but not GAL4, insertions (compare with Figure 14; also see Figure S4 and Figure S5).
A subset of genomic attP docking sites is flanked with gypsy insulators (Ni et al. 2009). Unexpectedly, GFP staining from crosses with GAL4 lines integrated into these sites shows a common background expression in a few cells in the lateral horn in addition to the expected pattern (Figure 14 and see also Figure S3). To test whether this common background was caused by the insulators themselves, we assayed the ability of the gypsy insulator alone to drive transcription of GAL4. We found that the lateral horn expression seen in docking sites su(Hw)attP1, su(Hw)attP2, su(Hw)attP5, su(Hw) attP6, and su(Hw)attP8 is reproduced when the insulator itself is used as an enhancer (data not shown). In contrast to the GAL4 drivers, UAS effectors integrated into these sites lacked the lateral horn background and showed reproducible expression patterns relative to attP2. However, we did not see a significant boost in reporter gene expression levels when either a UAS or GAL4 was inserted in insulated genomic attP sites.
DISCUSSION
Many experiments in biological research rely critically on the ability to express exogenous proteins or RNAs in transgenic animals in a manner that is regulated for level, timing, and cell type. Methods that seek to attain that goal in D. melanogaster have been developed in a number of laboratories. Although widely used, the characteristics and limitations of these methods have generally not been critically evaluated and are often not appreciated by end users. In the studies we report here, we have attempted to assess different methods, understand the variables that affect their performance, and use that knowledge to improve them.
We first studied the GAL4/UAS binary system (Brand and Perrimon 1993; Duffy 2002). Modifications in codon usage, activation domains, transcriptional terminators, and other features of the UTRs can result in substantial changes in both strength and pattern of expression. Varying the GAL4 activation domain has the single biggest effect on GAL4-driven expression, in terms of both expression level and number of cells observed in the pattern. The choice of GAL4 transcriptional terminator can also effect roughly twofold changes in reporter levels and alter the observed expression pattern, most likely by modifying the half-life of the GAL4 mRNA. We also systematically characterized the effects of promoters, introns, number of UAS-binding sites, tandem copies of UAS constructs, 3′-post-transcriptional modifiers, and protein tags on GFP expression levels from UAS reporter constructs. These data should facilitate optimal construct design for most future GAL4/UAS applications.
The ability to use distinct binary systems to independently target different cells or tissues in the same animal has many applications. The bacterial repressor LexA (Brent and Ptashne 1981; Little et al. 1981) binds DNA sequences distinct from those recognized by GAL4 and has been successfully used in diverse eukaryotic systems such as yeast (Estojak et al. 1995), Drosophila (Diegelmann et al. 2008), and zebrafish (Emelyanov and Parinov 2008). However, the utility of LexA/LexAop as a second binary system in Drosophila has been limited by the relative leakiness of the colE1-derived binding sites and weakness of the drivers compared to GAL4/UAS. We replaced the published GAL4 deletion (GADd) and VP16 activation domains with an extended GAL4 activation domain (GADfl) and the human p65 activation domain. We also screened LexA-binding sites from a variety of LexA target genes and found that 13 copies of a binding site derived from the sulA gene produced robust reporter expression in the presence of LexA, with no expression detectable by histochemical methods in its absence. However, sensitive assays using flippase suggest there may still be a very low level of leak under some circumstances (A. Nern, B. D. Pfeiffer and G. M. Rubin, unpublished results). Used together, our new LexA drivers and reporter constructs produce similar expression levels and signal-to-noise ratios to those of the GAL4/UAS system.
Potter et al. (2010) reported development of a third binary system for use in Drosophila based on the qa gene cluster of Neurospora crassa (Giles et al. 1991). This system shows great promise, but the apparent toxicity of the Q transcription factor (Potter et al. 2010) currently limits its widespread application. Once this issue is addressed, and with the improvements to the LexA system we report here, there will be three independent binary transcriptional activation systems available for use in Drosophila.
Even when single enhancers are used to drive GAL4 expression, few resultant patterns will be limited to a single cell type. In many cases, more restricted expression will be desired for use in behavioral, anatomical, or developmental studies. Using Split GAL4 to restrict expression to the overlap between two “parent” enhancers is one attractive option (Luan et al. 2006). However, the low level of expression obtained using the reconstituted GAL4 limited its utility. We were able to increase expression levels significantly by replacing the VP16 activation domain with the stronger activation domain from the human p65 protein.
Another option for refining expression patterns is to block GAL4 activity in regions where its expression overlaps with that of GAL80. GAL80 inhibits transcription of genes under UAS control by binding to a 30-amino-acid region on the C terminus of GAL4 (Johnston et al. 1987; Ma and Ptashne 1987b). Two factors have limited the use of this GAL80-based approach. The first is the lack of a method to visualize GAL80 expression, other than by its ability to suppress GAL4 activity. We can overcome this limitation by using an enhancer whose expression pattern has been previously determined by assaying its ability to drive GAL4 (Pfeiffer et al. 2008). Taking advantage of the modular nature of the vectors we describe here, we can then use that enhancer to drive GAL80. The second limitation is the stoichiometric requirement for GAL80. In cases where GAL80 is driven from a strong promoter, such as α-1 tubulin, a single transgene is sufficient to suppress GAL4-mediated transcription (Lee and Luo 1999; Vef et al. 2006); however, we found that when GAL4 and GAL80 are driven under the same enhancer, a single copy of GAL80 may be insufficient, especially when the vector utilizes the same, mRNA-destabilizing hsp70 3′-UTR as used in the GAL4 constructs. This problem can be fixed by using two copies of GAL80, either by generating a stock bearing GAL80 in two locations or by building a tandem construct with two copies of GAL80 in one insertion. Alternatively, the efficacy of a single copy of GAL80 can be improved by optimizing the codon usage of the GAL80 gene and including two post-transcriptional regulatory elements thought to increase the efficiency of mRNA transport, an intron and the WPRE. These changes allowed us to completely suppress GAL4 activity with a single copy of GAL80 when the two genes were expressed from the same enhancer in vectors designed to maximize the concurrence of the expression of the two proteins.
The local chromatin environment at the site of transgene insertion can alter both the pattern and the level of transgene expression (Spradling and Rubin 1983; Hiromi et al. 1985; Kirkpatrick et al. 1994). Markstein et al. (2008) used luciferase to quantify such position effects in 20 genomic attP docking sites and found a wide range of basal and inducible levels of expression. In addition, leakiness and inducibility varied not only across insertion loci, but also between tissues. These effects are not surprising, considering that chromatin changes are involved in tissue-specific gene regulation (Schulze and Wallrath 2007; Girton and Johansen 2008). Thus, identification of suitable genomic docking sites relies on empirical testing in the tissues and developmental stages of interest. We assayed for position effects affecting adult brain GFP expression in 16 attP genomic docking sites. Although there is no single ideal locus for all transgenes, we identified 5 sites that show reproducible levels of expression for enhancer–GAL4 constructs and 11 for UAS-mCD8∷GFP. As far as we have tested, these attP sites work well for other driver and responder combinations. These selected sites are also all permissive to integration, making them efficient for the production of transgenic lines.
In the course of this work we identified two areas that require further technology development. First, in a significant minority of driver lines, transgene expression is stochastic; that is, not all cells of a given type express the transgene, and the precise cells showing expression vary from animal to animal. We do not know the underlying cause of this variation, but we speculate that it most likely relates to the difficulty in overcoming chromatin blocks to initiating transcription. This phenomenon has been observed in a wide variety of transgenic systems (see, for example, Ahmad and Henikoff 2001; Skora and Spradling 2010). Detecting stochastic expression often requires careful examination and is most easily scored in lines that express in repeating patterns, for example, in a cell type that occurs in each of the 800 cartridges of the lamina or each of the segments of the larval ventral nerve cord. We observed that increasing the strength of the activation domain can reduce this variation; also, certain integration sites in the genome appear to favor more uniform expression. But we have not found a general mechanism to resolve this issue, and at this point we avoid using such lines in experiments that depend on nonstochastic expression, such as measuring the behavioral effects of inactivation of a cell class.
The second issue that has not yet been adequately addressed is intrinsic to any experiment involving expression of an exogenous protein or an endogenous protein at elevated levels: such expression can perturb cell function and cell structure. These effects can be obvious, such as cell death. However, sometimes they are subtle. Here the only solution is to assay the exogenous protein for its intended effects on the cell and use only the minimal level of expression sufficient for the experiment. The tools we have presented—for example, a series of vectors with different numbers of UAS sites—will be useful in achieving the desired expression level and also in normalizing expression levels when using enhancers of different strengths.
In conclusion, we report here an extensive set of experiments in which we empirically tested a wide range of modifications to the vectors and methods commonly used to direct exogenous gene expression in Drosophila. We have been able to modulate the level of transgene expression by varying the strength of the activation domain carried by the transcriptional activator as well as the number of copies of its binding site and other properties of the reporter construct. Additional engineering to increase expression levels is not warranted, as significant toxicity appears to result from the exogenous proteins at the high end of our current expression range. We also solved the problem of leakiness of the LexA operator in the absence of LexA protein and made the Split GAL4 and GAL80 intersectional strategies more robust. Given the widespread use of these methods, we expect our results will have considerable utility.
Acknowledgments
We thank Robin Harris and Aljoscha Nern for thoughtful advice and constructive criticism. We thank Bruce Baker, Robin Harris, Tzumin Lee, Aljoscha Nern, Andrew Seeds, and Julie Simpson for comments on the manuscript; David Anderson for alerting us to the WPRE element; Scott Sternson for FLEX DNA containing the WPRE; Gary Struhl for plasmid G610; Henry Chang for myr∷mRFP DNA; Chris Potter and Liqun Luo for communicating information about the Q system prior to publication; and Kevin McGowan for the suggestion of the name JFRC. Todd Laverty and the Janelia Farm Fly Core assisted with Drosophila genetics and stock maintenance. Rodney Simmons and Rose Rogall provided media and general lab support. Kevin McGowan, Melissa Ramirez, Torrey Gallagher, and Xiaorong Zhang performed DNA sequencing. Susan Zusman, Michael Tworoger, and GSI, Inc. generated the transgenic lines. Crystal Sullivan provided us with excellent administrative support.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.119917/DC1.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Ahmad, K., and S. Henikoff, 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104 839–847. [DOI] [PubMed] [Google Scholar]
- Atasoy, D., Y. Aponte, H. H. Su and S. M. Sternson, 2008. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28 7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., C. Wilson and W. J. Gehring, 1990. Dissecting the complexity of the nervous system by enhancer detection. BioEssays 12 199–204. [DOI] [PubMed] [Google Scholar]
- Bischof, J., R. K. Maeda, M. Hediger, F. Karch and K. Basler, 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., A. S. Manoukian and N. Perrimon, 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44 635–654. [DOI] [PubMed] [Google Scholar]
- Brent, R., and M. Ptashne, 1981. Mechanism of action of the lexA gene product. Proc. Natl. Acad. Sci. USA 78 4204–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent, R., and M. Ptashne, 1984. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature 312 612–615. [DOI] [PubMed] [Google Scholar]
- Brent, R., and M. Ptashne, 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43 729–736. [DOI] [PubMed] [Google Scholar]
- Butala, M., D. Zgur-Bertok and S. J. Busby, 2008. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, T., M. Huang, C. Gorman and R. Jaenisch, 1991. A generic intron increases gene expression in transgenic mice. Mol. Cell. Biol. 11 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. T., 1983. Characterisation of the promoter for the LexA regulated sulA gene of Escherichia coli. Mol. Gen. Genet. 189 400–404. [DOI] [PubMed] [Google Scholar]
- Diegelmann, S., M. Bate and M. Landgraf, 2008. Gateway cloning vectors for the LexA-based binary expression system in Drosophila. Fly 2 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, J. B., 2002. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34 1–15. [DOI] [PubMed] [Google Scholar]
- Duncker, B. P., P. L. Davies and V. K. Walker, 1997. Introns boost transgene expression in Drosophila melanogaster. Mol. Gen. Genet. 254 291–296. [DOI] [PubMed] [Google Scholar]
- Ebina, Y., Y. Takahara, F. Kishi, A. Nakazawa and R. Brent, 1983. LexA protein is a repressor of the colicin E1 gene. J. Biol. Chem. 258 13258–13261. [PubMed] [Google Scholar]
- Emelyanov, A., and S. Parinov, 2008. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 320 113–121. [DOI] [PubMed] [Google Scholar]
- Estojak, J., R. Brent and E. A. Golemis, 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15 5820–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, E. H., M. K. Vanhoven, A. Bendesky, G. Wang, R. D. Fetter et al., 2008. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57 353–363. [DOI] [PubMed] [Google Scholar]
- Fields, S., and O. Song, 1989. A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Fischer, J. A., E. Giniger, T. Maniatis and M. Ptashne, 1988. GAL4 activates transcription in Drosophila. Nature 332 853–856. [DOI] [PubMed] [Google Scholar]
- Gerlitz, O., D. Nellen, M. Ottiger and K. Basler, 2002. A screen for genes expressed in Drosophila imaginal discs. Int. J. Dev. Biol. 46 173–176. [PubMed] [Google Scholar]
- Giles, N. H., R. F. Geever, D. K. Asch, J. Avalos and M. E. Case, 1991. The Wilhelmine E. Key 1989 invitational lecture. Organization and regulation of the qa (quinic acid) genes in Neurospora crassa and other fungi. J. Hered. 82 1–7. [DOI] [PubMed] [Google Scholar]
- Girton, J. R., and K. M. Johansen, 2008. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv. Genet. 61 1–43. [DOI] [PubMed] [Google Scholar]
- Gordon, M. D., and K. Scott, 2009. Motor control in a Drosophila taste circuit. Neuron 61 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth, A. C., M. Fish, R. Nusse and M. P. Calos, 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, C., S. Govindarajan and J. Minshull, 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22 346–353. [DOI] [PubMed] [Google Scholar]
- Hiromi, Y., A. Kuroiwa and W. J. Gehring, 1985. Control elements of the Drosophila segmentation gene fushi tarazu. Cell 43 603–613. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac, S., and D. Bilder, 2008. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet. 4 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlicka, L., M. Gibson, A. Kiger, C. Micchelli, M. Schober et al., 2002. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis 34 51–57. [DOI] [PubMed] [Google Scholar]
- Huang, M. T., and C. M. Gorman, 1990. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S. A., and J. E. Hopper, 1982. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Neuron 22 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S. A., J. M. Salmeron, Jr. and S. S. Dincher, 1987. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell 50 143–146. [DOI] [PubMed] [Google Scholar]
- Keegan, L., G. Gill and M. Ptashne, 1986. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231 699–704. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, R. B., Z. Parveen and P. F. Martin, 1994. Isolation of silencer containing sequences causing a tissue-specific position effect on alcohol dehydrogenase expression in Drosophila melanogaster. Dev. Genet. 15 188–200. [DOI] [PubMed] [Google Scholar]
- Kitagawa, Y., E. Akaboshi, H. Shinagawa, T. Horii, H. Ogawa et al., 1985. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 82 4336–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, J. M., and B. E. Staveley, 2003. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res. 2 43–47. [PubMed] [Google Scholar]
- Lai, S. L., and T. Lee, 2006. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 9 703–709. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22 451–461. [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., and R. W. Carthew, 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30 322–329. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1950. The phenomenon of position effect. Adv. Genet. 3 73–115. [DOI] [PubMed] [Google Scholar]
- Little, J. W., and D. W. Mount, 1982. The SOS regulatory system of Escherichia coli. Cell 29 11–22. [DOI] [PubMed] [Google Scholar]
- Little, J. W., D. W. Mount and C. R. Yanisch-Perron, 1981. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. USA 78 4199–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, H., N. C. Peabody, C. R. Vinson and B. H. White, 2006. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and M. Ptashne, 1987. a Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48 847–853. [DOI] [PubMed] [Google Scholar]
- Ma, J., and M. Ptashne, 1987. b The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 50 137–142. [DOI] [PubMed] [Google Scholar]
- Markstein, M., C. Pitsouli, C. Villalta, S. E. Celniker and N. Perrimon, 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor, R. L., S. M. Parkhurst and V. G. Corces, 1986. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 6 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss, A., M. Tripodi, J. F. Evers and M. Landgraf, 2009. Midline signaling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 7 e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, K., 2000. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J. Mol. Biol. 301 1097–1112. [DOI] [PubMed] [Google Scholar]
- Ni, J. Q., M. Markstein, R. Binari, B. Pfeiffer, L. P. Liu et al., 2008. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 5 49–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J. Q., L. P. Liu, R. Binari, R. Hardy, H. S. Shim et al., 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi, Y., and T. Fukasawa, 1984. Nucleotide sequence of the yeast regulatory gene GAL80. Nucleic Acids Res. 12 9287–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane, C. J., and W. J. Gehring, 1987. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder, T., K. S. Yoon, B. W. White and H. Keshishian, 2001. A conditional tissue specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu, W., and V. Hartenstein, 2006. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 26 5534–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, K. L., S. J. Elledge, B. B. Mitchell, L. Marsh and G. C. Walker, 1985. umuDC and mucAB operons whose products are required for UV light- and chemical induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc. Natl. Acad. Sci. USA 82 4331–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, R. B., and S. Lindquist, 1989. Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul. 1 135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, B. D., A. Jenett, A. S. Hammonds, T.-T. Ngo, S. Misra et al., 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, C. J., B. Tasic, E. V. Russler, L. Liang and L. Luo, 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M., 1988. How eukaryotic transcriptional activators work. Nature 335 683–689. [DOI] [PubMed] [Google Scholar]
- Resh, M. D., 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451 1–16. [DOI] [PubMed] [Google Scholar]
- Roman, G., K. Endo, L. Zong and R. L. Davis, 2001. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98 12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski, I., J. Ma, S. Triezenberg and M. Ptashne, 1988. GAL4–VP16 is an unusually potent transcriptional activator. Nature 335 563–564. [DOI] [PubMed] [Google Scholar]
- Salmeron, J. M., Jr., S. D. Langdon and S. A. Johnston, 1989. Interaction between transcriptional activator protein LAC9 and negative regulatory protein GAL80. Mol. Cell. Biol. 9 2950–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron, J. M., Jr., K. K. Leuther and S. A. Johnston, 1990. GAL4 mutations that separate the transcriptional activation and GAL80-interactive functions of the yeast GAL4 protein. Genetics 125 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, M. L., and P. A. Baeuerle, 1991. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 10 3805–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarr, M., P. Oertel-Buchheit, M. Kazmaier and M. Granger-Schnarr, 1991. DNA binding properties of the LexA repressor. Biochimie 73 423–431. [DOI] [PubMed] [Google Scholar]
- Schulze, S. R., and L. L. Wallrath, 2007. Gene regulation by chromatin structure: paradigms established in Drosophila melanogaster. Annu. Rev. Entomol. 52 171–192. [DOI] [PubMed] [Google Scholar]
- Shang, Y., L. C. Griffith and M. Rosbash, 2008. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. USA 105 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora, A. D., and A. C. Spradling, 2010. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc. Natl. Acad. Sci. USA 107 7389–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. E., D. Kalderon, B. L. Roberts, W. H. Colledge, M. Edge et al., 1985. The nuclear location signal. Proc. R. Soc. Lond. B Biol. Sci. 226 43–58. [DOI] [PubMed] [Google Scholar]
- Spana, C., D. A. Harrison and V. G. Corces, 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2 1414–1423. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1983. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell 34 47–57. [DOI] [PubMed] [Google Scholar]
- Stoleru, D., Y. Peng, J. Agosto and M. Rosbash, 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431 862–868. [DOI] [PubMed] [Google Scholar]
- Suster, M. L., L. Seugnet, M. Bate and M. B. Sokolowski, 2004. Refining GAL4 driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis 39 240–245. [DOI] [PubMed] [Google Scholar]
- Szüts, D., and M. Bienz, 2000. LexA chimeras reveal the function of Drosophila Fos as a context-dependent transcriptional activator. Proc. Natl. Acad. Sci. USA 97 5351–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven, A., B. Jelicic and M. Sopta, 2006. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 7 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman, J. W., H. Schuppe, D. Shepherd and D. W. Williams, 2004. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131 5167–5184. [DOI] [PubMed] [Google Scholar]
- Vef, O., D. Cleppien, T. Löffler, B. Altenhein and G. M. Technau, 2006. A new strategy for efficient in vivo screening of mutagenized Drosophila embryos. Dev. Genes Evol. 216 105–108. [DOI] [PubMed] [Google Scholar]
- Venken, K. J., Y. He, R. A. Hoskins and H. J. Bellen, 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 1747–1751. [DOI] [PubMed] [Google Scholar]
- Villalobos, A., J. E. Ness, C. Gustafsson, J. Minshull and S. Govindarajan, 2006. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, J. T., N. B. Reppas, G. M. Church and K. Struhl, 2005. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 19 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, G. C., 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48 60–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, N., J. R. Jin, S. Green, M. Hollis and P. Chambon, 1988. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell 52 169–178. [DOI] [PubMed] [Google Scholar]
- Wilson, C., H. J. Bellen and W. J. Gehring, 1990. Position effects on eukaryotic gene expression. Annu. Rev. Cell Biol. 6 679–714. [DOI] [PubMed] [Google Scholar]
- Yun, S. J., Y. Hiraoka, M. Nishizawa, K. Takio, K. Titani et al., 1991. Purification and characterization of the yeast negative regulatory protein GAL80. J. Biol. Chem. 266 693–697. [PubMed] [Google Scholar]
- Zieler, H., and C. Q. Huynh, 2002. Intron-dependent stimulation of marker gene expression in cultured insect cells. Insect Mol. Biol. 11 87–95. [DOI] [PubMed] [Google Scholar]
- Zufferey, R., J. E. Donello, D. Trono and T. J. Hope, 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]