Abstract
The biological actions of heparin and heparan sulfate, two structurally related glycosaminoglycans, depend on the organization of the complex heparanome. Due to the structural complexity of the heparanome, the sequence of variably sulfonated uronic acid and glucosamine residues is usually characterized by the analysis of smaller oligosaccharide and disaccharide fragments. Even characterization of smaller heparin/heparan sulfate oligosaccharide or disaccharide fragments using simple 1D 1H NMR spectroscopy is often complicated by the extensive signal overlap. 13C NMR signals, on the other hand, overlap less and therefore, 13C NMR spectroscopy can greatly facilitate the structural elucidation of the complex heparanome and provide finer insights into the structural basis for biological functions. This is the first report of the preparation of anomeric carbon-specific 13C-labeled heparin/heparan sulfate precursors from the Escherichia coli K5 strain. Uniformly 13C- and 15N-labeled precursors were also produced and characterized by 13C NMR spectroscopy. Mass spectrometric analysis of enzymatically fragmented disaccharides revealed that anomeric carbon-specific labeling efforts resulted in a minor loss/scrambling of 13C in the precursor backbone, whereas uniform labeling efforts resulted in greater than 95% 13C isotope enrichment in the precursor backbone. These labeled precursors provided high-resolution NMR signals with great sensitivity and set the stage for studying the heparanome–proteome interactions.
Keywords: Glycosaminoglycan, Heparan sulfate, Heparin, Isotopes, Heparanomics, NMR spectroscopy, MS
1. Introduction
Heparan sulfate (HS), a member of the glycosaminoglycan (GAG) family, is a linear, sulfated polysaccharide composed of repeating disaccharide units comprising glucosamine and hexuronic acid (ido- or gluco-). Highly anionic HS is widely distributed on the cell surface and in the extracellular matrix. Interactions of HS sequences with various proteins have been implicated in many physiological and pathological processes such as their participation in cell–cell and cell–matrix interactions, cell proliferation, cell migration and cell differentiation, anti-coagulation, inflammation, tumor metathesis and various infections.1,2 However, the structure–function relationships of HS chains are difficult to establish because of their structural complexity arising from their highly variable length and composition.3 During HS biosynthesis, a number of enzymes seem to randomly and incompletely modify a nascent chain comprised of alternating glucuronic acid and N-acetylglucosamine residues. The resulting heterogeneity in HS fine structure, however, appears to be a key factor in determining the function of the HS chain, possibly because it confers specificity for interactions with HS binding proteins.
There are a number of methods through which HS–protein interactions can be studied, but one of the most powerful approaches is NMR spectroscopy. NMR spectroscopy provides crucial structural and conformational information useful in identifying precise contact points between interacting molecules and can also be used as an assay for identifying binding partners. Furthermore, NMR spectroscopy can also be used to detect modifications made by HS biosynthetic enzymes on precursor molecules and to study the biosynthetic process. However, high concentrations of HS chains are required for such studies due to the low natural abundance of nuclei with a net spin (13C and 15N). The preparation of HS biomolecules enriched with NMR-active nuclei would improve the detection and facilitate the analysis of structure–function relationships. Thus, the goal of this study is to prepare HS structures with atom-specific 13C labels at the anomeric centers of each sugar residue and also to prepare 15N-enriched and uniformly 13C-labeled HS precursors.
Escherichia coli K5 strain naturally synthesizes N-acetylheparosan (heparosan), a nonsulfated polysaccharide that resembles the unmodified, nascent HS chain.4 Thus, the preparation of isotope-enriched HS precursors can easily be accomplished by growing the E. coli strain K5 in a minimal media containing 15N-labeled ammonium salts and 13C-labeled glucose as the principal nitrogen source and carbon source, respectively. Several groups including our group recently published the preparation of isotope-enriched HS precursors for performing subsequent structural analysis.5–7 We report here the first preparation of atom specifically 13C-labeled HS precursor polysaccharides at the anomeric carbons. Differentially isotope-enriched HS precursor polysaccharides were isolated, quantified and characterized using NMR spectroscopy. They were also analyzed using ESI-qTOFMS to determine the extent of isotope incorporation.
2. Results and discussions
The intrinsically narrow range of proton chemical shifts has significant spectral overlap, particularly in GAG chains, and makes 1H NMR analysis challenging.8 On the other hand, 13C chemical shifts have a much wider range providing an opportunity to characterize complex HS/heparin chains using 13C NMR spectroscopy. However, NMR-active 13C nuclei are less abundant. Therefore, it is necessary to develop a strategy to prepare 13C isotope-enriched HS structures to fully exploit recent advances in NMR techniques to further our understanding of the structural basis for the biological actions of heparanome.
2.1 Preparation of differentially 13C-labeled HS precursors
N-Acetylheparosan, a capsular polysaccharide synthesized by the E.coli K5 strain, has the same structure as the non-sulfated and non-epimerized HS backbone. Therefore, this polymer has been used in the production of antithrombin III binding HS oligosaccharides and polysaccharides.9–12 Our group recently reported the use of the E.coli K5 strain for the production of 15N-labeled HS polysaccharide and oligosaccharide precursors.6 Zhang and co-workers produced uniformly 13C-, 15N-labeled HS polymers using a similar strategy for enzymatic modifications.5 They subsequently characterized the solution structures of these uniformly labeled HS polymers. However, one of the main disadvantages of uniformly 13C-labeled HS is that direct detection and assignment of 13C signals may not be straightforward due to the presence of one-bond 13C–13C couplings between adjacent carbons. One potential approach to overcome this difficulty is to produce atom-specific 13C-labeled HS precursors by utilizing an appropriate 13C-labeled glucose as a metabolic source for the backbone synthesis by the E.coli K5 strain. However, uniformly 13C-labeled glucose is less expensive than atom specific 13C-labeled glucose. Therefore, at the outset, we optimized the growth conditions of the E.coli K5 strain in a minimal medium containing uniformly 13C-labeled glucose and 15N-labeled ammonium sulfate or 15N-labeled ammonium chloride. We found that the overall yield of uniformly 13C-, 15N-labeled N-acetylheparosan is much higher when the E.coli K5 strain was grown in the presence of 15N-labeled ammonium sulfate (yield ~100 mg/L of culture) than in the presence of 15N-labeled ammonium chloride (yield ~30 mg/L of culture). After optimizing growth conditions to obtain the maximum amount of N-acetylheparosan polysaccharides using minimal media, we then turned our attention to prepare atom specifically 13C-labeled HS precursors by growing the E.coli K5 strain in a minimal medium containing [1-13C]-D-glucose as a carbon source and 15N-labeled ammonium sulfate as a nitrogen source. Differentially isotope enriched N-sulfoheparosan polysaccharides were obtained from the respective N-acetylheparosan polysaccharides by N-deacetylation and N-sulfonation as reported in the literature.13,14
2.2 Structural analysis of uniformly or atom-specifically 13C-labeled HS precursor polysaccharides
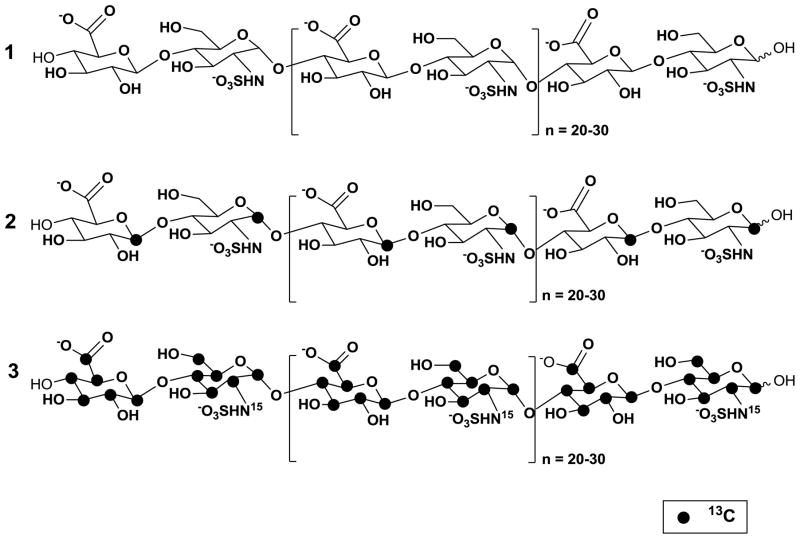
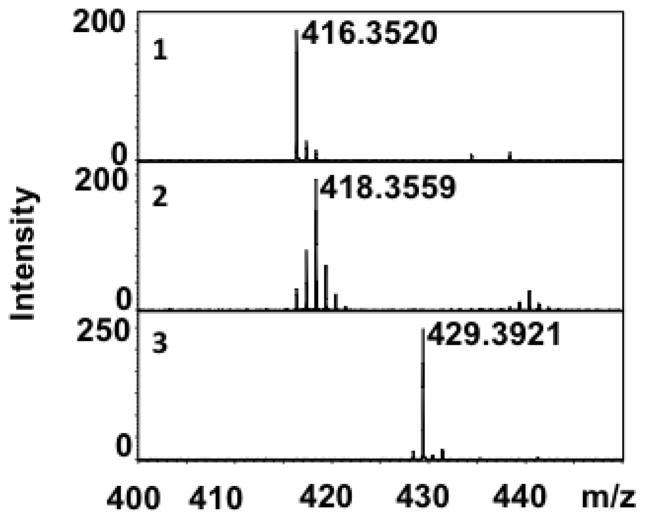
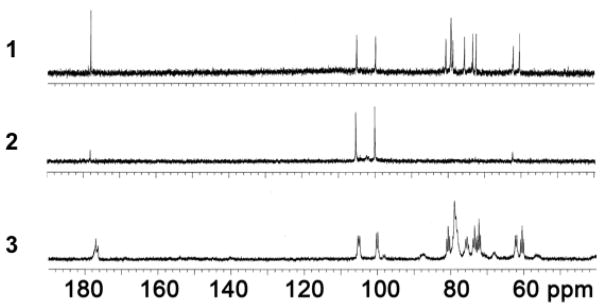
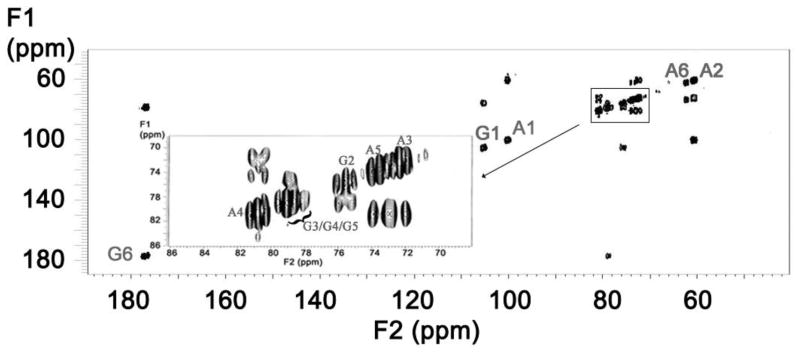
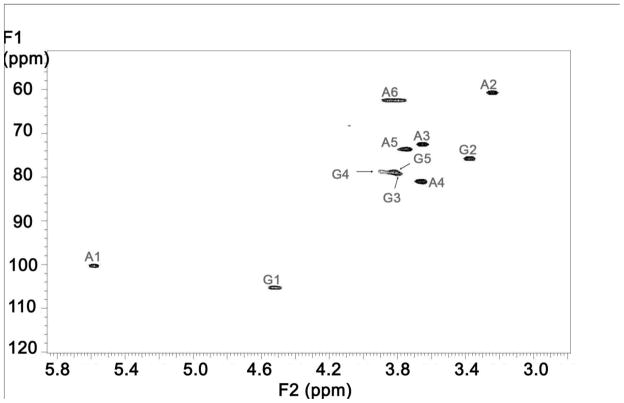
Differentially isotope-enriched N-sulfoheparosan polysaccharides and regular N-sulfoheparosan (1, 2, and 3, Figure 1) were exhaustively digested with recombinant heparitinase I. The disaccharide products so obtained were purified from the digestion mixture using 3000 MWCO Amicon centrifugal filter columns. The flow-through carrying the disaccharide components was dried in a speed-vac system and subsequently reconstituted in deionized water prior to mass spectrometric analysis. ESI-qTOFMS was utilized in the disaccharide analysis to determine the isotope enrichment.15 The mass spectrum of a given analyte typically consists of a sum of signals of various possible naturally occurring isotopic compositions. Thus, the monoisotopic patterns of the molecular ions of the given analyte provide further insights into the fine structure, the isotopic distribution, and the extent of isotopic enrichment. Therefore, it is important to have high-resolution mass spectra for which one needs a mass spectrometer with TOF or FT-ICR capability. MS analysis revealed the presence of significant peaks at m/z values of 416.3520, 418.3559 and 429.3921 for the molecular ions corresponding to the disaccharides derived from polysaccharides 1, 2 and 3, respectively (see Figure 1 and Figure 2). Based on the monoisotopic pattern, the isotopic purity of uniformly 13C-, 15N-labeled N-sulfoheparosan, polysaccharide 3, was found to be greater than 95%, whereas the isotopic purity of atom specifically 13C-labeled at the anomeric carbon of each sugar building block of N-sulfoheparosan, polysaccharide 2, was found to be ~70%. Thus, anomeric atom specific 13C-labeling efforts resulted in a partial isotopic loss that could not be overcome by altering the amounts of 13C-labeled glucose added to the minimal medium. Nevertheless, this is still significant enough to conduct NMR experiments to obtain critical structural information about glycosidic linkages which would not otherwise be possible with the polysaccharide 1. In addition to MS analysis, various NMR experiments were performed on these polysaccharides to obtain 1D 13C NMR (Figure 3), 2D [13C-13C] COSY (Figure 4) and [13C, 1H] HSQC NMR spectra (Figure 5). These NMR spectra confirmed the identity and the isotopic purity of differentially isotope-enriched N-sulfoheparosan polysaccharides 2 and 3. Our 13C and 1H chemical shift assignments are in good agreement with those previously reported in the literature (Table 1) considering the variations in NMR experimental conditions. Using intra-residue 13C-spin connectivities from 2D [13C-13C] COSY experiments, we assigned the peaks by a standard sequential assignment procedure.
Figure 1.
Structures of the HS precursor chains prepared without isotope enrichment (1) or with isotope enrichment (2, 3). The locations of 13C-isotopes are indicated by filled dark circles.
Figure 2.
ESI-Mass spectra of the disaccharides obtained from their corresponding heparan sulfate precursors with the natural isotope abundance 1 (m/z = 416.35), with atom-specific 13C-labels at anomeric carbon 2 (m/z = 418.36) and with uniform 13C and 15N-labels 3 (m/z = 429.39).
Figure 3.
Comparative 1D [13C] 100 MHz NMR spectra of the HS precursors with natural abundance (1), atom-specifically 13C-labeled HS precursors at anomeric carbons (2) and uniformly 13C- and 15N-labeled HS precursors (3). All NMR spectra were acquired in D2O at 25 °C. The values of 1H chemical shifts are referenced to the external 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) and 13C using an indirect reference.
Figure 4.
2D [13C-13C] COSY spectrum of the uniformly 13C- and 15N-labeled HS precursors. A represents glucosamine and G represents glucuronic acid.
Figure 5.
2D [13C-1H] HSQC spectrum of the uniformly 13C- and 15N-labeled HS precursors. A represents glucosamine and G represents glucuronic acid.
Table 1.
1H and 13C chemical shifts of HS precursors in D2O in 37 °Ca
| Position b | 1H Chemical Shift (ppm) | 13C Chemical Shift (ppm) |
|---|---|---|
| A1 | 5.58 | 100.30 |
| A2 | 3.24 | 60.77 |
| A3 | 3.65 | 72.48 |
| A4 | 3.65 | 81.11 |
| A5 | 3.75 | 73.61 |
| A6 | 3.84/3.79 | 62.65 |
| G1 | 4.52 | 105.33 |
| G2 | 3.38 | 75.83 |
| G3 | 3.80 | 79.96 |
| G4 | 3.89 | 78.70 |
| G5 | 3.82 | 79.06 |
| G6 | - | 176.85 |
Chemical shifts are given in ppm, 1H chemical shifts is measured indirectly with reference to DSS in D2O at 25°C. 13C is measured indirectly with reference to 1H using a chemical shift ratio of 0.25144.
A represents glucosamine and G represents glucuronic acid. The numerical suffix corresponds to the carbon position starting with the anomeric carbon as 1.
3. Concluding remarks
It is essential to elucidate both the primary sequence and the 3D conformation of HS structures of a biological origin to understand HS structure–function relationships. However, these structures are available only in a limited quantity, and this scarcity challenges our ability to characterize these complex molecules using traditional approaches. While MS analysis allows one to characterize oligosaccharide and disaccharide molecular weights, sulfation densities and sulfation patterns, NMR analysis aids the characterization of the intact heparanome at the polymer level in terms of conformation, microstructure, and dynamic solution properties. However, one of the major limitations of NMR spectroscopy is that its low sensitivity because of the low magnetogyric ratio and the low natural abundance of NMR active isotopes, 13C (1.1%) and 15N (0.37%). Therefore, the production of HS chains enriched with various isotopes will provide further insights into their sequence through MS-based fragmentation approaches and conformational dynamics through multi-dimensional NMR spectroscopy. Thus, the isotopic enrichment techniques described here for the production of HS chains along with the recent technological advances in NMR probe design, miniaturization of the separation techniques and softer MS ionization methods will overcome any remaining barriers in the structural analysis of HS in the coming decade. However, one note of caution is that the enzymatic approach does not have the capability to generate HS structures with discrete domain organizations as found in nature. Therefore, similar isotopic enrichment techniques need to be developed to prepare HS chains of a cellular or biological origin for structural biology studies.
In summary, uniform 13C labeling facilitated the assignment of 1H and 13C chemical shifts using [13C-13C] COSY and [13C- 1H] HSQC spectra. However, it is important to note that the measurements of exact distance in HS chains with uniform 13C labels will be significantly complicated because of the presence of several one-bond 13C–13C couplings between adjacent carbon atoms. Therefore, we also prepared atom-specific 13C-labeled HS chains that can facilitate the measurement of long distance weak couplings in the presence of strong interactions. Nevertheless, armed with differentially, uniformly or atom-specifically labeled HS chains, we can deduce the structural parameters required for the binding of a given HS structure to a protein target at a much higher molecular resolution than what is currently possible.
4. Experimental
4.1. Materials
Solid pronase from Streptomyces griseus and mung bean nuclease was purchased from Sigma–Aldrich Chemical Co. (St. Louis, MO, USA). Heparitinase I from Flavobacterium heparinum was a generous gift from Professor Robert D. Rosenberg (BIDMC, Harvard Medical School). Sodium salts of unsaturated disaccharide standards, ΔU-GlcNS and ΔU-GlcNAc were obtained from Iduron Co. (Manchester, England, UK). [1, 2, 3, 4, 5, 6- 13C6]-D-glucose, [1- 13C]-D-glucose, 15NH4Cl and (15NH4)2SO4 were obtained from Sigma–Aldrich Chemical Co. Water was purified using a Milli-Q filtration apparatus (Millipore Co., Bedford, MA, USA). Fast-flow DEAE-Sepharose was obtained from GE Healthcare (Amersham, Uppsala, Sweden). All other chemicals, including MS grade and NMR solvents, were obtained from Sigma–Aldrich Chemical Co. and Cambridge Isotope Laboratories, Inc. (Andover, MA, USA), respectively. Polypropylene chromatographic columns (2.5 × 20 cm) were purchased from Bio-Rad Life Sciences (Hercules, CA, USA). NMR tubes were purchased from Sigma–Aldrich Chemical Co. The E. coli strain K5 [O10:K5 (L):H4] was obtained from ATCC.
4.2. Isolation of differentially 13C-labeled N-acetylheparosan
[1, 2, 3, 4, 5, 6- 13C, 15N] N-Acetylheparosan was prepared from cultures of the E.coli K5 strain grown in a minimal media containing 4 g/L of [1, 2, 3, 4, 5, 6- 13C6]-D-glucose and 2 g/L (15NH4)2SO4 or 15NH4Cl. The bacterial primary cultures were grown in 100 mL of LB broth for 24 h at 37 °C with shaking (250 rpm) and harvested by centrifuging at 5000 rpm for 30 min. The pellet from the 100-mL primary cultures was used to inoculate 1 L of minimal medium in a 3-L Erlenmeyer flask containing 4 g/L [1,2,3,4,5,6-13C6]-D-glucose as the principal carbon source and 2 g/L (15NH4)2SO4 or 15NH4Cl as the principal nitrogen source. The cultures were incubated at 37 °C for 48 h with shaking (250 rpm) and then autoclaved. The autoclaved medium was adjusted to pH 7.0 and treated with protease for 12 h at 37 °C with gentle shaking (50 rpm). The insoluble material was removed by centrifugation (5000 rpm, 30 min), and the resulting supernatant was diluted to two times the original volume with deionized water. The diluted supernatant was then loaded onto a 50-mL DEAE-Sepharose column, previously equilibrated with wash buffer (20 mM NaOAc, 100 mM NaCl, pH 6). The column was washed three times (10 column volumes/wash) with wash buffer. The heparosan polysaccharide was eluted with 6 column volumes of elution buffer (20 mM NaOAc, 0.6 M NaCl, pH 6). The eluate was adjusted to 1.0 M NaCl, and 4 volumes of 99% EtOH were added to precipitate the desired polysaccharide. After 24 h at 4 °C, the resulting [13C, 15N] N-acetylheparosan was recovered by centrifugation (5000 rpm, 30 min, 4 °C) and removal of the supernatant, and was finally allowed to air-dry overnight. The amount of purified heparosan obtained was determined using a carbazole assay for uronic acids with the aid of glucuronolactone as a standard. The assay was performed in triplicate. In a similar manner, the anomeric carbon-specific 13C-labeled polysaccharide was obtained from the E.coli K5 strain grown in the minimal medium containing 1g/L [1-13C]-D-glucose.
4.3. Preparation of N-sulfoheparosan
Regular N-acetylheparosan, anomeric atom-specifically 13C-labeled heparosan or uniformly 13C, 15N-labeled heparosan was dissolved in 2.5 M NaOH, stirred for 12 h at 55 °C, then cooled to ice-cold temperature and adjusted to pH 7. The fully N-deacetylated product was N-sulfonated with the Et3N·SO3 as described before 13,14 and purified to obtain polysaccharides 1, 2 and 3 (Figure 1).
4.4. NMR Acquisition
1D [13C] and 2D [13C-13C] COSY spectra were recorded at 100 MHz on a Varian Mercury 400 MHz spectrometer. 2D [13C-1H] HSQC spectra were recorded at 125 MHz on a Varian Inova 500 MHz spectrometer (Varian, Palo Alto, USA). NMR measurements were performed at 25 °C for acquiring 1D [13C] spectra and 2D [13C-13C] COSY spectra whereas 2D [13C-1H] HSQC spectra were acquired at 37 °C. Samples (~10 mg) were lyophilized from deuterium oxide (D2O) and finally dissolved in 0.4 mL of D2O (99.9 % deuterium).
4.5. Isotopomer analysis of disaccharides using LC–ESI–TOFMS
MS analyses were performed on a Bruker ESI-TOFMS instrument. The samples containing disaccharides, obtained from the digestion of polysaccharides with heparitinase I, were processed and dissolved in 1:1 H2O–CH3CN containing 5 mM Bu2NH2OAc for the isotopomer analysis using mass spectrometry.15 Mass spectra were collected by the direct infusion of samples containing disaccharides at a flow rate of 5 μL/min. The electrospray interface was set in the negative-ionization mode with a collision energy of 7.0 eV, a capillary potential of 3500 V and a source temperature of 180 °C. Nitrogen was used as a drying gas (4 L/min) and nebulizer (0.4 bar). Mass spectra were processed using Data Analysis 2.2 software (Bruker Daltonics Inc., Billerica, MA).
Acknowledgments
This work was supported by the National Institutes of Health grant (GM075168), Mizutani Foundation for Glycoscience Award and American Heart Association National Scientist Development Award to B.K. We acknowledge the funding support from NIH, grant RR06262, for NMR instrumentation awarded to the University of Utah Health Sciences NMR Center. T.K.N.N. received a graduate fellowship from the Vietnam Education Foundation. We thank Karthik Raman for careful reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raman K, Kuberan B. Curr Chem Biol. 2010;4:20–31. doi: 10.2174/187231310790226206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings RD. Mol Biosyst. 2009;5:1087–104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 3.Raghuraman A, Mosier PD, Desai UR. J Med Chem. 2006;49:3553–62. doi: 10.1021/jm060092o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vann WF, Schmidt MA, Jann B, Jann K. Eur J Biochem. 1981;116:359–64. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, McCallum SA, Xie J, Nieto L, Corzana F, Jimenez-Barbero J, Chen M, Liu J, Linhardt RJ. J Am Chem Soc. 2008;130:12998–3007. doi: 10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sigulinsky C, Babu P, Victor XV, Kuberan B. Carbohydr Res. 2010;345:250–6. doi: 10.1016/j.carres.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mobli M, Nilsson M, Almond A. Glycoconjugate J. 2008;25:401–14. doi: 10.1007/s10719-007-9081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korir AK, Larive CK. Anal Bioanal Chem. 2009;393:155–69. doi: 10.1007/s00216-008-2412-2. [DOI] [PubMed] [Google Scholar]
- 9.Kuberan B, Beeler DL, Lawrence R, Lech M, Rosenberg RD. J Am Chem Soc. 2003;125:12424–5. doi: 10.1021/ja036737g. [DOI] [PubMed] [Google Scholar]
- 10.Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J Biol Chem. 2003;278:52613–21. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- 11.Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Nat Biotechnol. 2003;21:1343–6. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl U, Li JP, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emeis J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. J Med Chem. 2005;48:349–52. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- 13.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. Carbohydr Res. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 14.Casu B, Grazioli G, Razi N, Guerrini M, Naggi A, Torri G, Oreste P, Tursi F, Zoppetti G, Lindahl U. Carbohydr Res. 1994;263:271–84. doi: 10.1016/0008-6215(94)00172-3. [DOI] [PubMed] [Google Scholar]
- 15.Kuberan B, Lech M, Zhang L, Wu ZL, Beeler DL, Rosenberg RD. J Am Chem Soc. 2002;124:8707–18. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]