Abstract
Although evolutionary theories predict functional divergence between duplicate genes, many old duplicates still maintain a high degree of functional similarity and are synthetically lethal or sick, an observation that has puzzled many geneticists. We propose that expression reduction, a special type of subfunctionalization, facilitates the retention of duplicates and the conservation of their ancestral functions. Consistent with this hypothesis, gene expression data from both yeasts and mammals show a substantial decrease in the amount of gene expression after duplication. While the majority of the expression reductions are likely to be neutral, some are apparently beneficial to rebalancing gene dosage after duplication.
Gene duplication without functional divergence
Gene duplication is prevalent in all three domains of life and is the major source of new genes [1–2]. Immediately after gene duplication, the two daughter genes are usually functionally redundant, especially when the entire gene together with its regulatory region is duplicated. Thus, mutations that knock out one of the duplicates are invisible to natural selection. Consequently, usually only one daughter gene is stably retained in the genome while the other degenerates into a pseudogene that is eventually lost. Therefore, with the exception of a small number of genes for which an increased dosage is beneficial (e.g., ribosomal RNA and histone genes) [2], the two daughter genes cannot be stably maintained in the same genome unless they escape from the usual fate of pseudogenization by quickly diverging in function, which may occur by the acquisition of new functions (neofunctionalization) [1], subdivision of ancestral functions (subfunctionalization) [3], or a combination of the two [4]. Surprisingly, however, several studies in yeast and nematode have found many duplicate gene pairs with negative epistasis [5–9], meaning that deleting both gene copies produces a significantly larger defect than expected from the effects of individual deletions. Negative epistasis is caused by functional redundancy [10]. While one might think that most of these negatively epistatic gene pairs are young duplicates that have not had sufficient time to diverge in function, this is not the case [7–9]. In fact, many of them are quite old [7–9] and some originated as early as a billion years ago [7]. The long-term maintenance of functional redundancy of duplicate genes is unexpected and puzzling.
Reduced expression can lead to the maintenance of functional redundancy
Here we propose a simple mechanism for the stable maintenance of functional redundancy in duplicate genes. We propose that the amount of expression of each daughter gene is reduced compared to the expression of the progenitor gene. This expression reduction prevents the loss of either daughter gene because the loss would render the total expression level after duplication lower than that before duplication, which would be deleterious. The expression reduction, when it is sufficiently large, would require both daughter genes to retain all ancestral functions, preventing the occurrence of functional divergence. In our model, although the two daughter genes are functionally equivalent, they are not redundant in a strict sense, because the deletion of either copy is expected to cause a fitness reduction that is sufficiently large to be disfavored by natural selection. Negative epistasis between functionally equivalent duplicates, regardless of the definition of epistasis by non-multiplicative or non-additive fitness effects of individual mutations, results from the well established nonlinear relationship between gene expression level and fitness [11] (Fig. 1A). That is, the fitness effect of reducing the expression level by 50% is less than 50% [12]. This phenomenon is closely related to the observations that most genes are haplosufficient [12–13] and that most wild-type alleles are dominant to loss-of-function alleles [12–13]. Because subfunctionalization includes reductions in the joint levels or patterns of activity of the duplicate genes [3], expression reduction after gene duplication is a type of subfunctionalization in the joint levels rather than patterns of activity. So, all previous theoretical results on subfunctionalization should apply to our model. Note, however, that subfunctionalization in the joint patterns of activity cannot explain the long retention of genetic redundancy, because negative epistasis is not expected if the two daughter genes have non-overlapping protein functions or tissue/condition expressions.
Fig. 1.
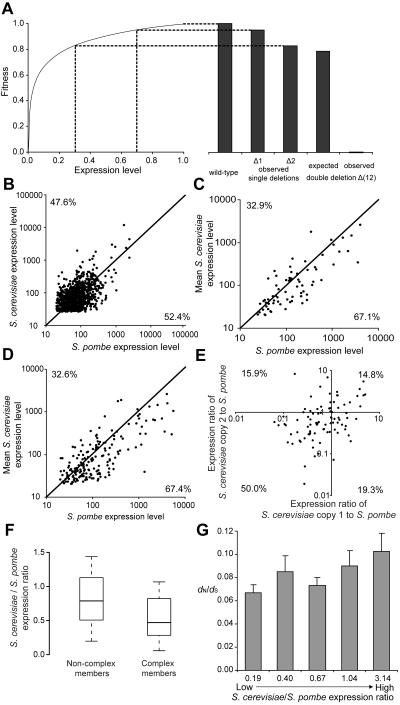
Expression reduction after gene duplication in yeasts. (A) Because fitness is a concave function of expression level, there is negative epistasis between duplicates with reduced expression. Synthetic lethality is observed in this hypothetical example. The fitness of the double deletion strain expected under no epistasis is calculated assuming multiplicative fitness effects of single deletions. (B) Expression levels of one-to-one orthologs in S. cerevisiae and S. pombe. Each dot represents a one-to-one orthologous pair. The diagonal line indicates identical expressions in the two species. Fractions of orthologs below and above the diagonal line are indicated. (C) Expression levels of two-to-one orthologs that are negatively epistatic in S. cerevisiae. Each dot represents a two-to-one ortholog. The expression level of the single gene in S. pombe and the mean expression of the duplicates in S. cerevisiae are presented. The fraction of dots below the diagonal is significantly greater than that in (B) (P = 0.006). (D) Expression levels of all two-to-one orthologs. The fraction of dots below the diagonal is significantly greater than that in (B) (P = 2×10−5). (E) Expression ratios between S. cerevisiae and S. pombe for all two-to-one orthologs. The fraction of dots in the lower-left quadrant is significantly greater than expected (P = 6×10−6). (F)S. cerevisiae duplicates involved in the same protein complexes have lower S. cerevisiae/S. pombe expression ratios (P < 0.05). The values of upper quartile, median, and lower quartile are indicated in each box, whereas the bars outside the box indicate semiquartile ranges. (G) Two-to-one orthologs with lower mean S. cerevisiae expression relative to S. pombe expression show lower nonsynonymous/synonymous rate ratios (dN/dS) between S. cerevisiae duplicates (P < 0.05 for unbinned data). The five bins are of equal sample size and the mean expression ratio and mean dN/dS value for each bin are presented. The error bars show one standard error.
Substantial expression reduction after gene duplication in yeasts
To test if the expression levels of duplicate genes are indeed decreased compared to their progenitor genes we examined gene expression levels measured by the next-generation sequencing based RNA-Seq method. RNA-Seq substantively outperforms microarray-based methods in the accuracy and dynamic range of the measurement [14]. We first identified one-to-one, two-to-one, and many-to-one orthologs between the baker's yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe (see Supplementary Methods). One-to-one orthologs are those genes with neither duplication nor gene loss in the two yeast lineages since their separation. Two-to-one (or many-to-one) orthologs had one (or multiple) duplications in the S. cerevisiae lineage, but experienced neither duplication nor gene loss in the S. pombe lineage. We focused on the duplicates in S. cerevisiae rather than S. pombe because epistasis between duplicate genes has been examined only in S. cerevisiae. Because of the different sequencing depths of the RNA-Seq data of the two yeasts, we adjusted the RNA-Seq depth in S. cerevisiae based on the assumption that one-to-one orthologs have the same average expression levels between the two species (see Supplementary Methods). Thus, one should regard our results from two-to-one and many-to-one orthologs as relative to those from one-to-one ortholgs. Our between-species expression comparison is meaningful because the RNA-Seq data from the two yeasts and the epistasis data from S. cerevisiae are all obtained under similar rich medium conditions.
There are 891 one-to-one orthologous genes with expression information from both yeasts. We found that 52.4% of them have lower expressions in S. cerevisiae than in S. pombe (Fig. 1B). In comparison, among the 70 two-to-one orthologs that are known to be negatively epistatic in S. cerevisiae and have expression data (see Supplementary Methods), 67.1% of the S. cerevisiae duplicate pairs have a lower mean expression than their single counterparts in S. pombe (Fig. 1C). The difference between two-to-one and one-to-one orthologs is highly significant (P = 0.006, one-tail Fisher's exact test). Using one-to-one orthologs as a control, we estimated that an excess of 0.671−(1−0.671)×(0.524/0.476) = 30.9% of duplicate gene pairs with negative epistasis experienced a decrease in mean expression after gene duplication. We calculated the S. cerevisiae/S. pombe expression ratio for each two-to-one ortholog, where the S. cerevisiae expression is the mean expression of the two paralogs. We found that this ratio (median = 0.74) is significantly lower than that from one-to-one orthologs (0.94; P = 0.001, one-tail Mann-Whitney U test), further supporting expression reduction after gene duplication.
To examine if the above observation is more widespread than the set of negatively epistatic duplicate genes, we examined all 227 two-to-one orthologs, 69% of which either have not been tested for epistasis or have no detectable negative epistasis. Note, however, that because negative epistasis is currently detected only when the overlapping function of a duplicate pair contributes substantially to cell growth in rich medium, duplicates that do not show detectable negative epistasis may still have overlapping functions. We found that 67.4% of two-to-one orthologs have lower mean expressions in S. cerevisiae than in S. pombe (Fig. 1D), significantly greater than that in one-to-one orthologs (Fig. 1B) (P = 2×10−5). The above result is supported even when only genes with significant expression reductions are considered (see Supplementary Methods). We estimated that an excess of 31.5% of duplicate gene pairs experienced mean expression reduction after gene duplication. The median expression ratio (S. cerevisiae/S. pombe) is 0.74 for all two-to-one orthologs, significantly lower than that (0.94) for one-to-one orthologs (P = 4×10−6). Similar results were obtained from 33 many-to-one orthologs between S. cerevisiae and S. pombe (Fig. S1).
Because ancestral S. cerevisiae experienced a whole genome duplication (WGD) ~100 million years ago [15], we separated duplicates based on whether they were generated by the WGD (Fig. S2A). We further separated the non-WGD group into four age groups (Fig. S2A). There is no significant variation in the prevalence or degree of expression reduction between WGD and non-WGD groups or among the non-WGD age groups (Fig. S2B).
In all of the above analyses, we calculated the mean expression level for paralogous genes of S. cerevisiae and then compared it with the expression level of the single copy ortholog in S. pombe. However, our hypothesis predicts that both daughter genes will have lower expressions than their progenitor gene. To verify this prediction, for each two-to-one ortholog, we examined the expression ratio between each of the two S. cerevisiae duplicates and its S. pombe ortholog (Fig. 1E). Because 52.4% of one-to-one orthologs have lower expressions in S. cerevisiae than in S. pombe, we expect that by chance (0.524)2=27.46% of two-to-one orthologs have lower expressions in S. cerevisiae than in S. pombe for both copies (i.e., in the lower-left quadrant of Fig. 1E). In fact, 50.0% lie in this quadrant (P = 6×10−6, binomial test), based on which we estimated that an excess of 31.1% of duplicates experienced expression reductions in both copies. These results also confirmed that the expression reduction phenomenon is not an artifact of condition-specific expressions of duplicate genes, because, under the latter scenario, only one daughter gene is expected to have lowered expression.
We also examined those genes that were duplicated in the S. pombe lineage since its separation from the S. cerevisiae lineage (one-to-two orthologs), and again found the phenomenon of expression reduction after gene duplication. For example, the S. cerevisiae/S. pombe expression ratio is significantly higher for one-to-two orthologs than for one-to-one orthologs (P = 0.03; U test).
Evolutionary mechanisms and consequences of expression reduction
The reduction of expression after gene duplication can occur simply by random fixation of neutral regulatory mutations that decrease gene expression, as long as the total expression of the two daughter genes is not below the required level for the wild-type function. Expression reduction may also be advantageous if the total gene expression upon duplication is higher than the optimal level. Excess of gene expression and protein production can be deleterious because they waste energy and raw materials [16] and result in additional misfolded protein molecules that are cytotoxic [17]. Further, the stoichiometry among different molecules in a cell may be broken by extra production of a protein. Specifically, the toxicity of dosage imbalance caused by the duplication of a gene that encodes a component of a stable protein complex is potentially high [18–19]. To explore the possibility of adaptive expression reduction after gene duplication, especially for rebalancing gene dosage, we focused on two-to-one orthologs. Because dosage balance should not be affected by WGD, we excluded from our analysis all duplicates that resulted from the WGD. By contrast, individual gene duplications are unlikely to occur simultaneously to multiple components of the same protein complex and thus may cause dosage imbalance. We found that the reduction in mean expression for paralogs involved in the same protein complexes is significantly greater than that for paralogs not involved in complexes (P < 0.05, one-tail U test, Fig. 1F). This finding suggests that, at least in some duplicate genes, expression reduction may have been beneficial and positively selected for, owing to its role in rebalancing gene dosage after duplication.
After a substantial reduction of expression in each daughter gene, protein function is less likely to change, because such a change would render the total activity of the products of the two daughter genes lower than that of the progenitor gene and be harmful. To test this prediction, for each two-to-one ortholog, we estimated the ratio of mean expression in S. cerevisiae and expression in S. pombe, as well as the ratio of the nonsynonymous to synonymous nucleotide substitution rates (dN/dS) between the two S. cerevisiae duplicates. We found that the dN/dS ratio decreases as the expression ratio decreases (Spearman's correlation ρ = 0.11, P = 0.047, one-tail t test; see Fig. 1G for the binned results). Because lower dN/dS ratios indicate slower functional changes, our observation suggests that the expression reduction after duplication indeed hampers functional divergence of duplicates. The above result is conservative, due to the strong negative impact of the absolute expression level of a gene on its rate of protein sequence evolution [17, 20]. Indeed, the partial correlation between dN/dS and the S. cerevisiae/S. pombe expression ratio becomes much stronger (ρ = 0.19, P = 0.001, one-tail t test) when the average expression level of S. cerevisiae duplicates is controlled for (see Supplementary Methods).
Widespread expression reduction in mammalian duplicate genes
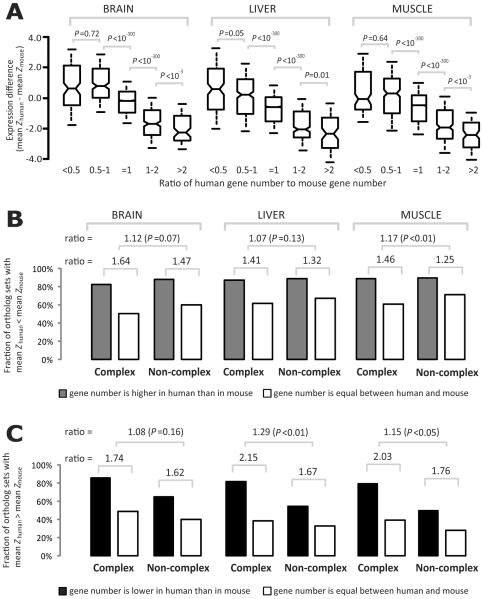
To examine whether the phenomenon of expression reduction after gene duplication also exists in other species, especially mammals, we analyzed RNA-Seq data from human and mouse. Because the gene expression distributions differ substantially between the two species (Fig. S3), it is inappropriate to compare the expression levels of human and mouse orthologs directly. Instead, we transformed the expression levels of human and mouse genes to Z-scores after a log2 transformation (see Supplementary Methods) and then compared the Z-scores of orthologous genes. For each gene that existed in the common ancestor of human and mouse, we identified all of its orthologs in extant human and mouse and referred to them as an orthologous set. We then calculated the difference in mean Z score between the human genes and the mouse genes in the orthologous set, as well as the human/mouse ratio of the gene number in the set. We found that when there are more human genes than mouse genes in an orthologous set, the mean expression level per gene tends to be lower in human than in mouse, and vice versa (Fig. 2A). This pattern is clear in each of the three tissues examined (brain, liver, and muscle) (Fig. 2A) and therefore is unlikely to result from the unique characteristics of particular tissues. Our conclusion is also supported by analyzing gene expression ranks rather than Z scores (See Supplementary Methods). Furthermore, duplicates involved in protein complexes tend to have more widespread expression reductions than those not involved in complexes (Fig. 2B and 2C). This trend exists in all three tissues examined, although some comparisons are not statistically significant due to small sample sizes (Fig. 2B and 2C).
Fig. 2.
Expression reduction after gene duplication in mammals. (A) Mean expression of a human-mouse orthologous set in a species reduces as the number of paralogs in that species increases. The values of upper quartile, median, and lower quartile are indicated in each box, whereas the bars outside the box indicate semiquartile ranges. P-values between boxes are from one-tail Mann-Whitney U tests. Spearman's rank correlation coefficients for the unbinned data of the three tissues are −0.178 (P < 10−106), −0.159 (P < 10−82) and −0.152 (P < 10−77), respectively. (B) Proteins functioning as subunits of complexes are subject to more widespread expression reductions after gene duplication, evident from orthologous sets in which human genes outnumber mouse genes. P-values are estimated from 10,000 parametric bootstrapping replications. (C) Proteins functioning as subunits of complexes are subject to more widespread expression reductions after gene duplication, evident from orthologous sets in which mouse genes outnumber human genes.
Concluding remarks and implications
In this work, we proposed that expression reduction after gene duplication, a special type of subfunctionalization, facilitates the long-term maintenance of duplicate genes and their functional redundancy. We showed in both yeasts and mammals that a substantial fraction of duplicate genes experienced expression reduction, which hampers functional divergence of duplicate genes. We further showed that the expression reduction in some genes may be adaptive for dosage rebalance, although it is probably neutral in most other cases. It has been proposed that functionally redundant duplicate genes are used to backup important functions in the event of a severe mutation much like the role of a spare tire in a car. However, theoretical population genetic analysis demonstrated that duplicates are unlikely to be maintained by the backup mechanism [21]. The present analysis further excludes the need of the backup hypothesis. Our finding is consistent with the recent discovery that only ~10% of duplicate genes are up-regulated when their paralogs are deleted [22]. Even in such cases, the apparent backup phenomenon could be an evolutionary byproduct [22]. These results echo the recent finding that the abundant functional redundancies caused by alternative pathways in metabolic networks need not and cannot be explained by the backup hypothesis [23]. Together, they suggest that the genetic robustness against mutations conferred by either duplicate genes or alternative pathways is a byproduct of other evolutionary processes [24]. A few other non-backup models have been proposed to explain the evolutionary maintenance of functional redundancy between duplicates [7, 25]. For instance, the piggyback hypothesis posits that two paralogs have some non-overlapping functions as well as some overlapping functions and the latter are kept as a byproduct of the former owing to strong structural constraints [7]. Our expression reduction model requires neither the existence of non-overlapping functions nor such functional constraints, and thus may be more widely applicable. An earlier hypothesis asserts that a substantial proportion of duplicate genes are fixed and retained due to the benefit of enhanced dosage [11]. This hypothesis is supported by neither human nor yeast genomic data [18]. The finding of expression reduction in approximately one third of duplicates further sets an upper limit for the fraction of duplicates whose retentions could possibly be explained by this hypothesis, because, under this hypothesis, the expression reduction would be deleterious and hence prohibited. While one might think that, functionally speaking, there is no change if the total expression level of two daughter genes is decreased to the level of their progenitor gene, we note that the stochastic variation in the total amount of product (i.e., expression noise) is lowered after duplication, which may be beneficial or deleterious depending on the genes concerned [26–27].
Supplementary Material
Acknowledgements
We thank Meg Bakewell, Mike Lynch, Calum Maclean, Csaba Pal, Jian-Rong Yang and three anonymous reviewers for valuable comments. This work was supported by US NIH research grants to J.Z. and Taiwan NHRI intramural funding to B.-Y.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ohno S. Evolution by Gene Duplication. Springer; Verlag: 1970. [Google Scholar]
- 2.Zhang J. Evolution by gene duplication-an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- 3.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLuna A, et al. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- 6.Musso G, et al. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vavouri T, et al. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24:485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Dean EJ, et al. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischler J, et al. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, et al. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat Genet. 2010;42:272–276. doi: 10.1038/ng.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Deutschbauer AM, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 16.Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- 17.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian W, Zhang J. Gene dosage and gene duplicability. Genetics. 2008;179:2319–2324. doi: 10.1534/genetics.108.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papp B, et al. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 20.Pal C, et al. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark AG. Invasion and maintenance of a gene duplication. Proc Natl Acad Sci U S A. 1994;91:2950–2954. doi: 10.1073/pnas.91.8.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLuna A, et al. Need-based up-regulation of protein levels in response to deletion of their duplicate genes. PLoS Biol. 2010;8:e1000347. doi: 10.1371/journal.pbio.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Zhang J. Abundant indispensable redundancies in cellular metabolic networks. Genome Biol. Evol. 2009;1:23–33. doi: 10.1093/gbe/evp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison R, et al. Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci U S A. 2007;104:2307–2312. doi: 10.1073/pnas.0607153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowak MA, et al. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Positive selection for elevated gene expression noise in yeast. Mol Syst Biol. 2009;5:299. doi: 10.1038/msb.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.