Abstract
Cell growth and differentiation during developmental processes require the activation of many inducible genes. However, eukaryotic chromatin, which consists of DNA and histones, becomes a natural barrier impeding access to the functional transcription machinery. To break through the chromatin barrier, eukaryotic organisms have evolved the strategy of using Poly(ADP-ribose) polymerase 1 (PARP1) to modulate chromatin structure and initiate the steps leading to gene expression control. As a structural protein in chromatin, enzymatically silent PARP1 inhibits transcription by contributing to the condensation of chromatin, which creates a barrier against gene transcription. However, once activated by environmental stimuli and developmental signals, PARP1 can modify itself and other chromatin-associated proteins, thereby loosening chromatin to facilitate gene transcription. Here we discuss the roles of PARP1 in transcriptional control during development.
INTRODUCTION
Poly(ADP-ribose) polymerase 1 (PARP1) is a multifunctional nuclear protein created by eukaryotes to manage the structure and function of high order chromatin. The PARP1 protein, which is conserved among eukaryotes [1] except in yeast [2], utilizes NAD+ as substrate to synthesize poly(ADP-ribose) polymer (pADPr) with the resulting sizes varying from 2 to 200 ADP-ribose units [3], The mammalian genome contains additional PARP superfamily members, PARP2-PARP17 [4], while the Drosophila genome encodes only two PARP superfamily members: a single nuclear PARP1 and a single cytoplasmic PARP5 (Tankyrase) [5]. PARP1 can modify target proteins by essentially attaching a poly(ADP-ribose) (pADPr) chain to itself through Glu/Asp [6] and/or lysine residues [7] in its automodification domain (Figure 1A). It is this accumulation of pADPr which leads to local chromatin loosening [8, 9] and facilitates transcription by RNA polymerase (Pol2) (Figure 1B). Although automodified PARP1 (pADPr-PARP1) loses its enzymatic capabilities in this process, it gains the ability to bind proteins through conserved pADPr-binding domains in a non-covalent manner [10–12] to further modulate chromatin [13, 14] and regulate RNA maturation steps [15, 16]. An antagonist of PARP1, Poly(ADP-ribose) glycohydrolase (PARG), degrades the pADPr polymer and regulates the level of poly(ADP-ribosyl)ated proteins within chromatin and nucleoplasm [17, 18] (Figure 1B). In this review, we focus on recent studies of PARP1 functions under normal physiological condition and explain how nuclei can utilize the ability of PARP1 protein to modulate chromatin for transcription and splicing control during development.
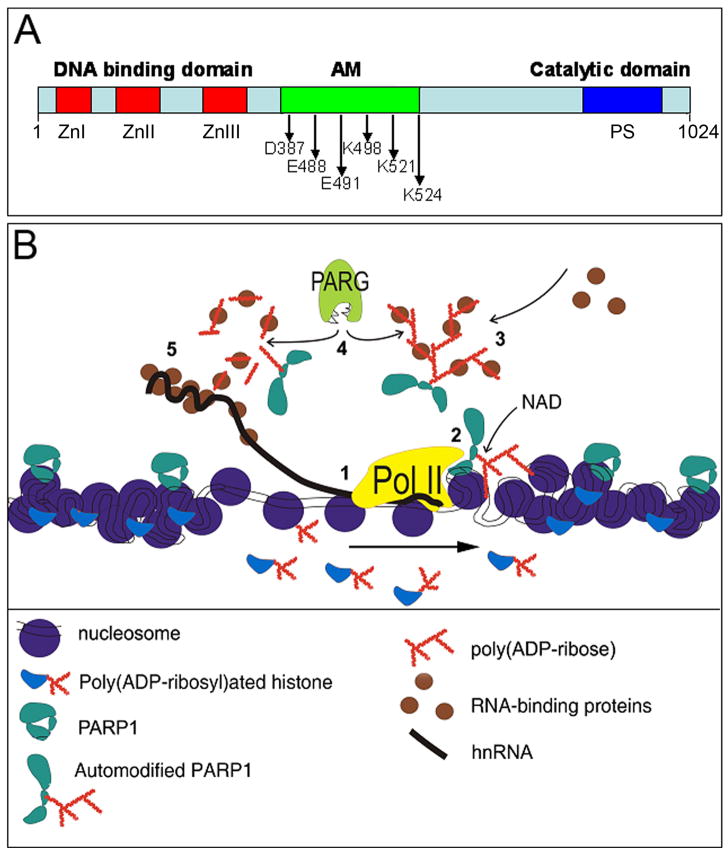
Figure 1. Schematic illustration showing PARP1 roles in chromatin gene expression control.
(A) The domain structure of the PARP1 protein. The PARP1 protein has the three functional domains: the DNA-binding domain in the N-terminus including three Zn-finger motifs (ZnI, ZnII, ZnIII); Automodification domain (AD) in the central region (374–524) having Glu/Asp and/or lysine residues as indicated for poly(ADP-ribosyl)ation; Catalytic domain in the C-terminus with PARP signature (PS), evolutionarily conserved PARP catalytic site. (B) PARP1 action for transcription control. Hypothetical order of events: 1. RNA polymerase 2 complex initiates transcription. 2. PARP1 is activated, destabilizes chromatin, and automodifies. 3. Automodified PARP1 recruits hnRNP protein from the nucleoplasm, creating high local concentration. 4. PARG proteins cleave pADPr and hnRNPs are released. 5. hnRNP proteins are utilized for hnRNP packaging, stabilization and splicing. Arrow indicates direction of transcription.
PARP1 as the chromatin protein for transcription inhibition
In steady state conditions, most PARP1 proteins are associated with chromatin and are accumulated in nucleoli [8, 9, 19]. Numerous studies suggest that PARP1 binds to the core histone proteins (H2A, H2B, H3 and H4) in the nucleosome [20, 21]. Specifically, the C-terminal domain of PARP1 preferentially interacts with H3 and H4, an event not mediated by DNA but negatively regulated by the N-terminal domain of PARP1 [20]. In contrast, PARP1 and H1 compete with each other for binding to the promoter regions of numerous genes [21]. As illustrated by in vivo and in vitro studies, extensive PARP1 binding to nucleosomes makes chromatin more dense and compact, thus preventing the transcription machinery from initiating gene transcription (Figure 2A). This phenomenon is variously supported in the literature. For example, adding recombinant PARP1 to purified chromatin in vitro changes the conformation of the chromatin from an open state to a more condensed state [22]. In vivo studies showed that PARP-1 is associated with both Drosophila and mouse Hsp70 genes before heat shock treatment when transcription is repressed [8, 23]. In addition, Drosophila PARP1 is required for silencing the transcription of the transposable elements and endogenous retroviruses in heterochromatin [5, 24]. When mouse PARP-1 was enriched, it was found to be necessary for stable gene silencing on the inactive X chromosome [25]. PARP1 was also localized to other constitutive heterochromatin regions, including the centromeres [26] and telomeres [27]. The heterochromatin protein HP1α also interacted with PARP1 and PARP2 on pericentric heterochromatin in mammalian cells [28]. Therefore, cumulative evidence suggests that PARP1 is an essential component of chromatin for the repression of transcription locally in euchromatin and more globally in the heterochromatin when its enzymatic activity is not activated (Figure 2A).
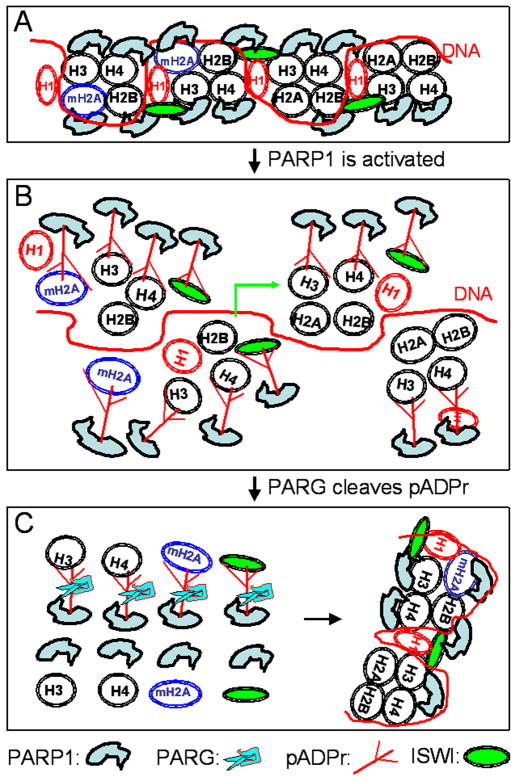
Figure 2. Transcription control by PARP1 through chromatin modulation.
(A) Transcription inhibition by PARP1 binding to H3 and H4 to compact chromatin. Histone variant, mH2A1, is enriched in the promoter region, which contains such genes as human hsp70, to inhibit PARP1 activity. Chromatin remodeling factor ISWI is also associated with PARP1 in chromatin. (B) Transcription induction by PARP1 activation to destabilize (or “loosen”) chromatin. Activated PARP1 modifies itself, histones (H3, H4) and its variants (mH2A1) and chromatin remodeling factors (ISWI) to destabilize nucleosome structure for transcription induction. (C) Chromatin reassembly by PARG after transcription induction. PARG degrades the pADPr polymer and releases the poly(ADP-ribosy)ated histones and chromatin remodeling factors (ISWI) into the free forms, which will be reassembled into chromatin. Note: histone tetramers is drawn in the nucelosome for simplification instead of histone octamer.
Interaction of activated PARP1 with histones for chromatin modulation
Many environmental and developmental signals can activate PARP1 during an organism’s development. Because poly (ADP-ribose) is highly negatively charged and has a high binding affinity for its associated proteins [29], automodifed PARP1 and subsequent interactions with histones and their variants dramatically change the structure of chromatin from a condensed state to a less concentrated (i.e., “loose”) state which facilitates gene transcription (Figure 2). Chromatin remodeling by PARP1 activation can best be illustrated in the hrp70 loci in Drosophila. Upon heat shock, PARP1 was potently activated, and pADPr signals were detected in the heat-shock induced puffs [8], which, in turn, resulted in PARP1 dissociation from the puffs [8]. This appears to be the main mechanism for the rapid loss of nucleosomes covering a larger region around the hrp70 genes upon heat shock [30]. Loss of nucleosomes induced by heat shock may result from automodified PARP1 dissociation from H3 and H4, which are binding sites for inactive PARP1 [20], further destabilizing nucleosome structure (Figure 2B).
Automodified PARP1 also acts as a platform to bind with histone variants for modulating chromatin structure (Figure 2). The histone variant macroH2A1 (mH2A1) has a homologous histone domain for assembly into the nucleosome and a unique macrodomain for interaction with other chromatin modifiers [31]. mH2A1 is enriched on the inactive X chromosome and many developmental genes on autosomes for repressing gene transcription by making chromatin more compact [32]. A dynamic interaction between mH2A1 and PARP1 appears to exist in the organism to modulate chromatin structure. On the one hand, mH2A1 inhibits PARP1 activity both in the promoter of the hsp70 gene [23] and on the inactive X chromosome in mice, which leads to transcription inhibition [25] (Figure 2A). On the other hand, once PARP1 is activated by, for example, DNA damage, automodifed PARP1 binds to mH2A1.1 through its macrodomain, thereby recruiting mH2A1.1 to PARP1 activation sites for chromatin transient condensation [13]. However, mH2A1 is dissociated from the promoter of the hsp70 gene upon heat shock when PARP1 is activated [23]. Thus, for developmentally regulated genes, it is more likely that poly(ADP-ribosyl)ation of mH2A facilitates the release of mH2A1.1 from repressive chromatin regions for transcription activation (Figure 2B).
Poly(ADP-ribosyl)ation of chromatin-remodeling factors for chromatin modulation
PARP1 can also modify several chromatin-remodeling factors, including Spt16 in the FACT (facilitates chromatin transcription) complex [33, 34] and the nucleosome remodeling ATPases, ISWI [35] and ALC1(amplified in liver cancer 1) [14, 36]. The FACT complex, a heterodimer of hSpt16 and SSRP1, is associated with the nucleosome and facilitates transcription elongation by removing one H2A-H2B dimer to enable the passage of pol II through the chromatin [37]. Upon DNA damage, poly(ADP-ribosyl)ation of Spt16 disrupts the association of FACT with the nucleosome and inhibits FACT-mediated exchange of histone variant H2AX with canonical H2A [33, 34]. In addition, ATP-dependent nucleosome remodeling complexes, including ISWI and ALC1, modulate chromatin structure by nucelosome sliding [14, 38]. An unbiased genetic screen showed that the Parp1 and Parg mutations enhanced the rough-eye phenotype caused by eye-specific expression of the dominant negative allele ISWIk159R, suggesting the genetic interaction between ISWI and PARP1 or PARG genes [35, 39]. Biochemical assays showed that ISWI can be poly(ADP-ribosyl)ated in vitro and that poly(ADP-ribosyl)ated ISWI can be detected in vivo (Figure 2B). It appears that poly(ADP-ribosyl)ation inhibits the ATPase activity of ISWI and further reduces its binding affinity for nucleosomes [35]. It has been shown in vivo that ISWI can promote histone H1 binding to chromatin and thus help to compact it [40]. However, since H1 also competes with PARP1 for binding to the gene promoters [21], poly(ADP-ribosyl)ation of ISWI may actually counteract H1 binding by relaxing chromatin after PARP1 activation (Figure 2B). In addition, two independent studies suggested that pADPr can bind with ALC1 (amplified in liver cancer 1) through its macrodomain by an in vitro binding assay [14, 36]. In contrast to the inhibition of ISWI ATPase activity and binding affinity for nucleosomes by poly(ADP-ribosyl)ation [35], PARP1-mediated pADPr binding with ALC1 strongly stimulated its ATPase and nucleosome-sliding activities and helped to recruit ALC1 to DNA damage sites, possibly for the purpose of relaxing chromatin [14, 36]. Notwithstanding these findings, the mechanism underlying the stimulation of ALC1 activity by automodified PARP is not entirely understood. However, since the nucleosome-sliding ability of ALC1 is dependent on H4 tail, we speculate that destabilization of mononucleosomes by automodified PARP may expose the histone H4 tail, thus allowing interaction with ALC1 for its activity in vivo [14]. Therefore, besides histone and its variants, it is clear that activated PARP1 also can interact with, and modulate the activities of, a number of nucleosome-remodeling factors to further destabilize chromatin and facilitate transcription activation (Figure 2B).
Poly(ADP-ribosyl)ation of the splicing proteins for splicing regulation
In addition to its direct effects on chromatin and transcription, as described above, PARP1 also mediates the follow-up steps of gene expression via regulation of proteins involved in RNA processing. Alternative splicing is used extensively to produce the different mRNA isoforms of a gene to increase the complexity of the transcriptome in higher eukaryotic genomes. It is generally believed that two groups of RNA-binding proteins, hnRNPs and serine-arginine-rich (SR) splicing factor, regulate alternative splicing by binding with exonic and intronic splicing silencers (ESEs and ISEs) and enhancers (ESSs and ISSs), respectively [41]. Our studies and those of others suggest that poly(ADP-ribose) also binds with hnRNPs and SR protein to regulate their functions [15, 16, 42] (Figure 3). A proteomic approach was used to identify hnRNPs as the most abundant pADPr-binding proteins through a conserved motif localized between two RNA-binding domains in human HeLa cells [42]. Using mass spectrometry analysis, Drosophila Hrp38, a homolog of hnRNP A1, was also found to be associated with PARP1 protein [20]. In addition, Drosophila PARP1 overexpression and PARG loss-of-function both result in increased amounts of hnRNPs (hrp38 and hrp40) bound to pADPr in vivo, suggesting that PARP and PARG activities can regulate pADPr binding to hnRNPs [15]. Interestingly, heat-shock treatment, which activates PARP1, also upregulates the ability of pADPr binding to hnRNPs and further causes the dissociation of hnRNPs from most of the transcripts in the nucleus. As a consequence, pADPr binding to hnRNPs inhibits the RNA-binding ability of hnRNPs and further modulates the alternative splicing pathway [15] (Figure 3A). It was also found that pADPr can bind with the alternative splicing factor/splicing factor 2 (ASF/SF2, a prototypical SR protein) through two functional domains (RRM1 and RS) [16].
Figure 3. Splicing regulation by poly(ADP-ribosyl)ation of the splicing proteins.
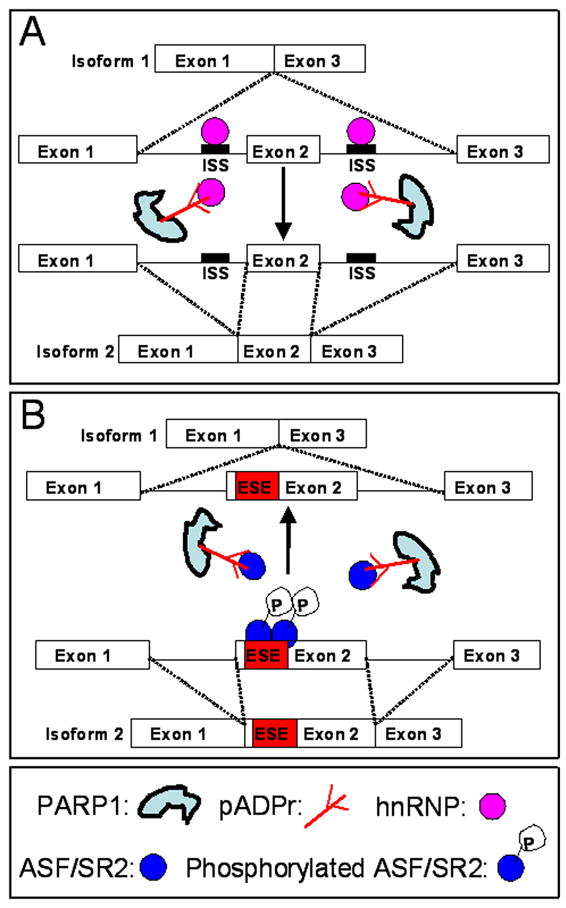
(A) Splicing enhancement by poly(ADP-ribosyl)ation of hnRNP A1. HnRNP A1, as the splicing repressor, binds to ISS (intronic splicing silencer) to inhibit the alternative splicing of exon 2 and thereby produce isoform 1. Poly(ADP-ribosyl)ation of hnRNP A1 by PARP1 causes the dissociation of hnRNP A1 from ISS, enhancing the splicing of exon 2 to produce isoform 2. (B) Splicing inhibition by poly(ADP-ribosyl)ation of ASF/SR2. Phosphorylated ASF/SR2 binds to ESE (exonic splicing enhancer) in order to promote the alternative splicing of exon 2 and thereby produce isoform 2. Poly(ADP-ribosyl)ation of ASF/SR2 by PARP1 inhibits phosphorylation of ASF/SR2 and may cause the dissociation of ASF/SR2 from ESE, which inhibits the splicing of exon 2 to produce isoform 1.
ASF/SF2 has an SR-rich C-terminal domain where serine residues are often phosphorylated by multiple kinases, including DNA Topoisomerase I, to regulate splicing [43]. It appears that pADPr inhibits ASF/SF2 phosphorylation by blocking the kinase activity of DNA [16]. Therefore, pADPr binding to SR proteins may also regulate alternative splicing by modulating phosphorylation of SR protein or directly influencing the affinity for RNA binding (Figure 3B). Indeed, an in silico study also revealed that mammalian Parp-1 knockout cells have an increased incidence of alternative splicing within a subset of inflammatory response genes [16]. Considering that transcription and splicing are closely coupled [44], poly(ADP-ribosyl)ation of the splicing proteins (hnRNPs and SR protein) by locally activated PARP1 may regulate alternative splicing pathway in a the spatial- and temporal-specific manner.
PARP1 controls developmental processes
Drosophila PARP1 loss-of-function has caused larval lethality [5], and mouse PARP-1 and PARP-2 double knockout mice died at the early embryonic stages [45], suggesting that poly(ADP-ribosyl)ation is essential for normal development. Although PARP1 is constitutively expressed, its enzyme activity is developmentally regulated. For example, the maximal accumulation of pADPr was observed at the prepupal stage in Drosophila [46]. Both exogenous stimuli, such as heat shock [8], and endogenous signals, such as hormone secretion [47], can induce PARP1 activation during development. Therefore, chromatin modulation by activated PARP1 regulates the transcription and splicing of genes in different cell types and at different developmental stages.
PARP1 controls metamorphosis in Drosophila
The developmental roles of PARP1 were illustrated in the observation that PARP1 enzymatic activity is required for chromatin loosening on ecdysone-inducible loci (E74 and E75) in Drosophila [8]. At the end of the wandering third larval stage in Drosophila, a pulse of ecdysone triggers puparium formation and the onset of metamorphosis by inducing the expression of E74, E75 and BR-C genes [48]. The transcriptional activation is achieved by ecdysone binding to a heterodimer of two nuclear receptors (EcR/USP), which further acts on the ecdysone response element of the induced genes [49]. PARP1 protein is present in the region surrounding the E74-75B loci, but high levels of poly(ADP-ribose) only appear after the inducible expression of the E74 and E75 genes, suggesting that PARP1 is activated for the transcription of ecdysone-inducible genes [8]. It appears that PARP1 interacts with the ecdysone receptor in vivo and is associated with the promoter region of one ecdysone-inducible gene (hsp27) [50]. In addition, Drosophila nucleosome remodeling factor (NURF)-specific subunit NURF301 is required for transactivation of the ecdysone-inducible genes [51]. A recent study showed that chromatin structure in the upstream region of the E75 gene exhibits a dramatic change, including H3 and nucleosome depletion upon ecdysone induction [52]. Therefore, by the interaction with histones and NURFs, activated PARP1 may destabilize nucleosome structure in the chromatin region of ecdysone-inducible genes, which, in turn, facilitates ligand-EcR/USP complex binding to EcR elements of ecdysone-inducible genes for transactivation.
PARPs in germline development
PARPs also play roles in germline development, including oogenesis and spermiogenesis. During meiosis, it appears that PARP1 had dynamic localization patterns in mouse oocytes, which correlates with transcription state [53]. PARP1 null oocytes showed meiotic defects, including persistent H2AX phosphorylation, suggesting a role of PARP1 for chromatin modification during ooctye maturation [53]. Because of the redundant function of PARP1 with PARP2, PARP1 null female mice are fertile [53], but PARP2 null male mice have lower fertility as a result of severe defects of spermiogenesis [54]. Interestingly, PARP2 null spermatocytes exhibited defective meiotic sex chromosome inactivation (MSCI) shown by retaining RNA polymerase II on the XY body and upregulation of X- and Y-linked gene expressions, suggesting that PARP2 is involved in the maintenance of MSCI [54]. Indeed, PARP2 and macroH2A1 are colocalized on the XY body [54]. In fact, it was found that mouse PARP-1 is enriched and is required for gene silencing on the inactive X chromosome where its enzymatic activity is inhibited by macroH2A1[25]. Thus, it will be interesting to further investigate the roles of PARP1 and PARP2 for the establishment of X chromosome inactivation.
PARP1 in cell differentiation
A number of studies have also demonstrated that PARP1 and poly(ADP-ribosyl)ation are involved in cell differentiation. After injection into nude mice, parp−/− embryonic stem (ES) cells can differentiate to form teratocarcinoma-like tumors with the characteristics of trophoblast giant cells, suggesting that PARP1 may inhibit ES cell differentiation into trophoectodermal cells in the wild type [55]. However, a recent study showed that PARP1, whose activity is upregulated during ES cell differentiation, poly(ADP-ribosyl)ates the transcription factor Sox2 to control the expression level of fibroblast growth factor 4 (FGF4) [56]. PARP1, PARP2 and their activities are also required for the differentiation of mouse embryonic carcinoma F9 cells into endodermal cells. Both PARP1 and PARP2 can poly(ADP-ribosyl)ate one isoform of heterochromatin protein 1(HP1α) to recruit the transcriptional intermediary factor (TIFIβ) to pericentric heterochromatin for the expression of the endoderm-specific gene during endodermal differentiation [28]. Therefore, PARP1 also plays important roles in cell differentiation.
SUMMARY
PARP1 performs a dual function in transcription control. First, as the essential component of chromatin, PARP1 represses transcription locally in euchromatin and more globally in the heterochromatin. Second, once activated by developmental cues, automodified PARP1 interacts with histone H3 and H4 and their variants, such as macroH2A1, to destabilize chromatin structure in order to allow the transcription machinery to operate. In addition, activated PARP1 also poly(ADP-ribosyl)ates several chromatin-remodeling factors to further modulate chromatin. As a coupled reaction with transcription, splicing is also regulated by PARP1 through poly(ADP-ribosyl)ation of the splicing proteins. The functions of PARP1 in transcription and splicing control are reflected in multiple roles during development, including metamorphosis, gametogenesis and cell differentiation. In the future, more effort should be directed to investigate the developmental roles of PARP1 in the context of chromatin modulation.
Acknowledgments
We thank Dr. Hua-Ying Fan for her critical reading of the manuscript and valuable comments. The expenses were defrayed by a grant from the National Institutes of Health (R01DK082623) (to A.V.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* - SPECIAL INTEREST
** - OUTSTANDING INTEREST
- 1.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6(1):139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins E, Sun D, Nguyen A, Tulac S, Francesco M, Tavana H, Nguyen H, Tugendreich S, Barthmaier P, Couto J, et al. Novel Inhibitors of Poly(ADP-ribose) Polymerase/PARP1 and PARP2 Identified Using a Cell-based Screen in Yeast. Cancer Research. 2001;61(10):4175–4183. [PubMed] [Google Scholar]
- 3.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 4.Amé JC, Spenlehauer C, Murcia GD. The PARP superfamily. BioEssays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 5.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes & Development. 2002;16(16):2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao ZP, Gao P, Liu HW. Identification of the ADP-Ribosylation Sites in the PARP-Automodification Domain: Analysis and Implications. Journal of the American Chemical Society. 2009;131(40):14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 7.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucl Acids Res. 2009;37(11):3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8**.Tulin A, Spradling AC. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. This paper first reported that PARP1 protein controls chromatin loosening and transcription. [DOI] [PubMed] [Google Scholar]
- 9.Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119(6):803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) Binds to Specific Domains inDNA Damage Checkpoint Proteins. Journal of Biological Chemistry. 2000;275(52):40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 11.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 13**.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16(9):923–929. doi: 10.1038/nsmb.1664. This paper described that automodifed PARP1 binds with histone macroH2A1.1 through its macrodomain as a pADPr-binding module. Once PARP1 is activated by DNA damage, pADPr binding to mH2A1.1 results in recruiting mH2A1.1 to PARP1 activation sites for chromatin rearrangement. A crystal structural analysis revealed that ADP-ribose binding causes the conformational changes in the macrodomin of mH2A1.1. [DOI] [PubMed] [Google Scholar]
- 14**.Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science. 2009;325(5945):1240–1243. doi: 10.1126/science.1177321. This paper showed that pADPr can bind with ALC1 (amplified in liver cancer 1) through its macrodomain. Automodified PARP1 binding with ALC1 strongly stimulated its ATPase and nucleosome-sliding activities, and helps recruiting ALC1 to DNA damage sites for chromatin modulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucl Acids Res. 2009;37(11):3501–3513. doi: 10.1093/nar/gkp218. This paper described that automodifed PARP1 binds with two hnRNPs (hrp38 and hrp40), which is regulated by endogenous PARP1 and PARG activity. Heat-shock treatment, which activates PARP1, also upregulates the ability of pADPr binding to hnRNPs and further causes the dissociation of hnRNPs from most of the transcripts in the nucleus. RNA EMAS showed that pADPr binding to hnRNPs inhibits the RNA-binding ability of hnRNPs, which further modulates the alternative splicing pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing actor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. Journal of Biological Chemistry. 2008;283(29):19991–19998. doi: 10.1074/jbc.M709495200. This paper showed that pADPr can bind with the alternative splicing factor/splicing factor 2 (ASF/SF2, a prototypical SR protein) through two functional domains (RRM1 and RS). pADPr inhibits ASF/SF2 phosphorylation by blocking the kinase activity of DNA. The in-silico study also revealed that mammalian Parp-1 knockout cells have an increased incidence of alternative splicing within a subset of inflammatory response genes. [DOI] [PubMed] [Google Scholar]
- 17.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. PNAS. 2004;101(1):82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila Poly(ADP-Ribose) Glycohydrolase Mediates Chromatin Structure and SIR2-Dependent Silencing. Genetics. 2006;172(1):363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118(1):211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 20.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal Core Histones Mediate Dynamic regulation of Poly(ADP-ribose) Polymerase 1 Protein Binding to Chromatin and Induction of Its enzymatic Activity. Journal of Biological Chemistry. 2007;282(44):32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 21.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal Binding of PARP-1 and Histone H1 at Promoters Specifies Transcriptional Outcomes. Science. 2008;319(5864):819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 22.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA Binding and Catalytic Domains of Poly(ADP-Ribose) Polymerase 1 Cooperate in the Regulation of Chromatin Structure and Transcription. Mol Cell Biol. 2007;27(21):7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes & Development. 2006;20(23):3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Kotova E, Jarnik M, Tulin AV. Uncoupling of the transactivation and transrepression functions of PARP1 protein. PNAS. 2010 doi: 10.1073/pnas.0914152107. In press. This paper described that a Drosophila mutant expressing a short isoform of PARP1 protein without Zn-finger 1, abolishes localization of PARP1 protein in heterochromatin and leads to the desilencing of retrotransposable elements. This observation demonstrates that the first zinc finger of PARP1 is required for PARP localization to heterochromatin, where it regulates transcriptional silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nusinow DA, Hernández-Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) Polymerase 1 Is Inhibited by a Histone H2A Variant, MacroH2A, and Contributes to Silencing of the Inactive X Chromosome. Journal of Biological Chemistry. 2007;282(17):12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- 26.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of Poly(ADP-Ribose) Polymerase 1 and Poly(ADP-Ribosyl)ation in Regulation of Centrosome Function. Mol Cell Biol. 2003;23(7):2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beneke S, Cohausz O, Malanga M, Boukamp P, Althaus F, Bürkle A. Rapid regulation of telomere length is mediated by poly(ADP-ribose) polymerase-1. Nucl Acids Res. 2008;36(19):6309–6317. doi: 10.1093/nar/gkn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quénet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F, Quenet D. The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1{beta} and the heterochromatin protein HP1{alpha} FASEB J. 2008;22(11):3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- 29.Fahrer J, Kranaster R, Altmeyer M, Marx A, Bürkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucl Acids Res. 2007;35(21):e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Petesch SJ, Lis JT. Rapid, Transcription-Independent Loss of Nucleosomes over a Large Chromatin Domain at Hsp70 Loci. Cell. 2008;134(1):74–84. doi: 10.1016/j.cell.2008.05.029. This paper showed that heat shock results in rapid loss of nucleosomes covering a larger region around the hrp70 genes. Consistent with the observation in [8], PARP1 RNAi or inhibition of PARP1 activity by PARP1 inhibitor resulted in a NHS (not heat shock) nucleosome profile around the hsp70 gene upon heat shock, suggesting that PARP1 is required for the loss of nucleosomes at Hsp70 after heat shock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural Characterization of the Histone Variant macroH2A. Mol Cell Biol. 2005;25(17):7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buschbeck M, Uribesalgo I, Wibowo I, Rué P, Martin D, Gutierrez A, Morey L, Guigó R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol. 2009;16(10):1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- 33.Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucl Acids Res. 2006;34(8):2398–2407. doi: 10.1093/nar/gkl241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-Mediated exchange of Histone Variant H2AX Regulated by Phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30(1):86–97. doi: 10.1016/j.molcel.2008.02.029. Together with [33], this paper described that PARP1 can poly(ADP-ribosl)ate Spt16 in the FACT (facilitates chromatin transcription) complex. Upon DNA damage, poly(ADP-ribosyl)ation of Spt16 disrupts the association of FACT with the nuclosome and inhibits FACT-mediated exchange of histone variant H2AX with canonical H2A. [DOI] [PubMed] [Google Scholar]
- 35**.Sala A, La Rocca G, Burgio G, Kotova E, Di Gesù D, Collesano M, Ingrassia AM, Tulin AV, Corona DF. The Nucleosome-Remodeling ATPase ISWI Is Regulated by Poly-ADP-Ribosylation. PLoS Biol. 2008;6(10):e252. doi: 10.1371/journal.pbio.0060252. This paper showed that PARP1 can modify ATP-dependent nucleosome remodeling factor ISWI by in-vitro and in vivo assay. Poly(ADP-ribosyl)ation of ISWI inhibits its ATPase activity and further reduces its binding affinity for nucleosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. PNAS. 2009;106(33):13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT Facilitates Transcription-Dependent Nucleosome Alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 38.Corona DFV, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 2004;1677(1–3):113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 39.Burgio G, La Rocca G, Sala A, Arancio W, Di Gesù D, Collesano M, Sperling AS, Armstrong JA, Van Heeringen Logie C, et al. Genetic Identification of a Network of Factors that Functionally Interact with the Nucleosome Remodeling ATPase ISWI. PLoS Genet. 2008;4(6):e1000089. doi: 10.1371/journal.pgen.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW. ISWI Regulates Higher-Order Chromatin Structure and Histone H1 Assembly In Vivo. PLoS Biol. 2007;5(9):e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matlin AJ, Clark F, Smith CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 42.Gagné JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochemical Journal. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamm S. Regulation of Alternative Splicing by Reversible Protein Phosphorylation. Journal of Biological Chemistry. 2008;283(3):1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 44.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Current Opinion in Cell Biology. 2005;17(3):251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Ménissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich A, LeMeur M, Sabatier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22(9):2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotova E, Jarnik M, Tulin AV. Poly (ADP-Ribose) Polymerase 1 Is Required for Protein Localization to Cajal Body. PLoS Genet. 2009;5(2):e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-Independent PARP-1 Activation by Phosphorylated ERK2 Increases Elk1 Activity: A Link to Histone Acetylation. Mol Cell. 2007;25(2):297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Fletcher JC, Burtis KC, Hogness DS, Thummel CS. The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development. 1995;121(5):1455–1465. doi: 10.1242/dev.121.5.1455. [DOI] [PubMed] [Google Scholar]
- 49.Thummel CS, Chory J. Steroid signaling in plants and insects: common themes, different pathways. Genes & Development. 2002;16(24):3113–29. doi: 10.1101/gad.1042102. [DOI] [PubMed] [Google Scholar]
- 50.Sawatsubashi S, Maki A, Ito S, Shirode Y, Suzuki E, Zhao Y, Yamagata K, Kouzmenko A, Takeyama K, Kato S. Ecdysone receptor-dependent gene regulation mediates histone poly(ADP-ribosyl)ation. Biochemical and Biophysical Research Communications. 2004;320(1):268–272. doi: 10.1016/j.bbrc.2004.05.157. [DOI] [PubMed] [Google Scholar]
- 51.Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes & Development. 2005;19(21):2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernardo TJ, Dubrovskaya VA, Jannat H, Maughan B, Dubrovsky EB. Hormonal Regulation of the E75 Gene in Drosophila: Identifying Functional Regulatory Elements through Computational and Biological Analysis. Journal of Molecular Biology. 2009;387(4):794–808. doi: 10.1016/j.jmb.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 53*.Yang F, Baumann C, Fuente DL. Persistence of histone H2AX phosphorylation after meiotic chromosome synapsis and abnormal centromere cohesion in poly (ADP-ribose) polymerase (Parp-1) null oocytes. Developmental Biology. 2009;331(2):326–338. doi: 10.1016/j.ydbio.2009.05.550. This paper showed that PARP1 has dynamic localization patterns in mouse oocytes during meiosis which correlate with their transcription state. PARP1 null oocytes showed meiotic defects, including persistent H2AX phosphorylation, suggesting a role of PARP1 for chromatin modification during ooctye maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Ménissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. PNAS. 2006;103(40):14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hemberger M, Nozaki T, Winterhager E, Yamamoto H, Nakagama H, Kamada N, Suzuki H, Ohta T, Ohki M, Masutani M, Cross JC. Parp1-deficiency induces differentiation of ES cells into trophoblast derivatives. Dev Biol. 2003;257(2):371–381. doi: 10.1016/s0012-1606(03)00097-6. [DOI] [PubMed] [Google Scholar]
- 56*.Gao F, Kwon SW, Zhao Y, Jin Y. PARP1 Poly(ADP-ribosyl)ates Sox2 to Control Sox2 Protein Levels and FGF4 Expression during Embryonic Stem Cell Differentiation. Journal of Biological Chemistry. 2009;284(33):22263–22273. doi: 10.1074/jbc.M109.033118. This paper describes that PARP1 poly(ADP-ribosy)lates the transcription factor Sox2 to control the expression level of fibroblast growth factor 4 (FGF4) during ES cell differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]