Abstract
Comparison of paralogous vitellogenins in 10 insect species representing six orders showed a remarkable degree of conservation of amino acid composition in spite of sequence differences. For example, the correlation between the percentages of the 20 amino acids in two vitellogenins from the beetle Tribolium castaneum was 0.975, even though the two amino acid sequences differed from each other at 49.4% of sites. There was a positive correlation between the frequency of occurrence of reciprocal pairs of amino acids in more distantly related paralogs, and this correlation was generally strongest when both of the amino acids in the pair were nutritionally essential. These results imply that conservation of amino acid composition occurs through amino acid replacements that result in a balanced loss and gain of each amino acid each amino acid residue. Thus insect vitellogenins seem to be subject to an unusual kind of purifying selection, where the amino acid content is conserved rather than the sequence per se, selection apparently arising from the nutritional needs of the developing embryo appears to be responsible for maintaining the balance of amino acids.
Keywords: amino acid composition, protein evolution, purifying selection, vitellogenin
1. Introduction
Proteins play a wide variety of roles in organisms, from structural components and enzymes to signaling molecules and receptors. Rather atypical among proteins are those whose functions involve the storage of amino acids for use in development of offspring, including the seed storage proteins of higher plants (Shewry and Halford 2002; Shutov et al. 2003) and the yolk storage proteins of animals (Tufail and Takeda 2008). In insects, there are two distinct families of yolk proteins. One of these families, confined to the order Diptera, includes the yolk proteins of Drosophila melanogaster, whose expression in the egg has provided an important model system for understanding hormonal regulation of gene expression (Bownes 1994). The other family of insect yolk proteins, known as vitellins, are processed from precursors known as vitellogenins, which are synthesized in the fat body and certain other tissues (Tufail and Takeda 2008). The latter family is known to occur not only in certain Diptera (though not in Drosophila) but also several other insect orders and in other arthropod classes (Hwang et al. 2010).
Alignments of insect vitellogenins have indicated a small number of primary sequence features conserved in most but not all members of the family: (1) a motif GLCG or GICG in the C-terminal region, conserved in most insect vitellogenins; (2) nine conserved cysteine residues C-terminal to the latter conserved motif; (3) an RXXR motif in the N-terminal region of most vitellogenins, where the protein is cleaved by proteases to form vitellins (Tufail and Takeda 2008). The functional role of the GLCG/GICG motif and the conserved cysteines is not known. In Hemimetabola (insects with incomplete metamorphosis), the vitellogenin is cleaved into several polypeptides, whereas in most Holometabola (insects with complete metamorphosis), vitellogenin is cleaved into just two polypeptides. In the wasps, bees, and ants (Hymenoptera: Apocrita), cleavage is absent (Tufail and Takeda 2008).
In spite of these apparent functional constraints, it might be predicted that most of vitellogenin protein will evolve in a very different fashion from typical proteins. Since the protein’s primary function is to provide a reserve of amino acids for use in development, it might be predicted that many amino acid replacements will be selectively neutral or nearly so, as long as the overall protein maintains approximately the proportions of the various amino acids needed by the developing embryo. A mutation that causes the proportion of each amino acid residue to deviate from the balance that meets the nutritional requirements of the developing embryo would be predicted to be slightly deleterious, while a mutation that restores the balance of amino acids would be slightly advantageous. The effect of each individual mutation might be expected to be slight because of the substantial size of the vitellogenin protein. Over evolutionary time, we might expect that such slightly deleterious and slightly advantageous mutations would balance each other, thereby preserving the nutritional value of the vitellogenin. The insect vitellogenins constitute a multi-gene family, with 2 or 3 members reported from a number of species. The present study takes advantage of comparisons between paralogous vitellogenins of 10 insect species, representing six orders, in order to test the hypothesis that amino acid composition is conserved between by a balance of slightly deleterious and slightly advantageous mutations.
2. Methods
2.1. Phylogenetic Analysis
Phylogenetic analyses employed 60 amino acid sequences of vitellogenins from 37 insect species and five species of non-insect arthropods. (For accession numbers, see Supplementary Table S1). Since most of these sequences were derived from unmapped genomes, it was not always possible to determine whether two database accessions from a given species represented two distinct loci, allelic sequences from the same locus, or alternative transcripts from the same locus. Therefore, as an operational rule of thumb, I used in analyses only one of any two sequences from the same species that differed from each other by less than 1% at amino acid sites. Amino acid sequences were aligned using the CLUSTAL X program (Thompson et al. 1997); in phylogenetic analysis, any site at which the alignment postulated a gap in any sequence was excluded from the computation of pairwise distances so that a comparable set of amino acid positions was used for each comparison. A phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei 1987) on the basis of the JTT distance (Jones et al. 1992), with the assumption that rate variation among sites followed a gamma distribution. The shape parameter of the gamma distribution (a = 2.19) was estimated by the TREE-PUZZLE program (Schmidt et al. 2002). The reliability of clustering patterns in phylogenetic trees was assessed by bootstrapping (Felsenstein 1985); 1000 bootstrap samples were used.
2.2. Paralogous Pair Comparisons
From the phylogenetic tree, pairs of paralogous vitellogenin sequences were chosen for 10 insect species; pairs of paralogous genes were chosen so that each pair was determined by the phylogenetic analysis to be to be phylogenetically (and thus statistically) independent of all other pairs. Note that not all of these pairs were reciprocally monophyletic; nonetheless, they were phylogenetically independent in that each sequence difference between the members of a given pair occurred independently of differences between the members of other pairs. These comparisons involved the following pairs (with numbers of aligned amino acid sites): (1) Rhyparobia maderae 1 and 2 (1910 sites); (2) Periplaneta americana 1 and 2 (1776 sites); (3) Plautia stali 1 and 2; (1146 sites); (4) Aedes aegypti B1 and B2 (1164 sites); (5) Culex quinquefasiatus A1 and A2 (1165 sites); (6) Ochlerotatus atropalpus B1 and B2 (1167 sites); (7) Pediculus humanus 1 and 2 (1124 sites); (8) Tribolium castaneum 1 and 2 (161 sites); (9) Nasonia vitripennis 1 and 2 (1469 sites); and (10) Solenopsis invicta 2 and 3 (1500 sites).
In analysis of the amino acid composition of these 10 pairs of sequences, I excluded any site at which the alignment postulated a gap in one of the two sequences relative to the paralogous sequence with which it was compared. The nutritionally essential amino acids for insects (F, H, I, K, L, M, R, T, V, and W) were defined according to Nation (2008). Chemical distances between amino acids were from Miyata et al. (1979). I compared the frequencies of reciprocal amino acid differences between the two members of each pair as follows. Let 1 and 2 designate the two sequences in the pair. If at a given site, sequence 1 has a certain amino acid (amino acid X) and sequence 2 has another amino acid (amino acid Z), then the amino acid pair for that site is XZ. The reciprocal amino acid difference (ZX) would occur at a site where sequence 1 has Z and sequence 2 has X. For example, the amino acid pairs IL (Ile-Leu) and LI constitute reciprocal amino acid differences. In the comparisons between the 10 pairs of paralogs, 180 of the 190 theoretically possible amino acid differences occurred at least once. For each of the 10 pairs of paralogs, I computed the correlation (rrec) between the frequency of each amino acid difference with that of its reciprocal amino acid difference.
3. Results
3.1. Phylogenetic Analysis
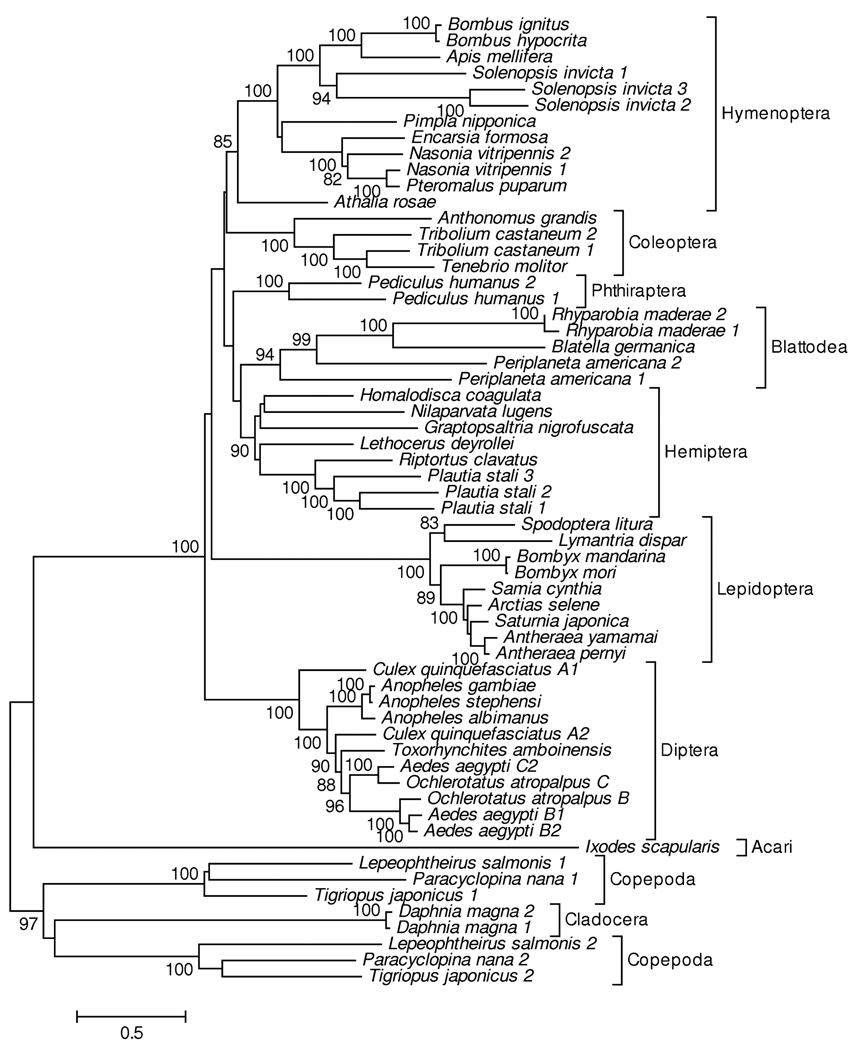
The phylogenetic tree of insect vitellogenin amino acid sequences was rooted with sequences from the tick Ixodes scapularis and from five species of Crustacea (Figure 1). Vitellogenin sequences from 7 insect orders were included, and in each case the sequences from a given order clustered together (Figure 1). The clusters of sequences from Coleoptera, Pthiraptera, Lepidoptera, and Diptera each received 100% bootstrap support; and that of sequences from Hymenoptera received 85% bootstrap support (Figure 1). In the case of Hymenoptera, a cluster of sequences from the suborder Apocrita received 100% bootstrap support, but a sequence from the coleseed sawfly Athalia rosae (suborder Symphyta) fell outside that cluster (Figure 1). Overall, deep branches within the phylogenetic tree were not well resolved; but the topology did not correspond to the known relationships of the insect orders (Kjer et al. 2006; Whiting 2002). In particular, the hemimetabolous orders Blattodea and Hemiptera did not cluster outside the holometabolous orders (Figure 1).
Figure 1.
NJ tree of insect vitellogenins (974 aligned amino acid sites), based on the JTT + gamma distance (a = 2.19). Numbers on the branches represent the percentage of 1000 bootstrap samples supporting the branch; only values ≥ 80% are shown.
When multiple sequences were available from a given species, those sequences clustered together or with sequences from closely related species. For example, three sequences from the imported fire ant Solenopsis invicta clustered together with 95% bootstrap support (Figure 1). Likewise, three sequences from the brown-winded green bug Plautia stali clustered together with 99% bootstrap support (Figure 1). Of two sequences from the American cockroach, one (Periplaneta americana 2) clustered with sequences from two other cockroach species with 98% bootstrap support, while the other (Periplaneta americana 1) fell outside that cluster (Figure 1). The three cockroach species included belong to three different families: Blattidae (P. americana), Blattellidae (Blatella germanica), and Blaberidae (Rhyparobia maderae). Thus, the topology supports the hypothesis that the two P. americana genes duplicated before Blattidae diverged from the latter two families.
Available sequences from the order Diptera represented three subfamilies from a single family, Culicidae (mosquitos): Anophelinae (Anopheles), Culicinae (Culex, Ochlerotatus, and Aedes) and Toxorhynchitinae (Toxorhynchites). The phylogenetic tree supported cases of both ancient and more recent duplication within this family. For example, Culex quinquefasciatus A1 clustered outside all other mosquito vitellogenins, including Culex quinquefasciatus A2; and this pattern received 100% bootstrap support (Figure 1). This topology supports the hypothesis that these two genes of C. quinquefasciatus duplicated before the Culicinae diverged from the other two subfamilies. On the other hand, the sequences from Aedes aegypti formed two clusters of two members each (designated, respectively, as B1 and B2 and C1 and C2; Figure 1). In the case of each of these two clusters, a sequence from Ochlerotatus atropalpus clustered outside the pair of Ae. aegypti genes; and in each case this topology received 100% bootstrap support (Figure 1). This topology supported the hypothesis that the duplication of Ae. aegypti B1 and B2 and the duplication of Ae. aegypti C1 and C2 occurred after Aedes diverged from Ochlerotatus.
3.2. Amino Acid and Nucleotide Usage
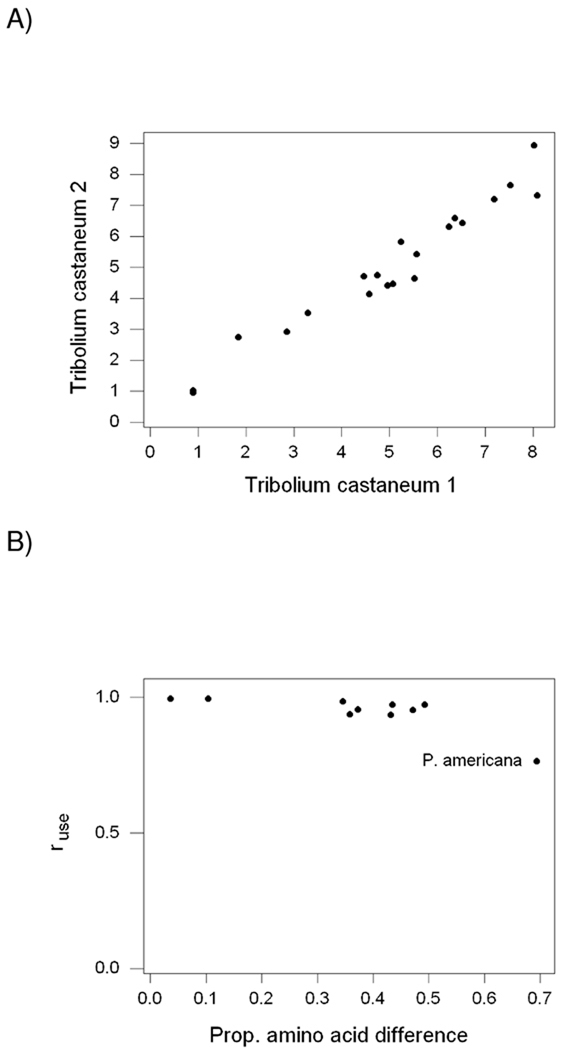
In comparisons between paralogous pairs of vitellogenins from 10 insect species, the frequencies of the 20 amino acids were highly positively correlated between the two pair members. For example, the correlation between the percentages of the 20 amino acids (ruse) in Tribolium castaneum 1 and 2 was 0.975 (P < 0.001; Figure 2A). This high ruse was observed even though the amino acid sequences of Tribolium castaneum 1 and 2 differed from each other at 49.4% of sites. There was a significant ruse in each of the 10 pairs (P < 0.001 in each case; Figure 2B). The value of ruse was greater than 0.93 in every case except Periplaneta americana, the species in which two paralogs showed the greatest amino acid sequence difference (69.4%); in the latter species, ruse was 0.765 (Figure 2B). When ruse was computed separately for the 10 nutritionally essential amino acids and for the 10 nonessential amino acids, the correlation coefficients were not significantly different for any of the 10 pairs of paralogs (not shown).
Figure 2.
(A) Plot of the percent usage of each of the 20 common amino acids in Tribolium castaneum 2 vs. that in Tribolium castaneum 1; the correlation coefficient (ruse) was 0.975 (P < 0.001). (B) Plot of ruse vs. proportion of amino acid difference pairs of paralogous vitellogenins from 10 insect species; the point for Periplaneta americana is indicated.
As a measure of nucleotide content bias, I examined percent G+C at third positions, where a majority of possible mutations are synonymous (Supplementary Table S2). The species of the hemimetabolous orders Blattodea (P. americana and R. maderae) and Hemiptera (P. stali) had mean % G+C less than 50%, as did the holometabolans S. invicta and Pediculus humanus (Supplementary Table S2). All other species showed mean % G+C greater than 50% (Supplementary Table S2). In 9 of the 10 species, % G+C at the two paralogous loci was very similar; the exception was Nasonia vitripennis, in which % G+C in the vitellogenin 1 gene (78.1%) was substantially higher than that in the vitellogenin 2 gene (49.0%; Supplementary Table S2). Thus, there was no evidence that patterns of amino acid usage represented simply the effects of mutational bias toward certain nucleotides.
3.3. Amino Acid Differences
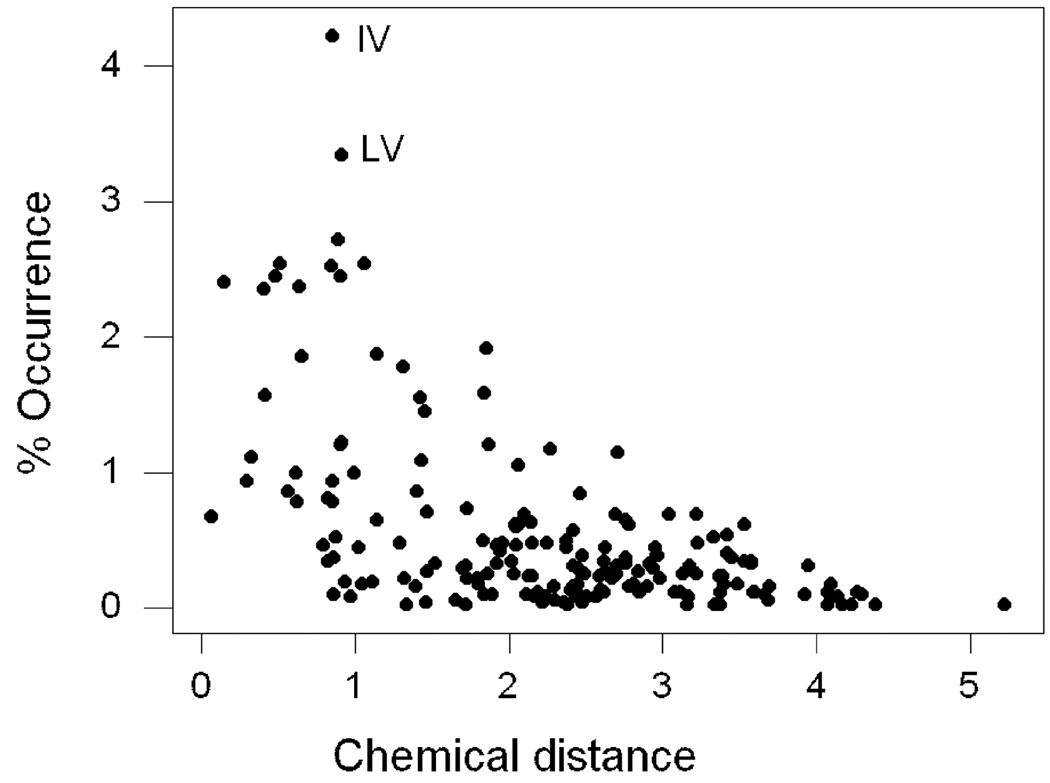
In comparisons between the 10 pairs of paralogs, there were a total of 5226 observed amino acid differences. Only six pairs of amino acids constituted more than 2.5% of the total number of differences: IV (4.23%); LV (3.35%); ST (2.72%); KQ (2.54%); AS (2.54%); and EQ (2.53%). Of these six, only IV was among the most frequently observed amino acid differences in a recent survey of mammalian orthologs (Hughes and Friedman 2009). There was a significant negative correlation between the frequency of occurrence of a given amino acid difference and the chemical distance between two amino acids (r = −0.555; P < 0.001; Figure 3). The correlation was similar when I considered separately only amino acid pairs in which both of the two amino acids were nutritionally nonessential (r = −0.550; P < 0.001; N = 43); pairs in which one amino acid was essential and the other nonessential (r = −0.519; P < 0.001; N = 95); and pairs in which both amino acids were essential (r = −0.640; P < 0.001; N = 42).
Figure 3.
Plot of percent occurrence of pairs of different amino acids occurring at aligned positions in paralogous vitellogenins vs. the chemical distance between the amino acid pair (r = −0.555; P < 0.001); the two most common amino acid pairs (IV and LV) are indicated.
3.4. Reciprocal Pairs
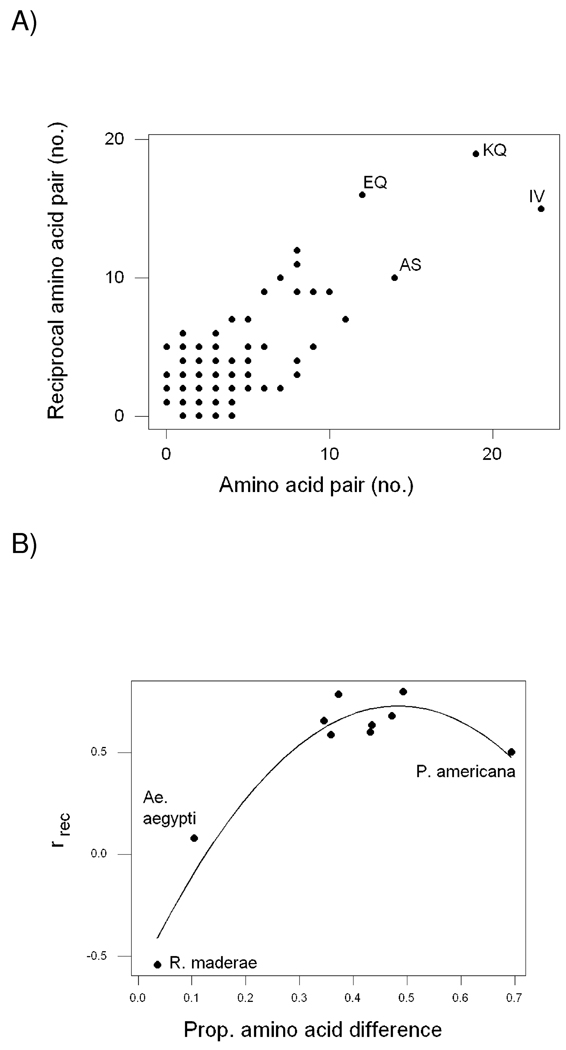
In order to examine how paralogous vitellogenins maintain similar amino acid usage despite sequence divergence, I compared each amino acid difference with the reciprocal amino acid difference. I designate the correlation between the number of occurrences of the two reciprocal amino acid differences rrec. For example, in the case of the two paralogs from T. castaneum, rrec was 0.799 (P < 0.001: Figure 4A).
Figure 4.
(A) Plot of numbers of reciprocal amino acid pairs in the comparison between Tribolium castaneum 2 and Tribolium castaneum 1; the correlation coefficient (rrec) was 0.799 (P < 0.001). Several of the more commonly occurring amino acid pairs are indicated. (B) Plot of ruse vs. proportion of amino acid difference pairs of paralogous vitellogenins from 10 insect species. The points for the species with the two lowest values of the proportion of amino acid difference (P. maderae and Ae. aegypti) are indicated, as is that for the species with the highest proportion of amino acid difference (P. americana). The line is a quadratic regression (Y = −0.600 + 5.50X − 5.69X2; R2 = 0.939; P < 0.001).
In 8 of the 10 paralogous pairs, rrec was positive and statistically significant (P < 0.001 in each case; Figure 4B). By contrast, in Rhyparobia maderae, the species with the lowest amino acid difference (3.6%) between the paralogs, rrec was statistically significant but negative (rrec = −0.543; P < 0.001; Figure 4B). The only pair of paralogs without a statistically significant rrec were Ae. aegypti B1 and B2, the second most similar pair of paralogs at the amino acid sequence level (10.4% difference; rrec = 0.078; n.s.; Figure 4B). The lowest rrec value among the remaining 8 pairs of paralogs was 0.504 (P < 0.001) in the case of P. americana (Figure 4B).
Thus, the relationship between rrec and proportion of amino acid sequence difference fit a quadratic regression (Y = −0.600 + 5.50X − 5.69X2; R2 = 0.939; P < 0.001; Figure 3B). Clearly the value for P. americana were influential in determining the shape of the quadratic regression (Figure 4B). However, it is of interest that, even when P. americana was excluded, there was still a highly significant quadratic relationship (Y = −0.656 + 6.40X − 7.50X2; R2 = 0.945; P < 0.001). The quadratic shape of the relationship between rrec and proportion of amino acid sequence difference thus replaced the tendency for rrec to level off at values between 0.5 and 0.8 as amino acid difference increased (Figure 4B).
Amino acid pairs were placed in three categories based on whether or not the amino acids involved are nutritionally essential for insects, and rrec was computed separately for each category in the eight species with significant positive rrec (Table 1). In seven of these eight species, rrec was higher when both amino acids in the pair were essential than when both amino acids were non-essential or when one amino acid was essential and the other was not essential (Table 1). The only exception to this trend was P. americana (Table 1). In four of the species, there were significant differences between rrec for pairs of essential amino acids and rrec for pairs of one essential and one non-essential amino acid (Table 1). In two of the species, there were significant differences between rrec for pairs of essential amino acids and rrec for pairs of non-essential amino acids (Table 1). Overall, there was a significant difference (P = 0.01; Friedman test) in median rrec among the three categories of amino acid pairs, with the highest value occurring when both pairs were essential (0.733; Table 1).
Table 1.
Correlation coefficients (rrec) between frequencies of reciprocal amino acid pairs categorized by nutritional essentiality of the amino acids.
| Species | Amino acid pair category | ||
|---|---|---|---|
| Nonessential- Nonessential |
Essential- Nonessential |
Essential- Essential |
|
| Periplaneta americana | 0.625 | 0.431 | 0.463 |
| Plautia stali | 0.697 | 0.422a | 0.806 |
| Culex quinquefasciatus | 0.549 | 0.566 | 0.678 |
| Ochlerotatus atropalpus | 0.596 | 0.674 | 0.748 |
| Pediculus humanus | 0.349a | 0.379b | 0.871 |
| Tribolium castaneum | 0.774 | 0.800 | 0.833 |
| Nasonia vitripennis | 0.833 | 0.581b | 0.915 |
| Solenopsis invicta | 0.298a | 0.446a | 0.815 |
| Median c | 0.573 | 0.552 | 0.733 |
Pairwise tests of the equality of individual rrec value to that for the essential-essential category in the same species:
P < 0.01;
P < 0.001.
Friedman test for equality of medians: P = 0.01.
3.5. Periplaneta americana Paralogs
In order to examine further the comparatively low ruse observed in Periplaneta americana (Figure 2B), studentized residuals were computed for the regression of amino acid use in P. americana 2 vs. that in P. americana 1. There was one significant studentized residual, indicating that the regression would be significantly improved if that point were removed; this significant studentized residual (t = 2.35; P < 0.05) corresponded to the amino acid V. The explanation for this result was a substantially higher frequency of V in P. americana 2 (8.3%) than in P. americana 1 (5.4%). The difference in percent V also played a major role in the relatively low rrec in P. americana (Figure 4B). There were 32 amino acid sites with V in P. americana 2 and L in P. americana 1, as opposed to only 9 sites with L in P. americana 2 and V in P. americana 1. Similarly, there were 23 sites with V in P. americana 2 and I in P. americana 1, as opposed to only 10 sites with I in P. americana 2 and V in P. americana 1; and there were 15 sites with V in P. americana 2 and A in P. americana 1, as opposed to only 6 sites with A in P. americana 2 and V in P. americana 1.
Discussion
Like a previous analysis (Tufail and Tukeda 2008), a phylogenetic tree of insect vitellogenins supported the hypothesis that the evolution of the insect vitellogenins has involved numerous independent gene duplications occurring at different evolutionary times in different lineages of insects. Because no orthogs were shared by insects belonging to different orders, none of the duplication events appeared to have preceded the divergence of the insect orders. Within orders, the tree suggested that certain duplications were quite ancient. Notably, the tree supported the hypothesis that two vitellogenis from the American cockroach Periplaneta americana duplicated before the Blattidae diverged from Blatellidae and Blaberidae, an event believed to have taken place in the Jurassic about 200 Mya (Grimaldi and Engel 2005). Others were much more recent, such as the duplication in Aedes aegypti that occurred after that species diverged from another member of the same mosquito subfamily (Culicinae), Ochlerotatus atropalpus. Deep branching patterns within the tree were not well resolved, as is to be expected in the case where conservation of amino acid sequence composition (see below) has led to numerous forward-and-backward and parallel amino acid replacements.
Given a pattern the pattern of independent duplications of vitellogenin genes over the history of the insects, it may seem surprising that the number of vitellogenin genes remains low, often only one, two, or three. One possible explanation for this pattern is that, in addition to lineage-specific duplications, there have also been repeated events of lineage-specific gene deletion. Thus, the vitellogenin gene family seems to have evolved by a pattern of multiple gene duplications and deletions, a pattern that characterizes a number of gene families and has been named a “birth-and-death” process (Hughes and Nei 1989; Nei and Hughes 1992; Nei and Rooney 2005).
Comparisons of paralogs from 10 insect species representing six orders showed that paralogous vitellogenins – except for the very anciently duplicated genes of P. americana – tended to retain very similar amino acid composition even when the degree of amino acid sequence divergence approached 50%. This represents an unusual kind of protein conservation, where the amino acid content is conserved rather than the sequence per se. There was a positive correlation between the frequencies of occurrence of reciprocal pairs of amino acids in paralogs with at least about 30% amino acid difference (Figure 4), supporting the hypothesis that the conservation of amino acid composition occurs through a balanced loss and gain of amino acid residues. This correlation was generally strongest when both of the amino acids in the pair were nutritionally essential (Table 1), consistent with the hypothesis that selection relating to the nutritional needs of the developing embryo is responsible for maintaining the balance of amino acids.
The mode of evolution of insect vitellogenins appears similar to that proposed in the case of vertebrate protamine P1, where a high proportion of arginine residues is conserved even though the position of those residues is not conserved (Rooney et al. 2000). A similar pattern was also seen in the TolA cell envelope protein of Proteobacteria, which are rich in alanine and lysine (Rooney 2003). Analysis of nucleotide content at third positions in vitellogenin genes showed no consistent relationship between the pattern of nucleotide usage and amino acid sequence conservation. Thus, as in TolA (Rooney 2003), the observed pattern of conservation of amino acid sequence conservation in paralogous vitellogenins is not simply the result of a mutational bias.
The conservation of amino acid composition without conserving amino acid sequence resembles the “minute effect selection” that has been proposed to act to maintain optimal codon usage in certain organisms (Kimura 1981). A typical mutation changing a single amino acid in vitellogenin (other than the few positions that are conserved for functional reasons) would be slightly deleterious if it caused a deviation from the optimal amino acid usage, or slightly advantageous if it restored the optimal amino acid usage. Whether deleterious or advantageous, a single such mutation might be expected to have a selection coefficient so low that it would be effectively neutral except when the effective population size was very large. Thus, genetic drift would likely be the primary determinant of the fate of such mutants. Nonetheless, even under these circumstances, purifying selection can be expected to maintain roughly constant frequencies of each amino acid because substantial deviations from the optimal pattern will be disfavored.
The two paralogous genes from the American cockroach Periplaneta americana were exceptions to the trends observed in other species. These two 200 million year-old paralogs were found to be unusually divergent in amino acid composition, particularly with respect to the percentage of valine. This example suggests that the accumulation of mutations can eventually lead to differences between insect vitellogenins with respect amino acid composition. Thus, over a very long evolutionary time, mutation and drift appear to be able to overcome the conservative effect of stabilizing selection on amino acid composition and give rise to a certain degree of functional differentiation between vitellogenin paralogs.
Supplementary Material
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health.
Abbreviations
- rrec
the correlation between the frequency of each amino acid difference with that of its reciprocal amino acid difference.
- ruse
the correlation between the percentages of the 20 amino acids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bownes M. The regulation of the yolk protein genes, a family of sex differentiation genes in Drosophila melanogaster. Bioessays. 1994;16:745–752. doi: 10.1002/bies.950161009. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the Insects. Cambridge: Cambridge University Press; [Google Scholar]
- Hughes AL, Friedman R. More radical amino acid replacements in primates than in rodents: support for the evolutionary role of effective population size. Gene. 2009;440:50–56. doi: 10.1016/j.gene.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol. Biol. Evol. 1989;6:559–579. doi: 10.1093/oxfordjournals.molbev.a040573. [DOI] [PubMed] [Google Scholar]
- Hwang D-S, Lee K-W, Han J, Park HG, Lee J, Lee Y-M, Lee J-S. Molecular characterization and expression of vitellogenin (Vg) genes from the cyclopoid copepod, Paracyclina nana exposed to heavy metals. Comp. Biochem. Physiol., Part C. 2010;151:360–368. doi: 10.1016/j.cbpc.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kimura M. Possibility of extensive neutral evolution under stabilizing selection with special reference to nonrandom usage of synonymous codons. Proc. Natl. Acad. Sci. USA. 1981;78:5773–5777. doi: 10.1073/pnas.78.9.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer KM, Carle FL, Litman J, Ware J. Phylogeny of the Hexapoda. Arthropod Syst. Phyl. 2006;64:35–44. [Google Scholar]
- Nation JL. Insect physiology and biochemistry. 2nd. Ed. Boca Raton FL: CRC Press; 2008. [Google Scholar]
- Miyata T, Miyazawa S, Yasunaga T. Two types of amino acid substitutions in protein evolution. J. Mol. Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- Nei M, Hughes AL. Balanced polymorphism and evolution by the birth-and-death process. In: Tuki K, Aizawa M, Sasazuki T, editors. Proceedings of the 11th Histocompatibility Workshop and Conference; Oxford: Oxford University Press; 1992. pp. 27–38. [Google Scholar]
- Nei M, Rooney A. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney AP, Zhang J, Nei M. An unusual form of purifying selection in a sperm protein. Mol. Biol. Evol. 2000;17:278–283. doi: 10.1093/oxfordjournals.molbev.a026307. [DOI] [PubMed] [Google Scholar]
- Rooney AP. Selection for highly biased amino acid frequency in the TolA cell envelope protein of Proteobacteria. J. Mol. Evol. 2003;57:731–736. doi: 10.1007/s00239-003-2530-6. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shewry PR. Cereal seed storage proteins: structures, properties and role in grain utilization. J. Exp. Bot. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Shutov AD, Bäumlein H, Blattner FR, Müntz K. Storage and mobilization as antagonistic functional constraints on seed storage blobulin evolution. J. Exp. Bot. 2003;54:1645–1654. doi: 10.1093/jxb/erg165. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail M, Takeda M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008;54:1447–1458. doi: 10.1016/j.jinsphys.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Whiting MF. Phylogeny of the holometabolous insect orders: molecular evidence. Zool. Scripta. 2002;31:3–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.