SUMMARY
Genomic instability, such as copy-number losses and gains, occurs in many genetic diseases. Recent technology developments enable researchers to measure copy numbers at tens of thousands of markers simultaneously. In this paper, we propose a non-parametric approach for detecting the locations of copy-number changes and provide a measure of significance for each change point. The proposed test is based on seeking scale-based changes in the sequence of copy numbers, which is ordered by the marker locations along the chromosome. The method leads to a natural way to estimate the null distribution for the test of a change point and adjusted p-values for the significance of a change point using a step-down maxT permutation algorithm to control the family-wise error rate. A simulation study investigates the finite sample performance of the proposed method and compares it with a more standard sequential testing method. The method is illustrated using two real data sets.
Keywords: Array-based comparative genomic hybridization, Change point, Copy number variation, Multiple comparison, Multiscale product, p-value, Wavelet
1. Introduction
The integrity and stability of chromosomes enable the cell to transmit accurately its genetic information and function properly physiologically. Aberrations in chromosomes such as rearrangements, deletions, amplifications and other types of copy number changes occur in many genetic diseases including, for example, Down syndrome which is a well-known developmental abnormality caused by trisomy (triplication) of the 21st chromosome. Studies of such aberrations, commonly referred to as genomic instability, can help understand the underlying mechanism of disease initiation and progression (Pinkel and Albertson, 2005).
A popular approach for assessing genomic instability is to measure genome-wide copy number variations using array-based comparative genomic hybridization (array CGH) (e.g., Snijders et al., 2001; Hodgson et al., 2001; Pollack et al., 2002). More recently, high-density single nucleotide polymorphism (SNP) arrays are used to genotype up to one million markers (e.g. Illumina Human 1M BeadChip). Although the two array technologies are different, data generated by them share common features. For these arrays, a set of genomic markers with known locations are chosen, and DNA copy numbers are measured as fluorescent intensity at each marker, so the data can be viewed as a sequence of copy number measurements ordered by the marker locations along the chromosome. With proper normalization (not the focus of this paper), an increase in the magnitude of positive or negative log ratios of test versus reference samples corresponds to possible copy number changes such as amplifications or deletions, respectively.
A variety of methods have been developed to segment chromosomes into regions of markers having the same underlying copy number. Although such a segmentation can also be used for identifying change points, little work has been done directly on the detection of change points and the corresponding statistical inference. This is the focus of our presentation.
A review of current approaches for copy number segmentation places them broadly into three main categories. The first category uses model-selection procedures, penalizing the number of segments (parameters) to avoid over segmenting an array CGH profile. These include Gaussian likelihood under a piecewise-constant model with various penalty parameters (Jong et al., 2003; Picard et al., 2007; Zhang and Siegmund, 2007), unsupervised Hidden Markov Models using Bayesian information criterion (BIC) or Akaike information criterion (AIC) (Fridlyand et al., 2004) and penalized least squares regression (Huang et al., 2005). More recently, Bayesian techniques have also been used in regularizing parameter estimation (Lai et al., 2008; Guha et al., 2008).
A second category consists of nonparametric function estimation to infer underlying true copy numbers, including a quantile smoothing method (Eilers and Menezes, 2005), a “fused lasso” method (Tibshirani and Wang, 2007) and a wavelet-based denoising method (Hsu et al., 2005). Wavelet methods and other nonparametric techniques are also suited to detect sharp changes as often observed in array CGH data, but to recover a piecewise constant function, an additional clustering of adjacent values may be needed.
A third category selects segments by controlling the overall type I error rate. For example, Olshen et al. (2004) proposed a circular binary segmentation (CBS) method using a sequential testing procedure. They considered all possible locations and widths of step functions and calculated the maximum t statistic for all combinations. The genome is then partitioned according to the maximum t statistic which exceeds the critical value at a pre-specified significant level. Each segment is then subjected to the same testing procedure. This procedure continues until no test statistic in each segment is significant. The approach of Wang et al. (2005) selects “significant” clusters formed along the chromosome by controlling false discovery rate (FDR). Cheng et al. (2003) proposed to detect copy number changes in a regression framework by pooling information across multiple samples.
Lacking among these methods are measures of significance for individual change points. The methods of Olshen et al. (2004) and Wang et al. (2005) provide some control on the overall type I error rate, although the latter requires an external set of normal samples to obtain the null distribution, and in the former, the false positive rate increases with the number of true change points. Both methods require one to pre-specify a level of significance. When individual change points are of interest, this is relatively inefficient as it requires one to re-run the segmentation procedures at different significant levels. We consider an alternative to this pre-specification of significance by estimating a p-value for each marker. This allows investigators to examine change points at their own significant levels without repetitive implementation of the same procedure. The method of Cheng et al. (2003) provides adjusted p-values by controlling family-wise error rate. However, it relies on multiple samples to estimate, and make inference about, the average intensity ratio for each marker. Like most literature in this field, our proposed method is designed to detect change points for each individual sample (i.e., in one array).
The paper is organized as follows. Section 2 describes the method for detecting change points which is based on a cross-scale product in a multiscale decomposition of the array signal. The wavelet transform used to produce this is introduced in 2.1. The test statistic and multiple comparison adjustment procedure are given in Section 2.2 and 2.3, respectively. Two real data sets are used to illustrate the proposed method and the results are shown in Section 3. Section 4 describes the results from a simulation study for examining the performance of the proposed approach and the method proposed by Olshen et al. (2004). Some final remarks are given in Section 5.
2. Detecting change points by multiscale products
Let Yi be the observed log-relative intensity ratio for the ith marker location, xi, for i = 1, …, n. Assuming additive measurement error for the log-relative intensities, the observed data can be modeled as
| (1) |
where f is a piecewise constant function reflecting the discreteness in copy number; the εi, i = 1, …, n, are independent and identically distributed with mean 0 and variance σ2.
Given R change points, there are R + 1 non-overlapping segments (0 ≤ R ≤ n − 1), each representing a region of amplification, deletion or no change in gene copy numbers. For the rth segment, let cr denote the index of the end marker in the segment and μr be the corresponding copy number. Assume c1 < c2 < … < cR and define c0 = 0, cR+1 = n. The collection {xcr : r = 1, …, R} is the set of change point locations. Rewriting model (1) in these terms, Yi = μr + εi for cr − 1 < i ≤ cr. For identifiability of the change points, we assume μr ≠ μr+1. Under the null hypothesis of no change, f(xi) = μ0. Without loss of generality, we assume μ0 = 0.
2.1 Multiscale wavelet products
We are interested in a test statistic that reflects significant step-like changes in copy number along the chromosome as modeled by f in (1). One way to focus attention on the step-like changes is to quantify the difference of adjacent averages in a neighborhood of each location xi, such as , where B is the number of markers in the neighborhood. Since the aberration size is unknown, it would be prudent to examine various neighborhood sizes.
A rigorous and efficient way to calculate these differences is by a wavelet transform of f: let , and ψ = 0 otherwise. The collection of all translations and dilations, , forms a family {ψs,x : s ∈ ℝ+, x ∈ ℝ} of functions that define a wavelet transform of f defined as W f(s, x) ≔ ∫ ψs,x(u) f(u)du. Sampling at every marker location, xi, i = 1, …, n, and at dyadic scales, s = 2j, j = 1, …, J = ⌊log2(n)⌋ (the greatest integer less than log2(n)), gives coefficients Wj,i ≔ W f(2j, xi) of the maximal overlap discrete wavelet transform (MODWT). Of particular interest is the fact that , B = 2j − 1, which quantifies the difference of adjacent averages in varying sized neighborhoods of xi and reflects precisely-located changes in f that occur at scale 2j. We use the terminology “level j” interchangeably with “scale 2j”. In particular, let denote the level j coefficient function. See Percival and Walden (2000) regarding details of the MODWT.
We note that our use of a wavelet analysis differs from the more common goal of function estimation or denoising. In fact, we make no use of the inverse transform or thresholding techniques. Instead, our proposed test statistic is based on the coefficient functions Wj and whether, at a given location, they are more extreme than expected under the null (for a given amount of noise in the data). A useful property of these coefficient functions is that a change in f at a location xi propagates across scales: the values |Wj,i′|, …, |Wj+k,i′| are increased for all i′ in some neighborhood of i, where j and k depend on the sharpness of the change and the width of the feature. We exploit this persistence across scales by considering the pointwise product Wj,i Wj+1,i of adjacent coefficient functions. This reinforces signal while canceling noise since coefficients related to high-frequency noise do not persist across scales and are diminished in the product.
A general multiscale product at the ith location is of the form Πj∈D Wj,i, where D is a subset of all the possible levels {1, …, J}. If location i is indeed a change point, the wavelet coefficients at the adjacent levels are most correlated and so we focus attention on the product of two adjacent levels Wj,i Wj+1,i. We have restricted attention to two levels since for small aberrations and short segments, coarse levels that include markers not related to the aberration are less effective in detecting abrupt changes. We create, however, a test that is adaptive to varying sizes of aberration by considering the maximum of this product across levels at each location. That is, define Mi ≔ maxj∈{2,…,J0} {Wj,i Wj + 1,i} for some J0 < J. Then the problem of detecting change points becomes one of testing
for each location i. Note that the test statistic Mi is always positive if location i is indeed a change point, so a one-sided test is used here.
For a genomic profile with different aberration sizes, the optimal scales used in Mi for detecting a change in copy number will depend on the properties of the chromosome aberrations and the density of markers. The choice of J0 loosely depends on n since J0 < J = ⌊log2(n)⌋. However, averaging over long segments induces high spatial correlation among the Mi making it more difficult to precisely locate a true change point. Hence, when markers are sufficiently dense, J0 is typically substantially less that J; setting J0 = 6 or 7 generally provides adequate power for detecting change points.
Regarding the historical use of wavelet products, Bao and Zhang (2003) used two-scale products in a method for wavelet thresholding in signal recovery. In a work more closely related to ours, Sadler and Swami (1999) used two- and three-scale products aimed at detecting discontinuities. Their presentation considered theoretical and empirical distributions for these products, but failed to control an overall type I error rate and did not adjust for multiple comparisons. We are not aware of any work aimed at statistical inference based on multiscale products, one of the focuses of this presentation.
2.2 Local maximum
Given the two-scale product statistic at each marker, one might simply apply a multiple testing procedure to all n test statistics in an attempt to control the overall type I error rate. However, the MODWT coefficients within each level are locally correlated and elevated in an entire neighborhood surrounding a change point. The width of this neighborhood depends on the width of the aberration and the levels used in Mi. To circumvent this, we test only locations at which local maxima occur in M (as a function of i). Then for each local maximum, its location is designated to be a change point if the adjusted p-value is less than a pre-specified significance level. The notation i* is used to denote that a local maximum is detected at the ith location.
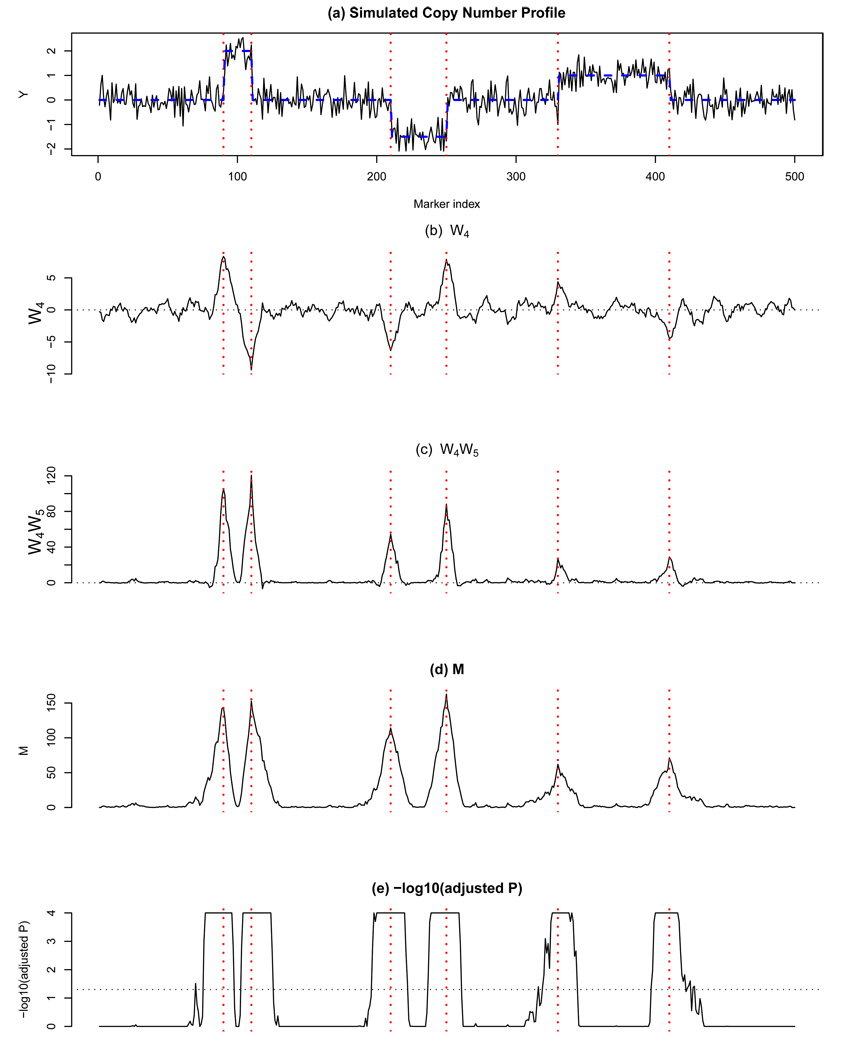
Figure 1 illustrates these ideas with a simulated copy number profile, the corresponding W4, W4W5, M and the associated significance values, − log10(p); each is plotted against i. We are only interested in the local maxima in M since they correspond to potential change points, yet as seen in Figure 1 the values of Mi tend to be elevated in an entire neighborhood of a change point since the Wj’s reflect neighborhood changes in f. It would be easy to obtain these if the copy number signal (hence M) was smooth, but any discrete noisy function will exhibit small local maxima that are not relevant since they don’t occur at a scale of interest. Although one could smooth M (say, with a kernel smoother using a particular bandwidth) and then seek local maxima, we have chosen to do this in one step through an efficient procedure based on the following fact: the level j transform of f is the first derivative of f after it has been smoothed with a kernel of scale 2j (see Mallat, 1999). Therefore, the search for local maxima in M is achieved simply by performing a MODWT of M, at level j, and recording its zero crossings.
Figure 1.
Illustration of the proposed method. (a) A simulated copy number profile with 500 markers (f in a dotted line) and 6 change points (dotted vertical lines); (b) W4; (c) W4W5; (d) M; (e) −log10 of adjusted p-values truncated at 4. The horizontal dotted line indicates p = 0:05.
The locations of these local maxima in M are a small subset, {xi* : i* ∈ K}, of the entire set of marker locations, where K is a set of indices for local maxima. A focus on only this subset of markers dramatically reduces the number of tests for change points. As a result, computing time for the proposed method does not substantially increase even as the number of markers increases to tens of thousands. We also show that the estimated change points converge to the true change point locations as the number of markers goes to infinity (see the Web Appendix).
2.3 Multiple testing
We describe a procedure for obtaining adjusted p-values at local maxima while accounting for multiple comparisons. We focus on controlling the family wise error rate (FWER) and note that the same test statistics can be used for obtaining other measures of statistical significance, such as q-values, using the algorithms described in Ge et al. (2003), which provides a comprehensive review of this topic.
Since both the marginal and joint distributions of the test statistics Mi* are unknown, we use resampling methods to estimate both raw and adjusted p-values. It is not obvious how to estimate a null distribution since the true function f is unknown. We consider two approaches for generating a null distribution.
The first approach is to permute ε̂i = Yi − f̂i where f̂ s a robust estimator of f using lowess, a locally weighted regression (Cleveland, 1979). A simple permutation of the observed Yi without subtracting f̂ would work if non-zero segments are only a small proportion of the whole region. Unfortunately the empirical distribution of errors is highly dependent on the estimated f̂. The key parameter in lowess smoothing is the width of the smoothing window; the larger the window size, the smoother f̂. We assessed the performance of the multiscale method by simulation using two different window sizes, 0.05 and 0.1, and found that the two sizes gave comparable results (results not shown). In the following sections, we used a window size of 0.1 and present results for window size 0.05 whenever a difference is observed.
The second approach is to permute the wavelet coefficients W1 at the finest level. Since the W1,i denotes the scaled difference Yi − Yi−1, this gives a close approximate estimation of the null distribution as R is relatively small compared to n. This approach, however, may overestimate R if the true error distribution is heavily tailed. To see this, let ε1 and ε2 be i.i.d. with mean 0 and variance σ2, and W = ε1 − ε2. In general, W doesn’t have the same distribution as ε1 and ε2 unless ε1 and ε2 are normally distributed. To see the relationship between W and ε1 (or ε2), consider the first four moments: mean, variance, skewness and kurtosis for W and ε1, one can show that the skewness of W is 0 and the kurtosis of W is one half of the kurtosis of ε1 (or ε2). For distributions with extremely heavy tails, permuting W1 yields more false positives than expected. In this case, the first approach (permuting ε̂) is more appropriate.
After the null distribution of ε is estimated, the adjusted p-values can be computed using the step-down maxT permutation algorithm proposed by Westfall and Young (1993).
The following summarizes the proposed algorithm for detecting change points.
Compute the coefficient functions Wj for levels j = 1, …, J0 + 1. Estimate the standard deviation σ using the MAD of W1: (the divisor provides asymptotically normal consistency). Standardize each level as .
Calculate point-wise products of standardized wavelet coefficients at all adjacent levels, WjWj+1, j = 2, … J0. The test statistic for location i is Mi = maxj∈{2,…,J0} {Wj,i Wj + 1,i}.
Obtain the local maxima {Mi* : i* ∈ K} in M using level 4 wavelet transform.
Estimate the null distribution of Mi* by permuting W1 (or ε̂) and obtain an adjusted p-value for each Mi* using the step-down maxT algorithm. Estimated change points are those whose corresponding adjusted p-values are smaller than a pre-specified threshold.
3. Results from real data
We illustrate the method using two real data sets: Coriel cell lines data (Snijders et al., 2001) and Illumina Human 1M SNP data (Peiffer et al., 2006). The proposed method is compared with the CBS method (Olshen et al., 2004). The CBS method is singled out because it performed consistently well based on a comprehensive comparison study by Lai et al. (2005). Since the CBS method detects change points by controlling the overall type I error rate, it makes the comparison with our proposed method more equitable than those based on model selection for which choosing tuning parameters is often an issue. The CBS method is used here without the extra preprocessing or pruning step because these steps are not part of the hypothesis testing and the number of estimated change points is very sensitive to the pruning parameters, the choice of which is rather subjective.
3.1 Coriel cell lines data
In 2001, Snijders et al. studied the DNA copy number changes for 15 Coriel cell lines using array CGH technology. Each array contained 2276 mapped BAC clones (markers) spotted in triplicate. The Coriel cell line data have been analyzed by many methods (e.g., Hsu et al., 2005; Olshen et al., 2004) and can be freely downloaded at http://www.nature.com/ng/journal/v29/n3/suppinfo/ng754_S1.html. This data is considered here primarily for proof-of-principle since the copy number alterations are known, having been identified by spectral karyotyping. Of 15 cell lines, six cell lines have whole chromosome amplifications only and nine cell lines have partial chromosome amplifications or deletions. We applied the proposed method and the CBS method to all 15 Coriel cell lines.
We analyzed the whole genome (all 23 chromosomes) simultaneously. The main consideration is that the total number of markers is 2276 and the density of markers varies greatly from chromosome to chromosome. For example, chromosomes 19, 21 and 22 have 35, 30 and 15 markers, respectively, and only seven chromosomes have more than 100 markers. The estimation of variance will be more accurate with the whole genome data than with each individual chromosome. In addition, it allows us to detect whole chromosome aberrations as the rest of the genome provides a good baseline reference. Finally, a genome-wide analysis allows us to control type I error rate for the whole genome, instead of at the chromosome level. A significance level of 0.01 was used.
The noise level of the Coriel data was very low with σ̂ ranging from 0.06 to 0.10 (a median of 0.07). The data appears approximately normally distributed based on Q-Q plots and histograms (results not shown), so both permutation procedures (W1 and ε̂) were used to estimate the adjusted p-values and they gave the exactly same results for all cell lines except for GM02948, where permuting W1 gave an additional false positive on Chromosome 20 at 65.3Mb. To save space only results from permuting ε̂ will be presented.
The Coriel data had strong signals and low background noise with median of the signal-to-noise ratio (SNR) 6.83 (first and third quantiles are 5.73 and 8.46, respectively). We observed that the CBS method detected many small segments for a total of 95 false positives. The multiscale method had no false positives (Table 1). Both methods missed the singleton on cell lines GM01535 in which only one altered marker exists on chromosome 12qtel. Note that by our convention in defining a change point in Section 2, the count of change points for a singleton is two. The adjusted p-values for the two false negatives on GM01535 are 0.231 and 1.000, respectively.
Table 1.
Summary of the number of false positives (FP) and false negatives (FN) on 15 Coriel cell lines using the multiscale and CBS methods. A significance level 0.01 was used for both methods.
| Multiscale | CBS | |||
|---|---|---|---|---|
| Cell line | FP | FN | FP | FN |
| GM00143 | 0 | 0 | 26 | 0 |
| GM01524 | 0 | 0 | 11 | 0 |
| GM01535 | 0 | 2 | 0 | 2 |
| GM01750 | 0 | 0 | 2 | 0 |
| GM02948 | 0 | 0 | 7 | 0 |
| GM03134 | 0 | 0 | 0 | 0 |
| GM03563 | 0 | 0 | 10 | 0 |
| GM03576 | 0 | 0 | 1 | 0 |
| GM04435 | 0 | 0 | 2 | 0 |
| GM05296 | 0 | 0 | 0 | 0 |
| GM07081 | 0 | 0 | 0 | 0 |
| GM07408 | 0 | 0 | 2 | 0 |
| GM10315 | 0 | 0 | 10 | 0 |
| GM13031 | 0 | 0 | 0 | 0 |
| GM13330 | 0 | 0 | 23 | 0 |
| Total | 0 | 2 | 95 | 2 |
3.2 Illumina’s Human 1M SNP data
The proposed method was also applied to detect copy number variation (CNV) using Illumina’s Human 1M SNPs data (http://www.illumina.com).To save space, we refer interested readers to Peiffer et al. (2006) for the normalization. The key point here is that, as in array CGH data, an increase in the magnitude of positive or negative log intensity ratios corresponds to possible insertion or deletion events, respectively. Therefore, segmentation methods developed for array CGH data would be applicable here.
The data here included 8 HapMap individuals whose high-resolution SNP intensity data, including normalized log intensity ratios, were freely available from Illumina web site. The reason we chose to work on these 8 HapMap individuals is that the CNV deletion events were detected and validated by two independent molecular experiments, fosmid-ESP assay and complete fosmid sequencing (Kidd et al., 2008). The data of the validated CNV events were downloaded from the supplementary material by Kidd et al. (2008) and Cooper et al. (2008).
We focused on deletion events that are covered by 10 or more SNPs to be consistent with Cooper et al. (2008)’s definition for detectable deletion events on the Illumina 1M array. This yielded 97 deletion events (i.e. 194 change points) which were twice validated experimentally and have sufficient probe coverage. We notice that the start and end locations of the selected 97 events detected by the two experiments were not identical, differing by up to 100K base pairs. Therefore, in the analysis we used the change point locations detected by complete fosmid sequencing as a gold standard and allowed for a 5-SNP difference on either side. Larger than 5 SNPs would cause ambiguity in the definition of change points for small-sized deletion events.
We analyzed one chromosome at a time due to the high-density markers. The number of SNPs per chromosome ranged from 15,408 to 98,752 with an average of 53,042. Two adjacent SNPs spanned on average 2.6kbp. The noise level of the SNP data was low with σ̂ ranging from 0.08 to 0.13 (a median of 0.09). We analyzed the data using a permutation of ε̂ because the error distribution was heavily tailed and permuting W1 yielded more false positives (results not shown). We used a significance level 0.05, which differed from that used in the Coriel cell line data. This is because signals in the Illumina 1M SNP data are rather weak. A more relaxed significance level than 0.01 would allow us to detect more change points, and thus better discern the performance of the two approaches.
We observed that the CBS method falsely detected a total of 324 change points while the multiscale method had 217 false positives. The sensitivities were low for both CBS and multiscale methods. Of 194 change points which were twice validated by molecular experiments, the multiscale method detected 25 (12.9%) while CBS 19 (9.8%) (Table 2). All but two true change points detected by CBS were detected by the multiscale method. The two change points missed by the multiscale method were on chromosome 16 for NA19129 and had adjusted p-values 0.237 and 0.206 and raw p-values 0.003 and 0.002, respectively (see Web Figure 1). On the other hand, the CBS method missed eight change points that the multiscale method was able to detect. Figure 2 gives an example of a profile in which a deletion event is detected by the multiscale method but not by CBS. This is probably because the outliers in the data, as well as the large number of markers, increased the critical value for calling the significance of change points, which consequently reduced the power of CBS to detect these change points. In contrast, the multiscale method which is based on multiple scales and local maxima is more robust to outliers and more amenable to a large number of markers since the number of local maxima increases far less quickly than the number of markers.
Table 2.
Summary of the number of false positives (FP) and false negatives (FN) on 8 HapMap individuals using the multiscale and CBS methods. A significance level 0.05 was used for both methods.
| Multiscale | CBS | # change points | |||
|---|---|---|---|---|---|
| Cell line | FP | FN | FP | FN | |
| NA12156 | 28 | 18 | 76 | 20 | 22 |
| NA12878 | 52 | 33 | 66 | 35 | 38 |
| NA18507 | 21 | 18 | 26 | 18 | 20 |
| NA18517 | 14 | 11 | 16 | 12 | 12 |
| NA18555 | 22 | 18 | 51 | 21 | 22 |
| NA18956 | 19 | 17 | 27 | 17 | 22 |
| NA19129 | 30 | 26 | 32 | 24 | 28 |
| NA19240 | 31 | 28 | 30 | 28 | 30 |
| Total | 217 | 169 | 324 | 175 | 194 |
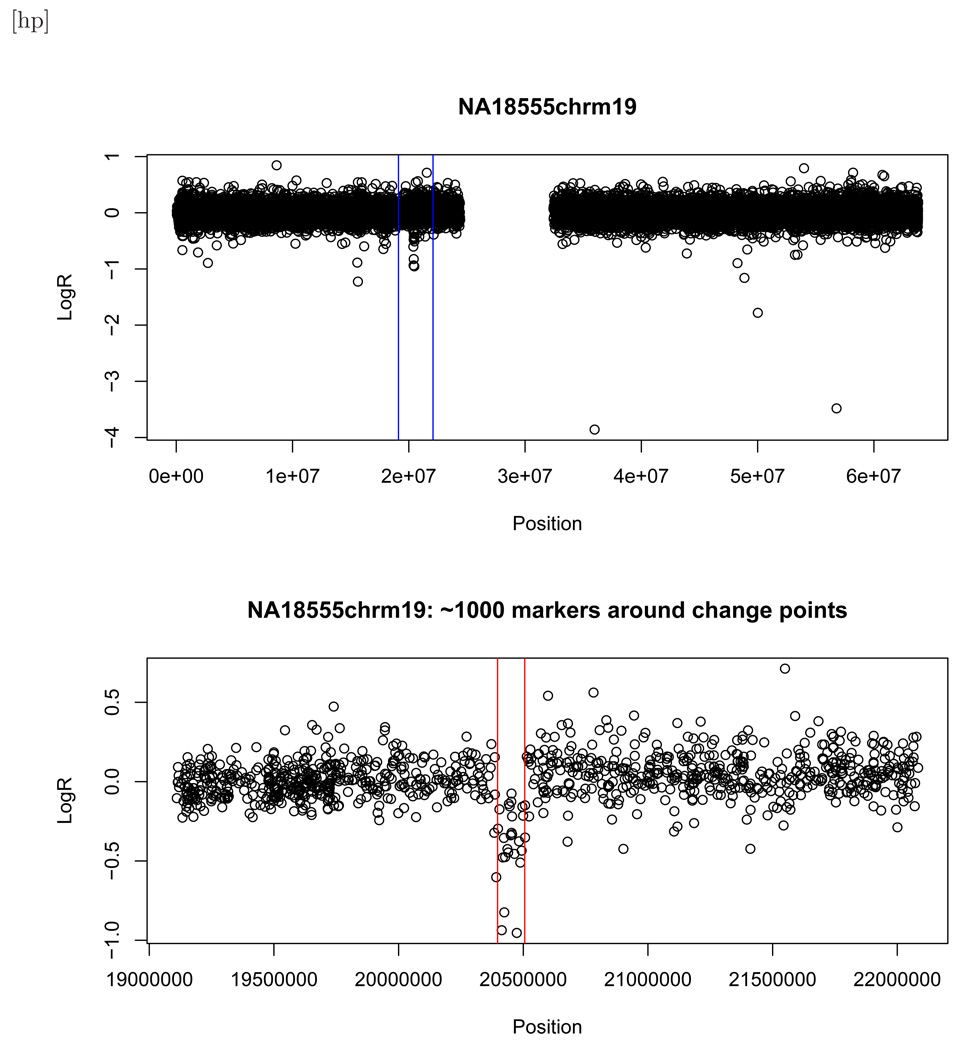
Figure 2.
Top panel: Scatter plot for NA18555 chromosome 19 (SNR = −3.71). The vertical lines indicate a small region that surrounds the validated change points. The blank spot is the centromere. Bottom panel: Zoomed-in scatter plot of the region that surrounds the change points. The vertical lines indicate the validated change points.
To understand why both methods had such low sensitivity we examined each profile manually. The reason appears to be that the signals for deletion events are weak for most profiles. The median SNR was −0.29 with first and third quartiles were −3.66 and 0.049, respectively; only one third (33) of 97 deletion events had SNR ≤ −1. Among the 64 events that had SNP > −1, 28 even had positive mean log intensity ratios and the maximum SNR was 1.24. This implies the measurement error, even though small in absolute value, is still quite large compared to the signal in the copy number data. For an example of a profile in which both methods failed see Web Figure 2.
It has been suggested that the use of genotyping information may help in detecting allele-specific CNV from these data (Dr. Adam Olshen, personal communications). Additional work along this line is clearly warranted, but is beyond the scope of this paper and not considered further.
4. A simulation study
This section discusses the finite-sample performance of the proposed method and contrasts it with the CBS method. Performance was measured by the number of estimated change points, true positive rate (TPR), false discovery rate (FDR) and number of exact detections. The TPR is the proportion of true change points rejected at a pre-specified significance level. The FDR is the proportion of false rejections among the total rejections. The number of exact detections is the number of simulated data sets in which all change points are correctly detected without any false positives and false negatives. Simulations were performed for both normally and non-normally distributed ε. For each simulation scenario, a total of 500 datasets were simulated with each dataset having 500 markers. A significance level 0.01 was used throughout, unless otherwise stated.
4.1 Simulation under normal distribution
Assume ε is normally distributed and consider data simulated according to the model (1). The performance of the proposed and CBS methods were evaluated under a variety of underlying mean functions, f, and noise levels, σ. For the complete null case, i.e., f = 0, the type I error rates were 0.008 and 0.002 for the proposed and CBS method, respectively. Permuting W1 and ε̂ gave very comparable results.
Next, for the presence of change points, the data sets were generated from a model created by Olshen et al. (2004) as follows:
where εi are i.i.d. N(0, σ2), n = 500, and the second term is a sinusoid trend component to make the simulated data set more realistic and challenging. The noise parameter σ was set to be 0.1 or 0.2, and the trend parameter a was chosen to be 0, 0.01 or 0.025, corresponding to no trend and local trend with long and short periods, respectively. There were seven segments along the chromosome. The means of log intensity ratios within segments were given by:
| i | 1–130 | 131–220 | 221–240 | 241–300 | 301–310 | 311–350 | 351–500 |
| f(i) | −0.40 | 0.08 | 1.20 | −0.50 | 0.30 | −0.70 | −0.20 |
An example of a simulated data set using the trend model is given in Web Figure 3. We found that the proposed method using permutation of W1 outperformed the CBS method in each of the no-trend, long- and short-period trend models (Table 3). The proposed method was robust to local trend in the sense that FDR, TPR and the number of exact detections did not appear to change with the trend parameter a. The CBS method tended to overestimate the number of change points in the presence of a local trend. The FDR increased with a and the number of exact change points detected decreased with a. However, permuting ε̂ was less powerful under the trend model because the default window size (0.1) over-corrected the trend. Therefore, under the normal (or near normal) situation, permuting W1 is recommended because it is robust to trend and does not require any tuning parameter as for the approach based on ε̂.
Table 3.
Summary of results under the trend model. σ =noise level, a =trend parameter. FDR=# false rejections/# total rejections, TPR= # true rejections/R, Exact=# data sets with correct locations and number of estimated change points. A significance level 0.01 was used.
| R̂ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| σ | a | Method | 4− | 5 | 6 | 7 | 8+ | FDR (%) | TPR(%) | #Exact |
| 0.1 | 0 | Permute W1 | 0 | 0 | 490 | 10 | 0 | 0.3 | 100 | 490 |
| Permute ε̂(0.10) | 2 | 100 | 398 | 0 | 0 | 0 | 96.5 | 398 | ||
| CBS | 0 | 0 | 472 | 11 | 17 | 1.2 | 100 | 472 | ||
| 0.01 | Permute W1 | 0 | 0 | 486 | 14 | 0 | 0.4 | 100 | 486 | |
| Permute ε̂(0.10) | 2 | 107 | 391 | 0 | 0 | 0 | 96.3 | 391 | ||
| CBS | 0 | 0 | 407 | 35 | 58 | 4.1 | 100 | 407 | ||
| 0.025 | Permute W1 | 0 | 0 | 487 | 13 | 0 | 0.4 | 100 | 487 | |
| Permute ε̂(0.10) | 2 | 72 | 426 | 0 | 0 | 0 | 97.4 | 426 | ||
| CBS | 0 | 0 | 394 | 22 | 84 | 5.1 | 100 | 394 | ||
| 0.2 | 0 | Permute W1 | 0 | 0 | 436 | 57 | 7 | 2.3 | 99.7 | 428 |
| Permute ε̂(0.10) | 0 | 85 | 410 | 5 | 0 | 0.8 | 96.6 | 405 | ||
| CBS | 0 | 0 | 450 | 34 | 16 | 2.3 | 99.5 | 439 | ||
| 0.01 | Permute W1 | 0 | 6 | 437 | 52 | 5 | 2.2 | 99.3 | 423 | |
| Permute ε̂(0.10) | 5 | 150 | 343 | 2 | 0 | 0.8 | 94.0 | 328 | ||
| CBS | 0 | 0 | 390 | 54 | 56 | 4.8 | 99.7 | 384 | ||
| 0.025 | Permute W1 | 0 | 7 | 432 | 54 | 7 | 2.2 | 99.4 | 422 | |
| Permute ε̂(0.10) | 2 | 75 | 413 | 10 | 0 | 0.5 | 97.1 | 408 | ||
| CBS | 0 | 0 | 376 | 42 | 82 | 6.0 | 99.5 | 364 |
4.2 Simulation under non-normal distribution
In this section we contrast the performance of the proposed method with the CBS method when ε was not normally distributed. We evaluated both approaches of permuting W1 and ε̂. We started with the complete null situation to examine the type I error rate and then followed with a power evaluation under an evenly-spaced change point model.
Under the complete null we generated the errors i.i.d. from t distributions with degrees of freedom (df) 1, 2, and 3, respectively. Web Table 1 shows the summary of simulation results. The CBS method performed consistently well and the family-wise type I error rates were below or close to the pre-specified level. The proposed method had the correct type I error rates when the null distribution was estimated by permuting ε̂. The null distribution estimated by permuting W1 underestimated the tail probabilities and yielded increased type I error rates as the df for t distribution decreased, i.e., tails get heavier. However, the type I error rates were already below the pre-specified level when the df > 3 (results not shown).
To investigate the power of our method, we generated chromosome profiles with different numbers of evenly-spaced change points and aberration width under a t distribution with 3 degrees of freedom. The function f(i) was set to be either 0 or 1 corresponding to no change or copy number gains. The number of change points (R) varied among 2, 4, and 6. The width of each aberration region (width) increased from 20, 40 to 80 markers. Both the multiscale and CBS method were robust to this non-normal distribution and comparable under all but one setting examined (Table 4). This scenario, in which CBS appears more powerful, has only one large aberration segment: width = 80 and R = 2. As expected, permuting W1 tended to overestimate the number of change points compared to permuting ε̂.
Table 4.
Summary of results for when ε is i.i.d. t(df=3). FDR=# false rejections/# total rejections, TPR= # true rejections/R, Exact=# data sets with correct locations and number of estimated change points. A significance level 0.01 was used.
| R | Method | ≤R−2 | R−1 | R | R+1 | ≥R+2 | FDR(%) | TPR(%) | #Exact |
|---|---|---|---|---|---|---|---|---|---|
| width=20 | |||||||||
| 2 | Permute W1 | 22 | 8 | 432 | 37 | 1 | 3.6 | 94.0 | 427 |
| Permute ε̂(0.10) | 24 | 17 | 451 | 7 | 1 | 2.2 | 92.4 | 446 | |
| CBS | 40 | 0 | 449 | 5 | 6 | 1.4 | 91.5 | 445 | |
| 4 | Permute W1 | 27 | 7 | 433 | 32 | 1 | 3.5 | 94.1 | 419 |
| Permute ε̂(0.10) | 28 | 14 | 444 | 13 | 1 | 2.8 | 93.6 | 430 | |
| CBS | 44 | 0 | 441 | 9 | 6 | 1.4 | 90.8 | 428 | |
| 6 | Permute W1 | 30 | 4 | 433 | 30 | 3 | 2.4 | 94.9 | 423 |
| Permute ε̂(0.10) | 26 | 7 | 434 | 29 | 4 | 2.8 | 95.4 | 423 | |
| CBS | 48 | 0 | 438 | 10 | 4 | 0.7 | 90.6 | 432 | |
| width=40 | |||||||||
| 2 | Permute W1 | 8 | 3 | 446 | 42 | 1 | 3.7 | 97.3 | 439 |
| Permute ε̂(0.10) | 10 | 7 | 474 | 8 | 1 | 1.6 | 96.4 | 466 | |
| CBS | 13 | 0 | 479 | 3 | 5 | 1.6 | 96.5 | 470 | |
| 4 | Permute W1 | 12 | 4 | 456 | 27 | 1 | 2.1 | 97.1 | 442 |
| Permute ε̂(0.10) | 12 | 0 | 473 | 14 | 1 | 1.8 | 97.2 | 458 | |
| CBS | 16 | 0 | 471 | 3 | 10 | 1.5 | 96.1 | 457 | |
| 6 | Permute W1 | 12 | 3 | 462 | 22 | 1 | 1.5 | 97.3 | 441 |
| Permute ε̂(0.10) | 12 | 2 | 467 | 18 | 1 | 1.5 | 97.6 | 446 | |
| CBS | 25 | 0 | 458 | 11 | 6 | 1.1 | 94.8 | 444 | |
| width=80 | |||||||||
| 2 | Permute W1 | 5 | 7 | 448 | 38 | 2 | 3.3 | 97.7 | 443 |
| Permute ε̂(0.10) | 8 | 7 | 475 | 9 | 1 | 1.2 | 97.2 | 470 | |
| CBS | 5 | 0 | 492 | 1 | 2 | 0.9 | 98.4 | 486 | |
| 4 | Permute W1 | 10 | 1 | 458 | 29 | 2 | 2.1 | 97.4 | 442 |
| Permute ε̂(0.10) | 9 | 5 | 473 | 12 | 1 | 1.6 | 97.4 | 456 | |
| CBS | 9 | 0 | 478 | 5 | 8 | 1.7 | 97.3 | 459 | |
| 6 | Permute W1 | 10 | 5 | 455 | 29 | 1 | 1.5 | 97.6 | 438 |
| Permute ε̂(0.10) | 9 | 3 | 465 | 22 | 1 | 1.5 | 97.8 | 447 | |
| CBS | 15 | 0 | 453 | 16 | 16 | 2.1 | 96.2 | 428 | |
We also examined a smaller window size, 0.05, and found that it was slightly more conservative and thus less powerful in detecting the change points than smoothing with window size 0.1 (results not shown). Based on the settings we examined here, a window size 0.10 for obtaining ε̂ appears to be a reasonable choice when ε is not normally distributed.
5. Discussion
We have proposed a non-parametric approach for detecting change points in genomic copy number data by seeking local changes that occur at multiple scales. We provide multiple comparison adjusted p-values for each potential change point. The p-values provide flexibility for investigators to call change points at their chosen level of significance. These p-values can be computed using re-sampling approaches by permuting either W1 or ε̂. Which approach to use will depend on the error distribution, which, unfortunately, is not usually known. In practice, we suggest to visually examine the residuals from the lowess smoothing to determine if the errors are roughly normal or have heavy tails. Under the normal or near normal situation, permuting W1 is recommended because it is robust to trend and does not require any tuning parameter as for permuting ε̂. For distributions with extremely heavy tails, permuting W1 yields more false positives. In this case, permuting ε̂ is more appropriate than permuting W1 and we recommend to use a smoothing window of width 0.1.
The proposed method performed well in most settings that we examined. It has the correct type I error rates under the null and is robust to background trend in the data and non-normal errors. However, because the test statistics are based on local variations the method has low power in detecting change points when the noise level is very high and the aberration region is narrow. This weakness may be overcome by improvements in array technologies, DNA extraction methods and increasing marker density. The CBS method, on the other hand, performs relatively well when the noise level is high and the aberration region is narrow. Like the proposed method, the CBS method also performs well when the error distribution is not normal. However, the CBS method tends to overestimate the number of change points when there exist multiple change points or when there is a background trend. This overestimation occurs even when the noise level is low, as shown in the Coriel cell line data and Illumina 1M SNP data.
All computations were done in R; the code is available from the authors upon request.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr. Lenora Loo for helpful discussions and Benjamin Ely for assembling the Illumina data. The authors also thank the referees and the editors for their many constructive comments and suggestions, which have improved the manuscript substantially. This work is supported in part by NIH grants R01 AG14358 (L.H.), R01 CA126205 (T.R.) and P01 CA053996 (L.H. and T.R.).
Footnotes
Supplementary materials
Web appendix, tables, and figures referenced in Sections 2, 3.2 and 4 are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
REFERENCES
- Bao P, Zhang L. Noise reduction for magnetic resonance images via adaptive multiscale products thresholding. IEEE Trans. Medical Imaging. 2003;22:1089–1099. doi: 10.1109/TMI.2003.816958. [DOI] [PubMed] [Google Scholar]
- Cheng C, Kimmel R, Neiman P, Zhao LP. Array rank order regression analysis for the detection of gene copy-number changes in human cancer. Genomics. 2003;82:122–129. doi: 10.1016/s0888-7543(03)00122-8. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Robust locally weighted regression and smoothing scatterplots. Journal of American Statistical Assocication. 1979;74 [Google Scholar]
- Cooper G, Zerr T, Kidd J, Eichler E, Nickerson D. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nature Genetics. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers PHC, Menezes RXd. Quantile smoothing of array CGH data. Bioinformatics. 2005;21:1146–1153. doi: 10.1093/bioinformatics/bti148. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders A, Pinkel D, Albertson D, Jain A. Hidden Markov models approach to the analysis of array CGH data. Journal of Multivariate Analysis. 2004;90:132–153. [Google Scholar]
- Ge Y, Dudoit S, Speed TP. Resampling-based multiple testing for microarray data analysis. Test. 2003;12:1–77. [Google Scholar]
- Guha S, Li Y, Neuberg D. Bayesian hidden markov modeling of arrasy cgh data. Journal of the American Statistical Association. 2008;13:485–497. doi: 10.1198/016214507000000923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson G, Hager J, Volik S, Hariono S, Wernick M, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nature Genetics. 2001;929:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Hsu L, Self SG, Grove D, Randolph T, Wang K, Delrow J, Loo L, Porter P. Denoising array-based comparative genomic hybridization data using wavelets. Biostatistics. 2005;6:211–226. doi: 10.1093/biostatistics/kxi004. [DOI] [PubMed] [Google Scholar]
- Huang T, Wu B, Lizardi P, Zhao H. Detection of DNA copy number alterations using penalized least squares regression. Bioinformatics. 2005;21:3811–3817. doi: 10.1093/bioinformatics/bti646. [DOI] [PubMed] [Google Scholar]
- Jong K, Marchiori E, van der Vaart A. Chromosomal breakpoint detection in array comparative genomic hybridization data. Applications of Evolutionary Computing: Evolutionary Computation and Bioinformatics. 2003;2611:54–65. [Google Scholar]
- Kidd J, Cooper G, Donahue W, Hayden H, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Xing H, Zhang N. Stochastic segmentation models for array-based comparative genomic hybridization data analysis. Biostatistics. 2008;9:290–307. doi: 10.1093/biostatistics/kxm031. [DOI] [PubMed] [Google Scholar]
- Lai W, Johnson MD, Kucherlapati R, Park P. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:37633770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallat S. A Wavelet Tour of Signal Processing. Academic Press; 1999. [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based dna copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Peiffer D, Le J, Steemers F, Chang W, et al. High resolution genomic profiling of chromosomal aberrations using infinium whole-genome genotyping. Genome Research. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival DB, Walden AT. Wavelet Methods for Time Series Analysis. Cambridge University Press; 2000. [Google Scholar]
- Picard F, Robin S, Lebarbier E, Daudin J. A segmentation/clustering model for the analysis of array CGH data. Biometrics. 2007;63:758–766. doi: 10.1111/j.1541-0420.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Albertson D. Array comparative genomic hybridization and its applications in cancer. Nature Genetics. 2005;37:S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- Pollack J, Sorlie T, Perou C, Rees C, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proceedings of the National Academy of Sciences. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler B, Swami A. Analysis of multiscale products for step detection and estimation. IEEE Trans. Inform. Theory. 1999;45:1043–1051. [Google Scholar]
- Snijders A, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G. Assembly of microarrays for genome-wide measurement of DNA copy numbers. Nature Genetics. 2001;29:263264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Wang P. Spatial smoothing and hot spot detection for CGH data using the fused lasso. Biostatistics. 2007;8:1–12. doi: 10.1093/biostatistics/kxm013. [DOI] [PubMed] [Google Scholar]
- Wang P, Kim Y, Pollack J, Narasimhan B, Tibshirani R. A method for calling gains and losses in array CGH data. Biostatistics. 2005;6:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]
- Westfall P, Young S. Resampling-based multiple testing: Examples and methods for p-value adjustment. John Wiley & Sons; 1993. [Google Scholar]
- Zhang NR, Siegmund DO. A modified bayes information criterion with applications to the analysis of comparative genomic hybridization data. Biometrics. 2007;63:22–32. doi: 10.1111/j.1541-0420.2006.00662.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.