Abstract
The X-ray crystal structure of recombinant isoprene synthase from grey poplar leaves (Populus x canescens) (PcISPS) has been determined at 2.7 Å resolution, and the structure of its complex with three Mg2+ ions and the unreactive substrate analogue dimethylallyl-S-thiolodiphosphate has been determined at 2.8 Å resolution. Analysis of these structures suggests that the generation of isoprene from substrate dimethylallyl diphosphate occurs via a syn-periplanar elimination mechanism in which the diphosphate leaving group serves as a general base. This chemical mechanism is responsible for the annual atmospheric emission of 100 billion kilograms of isoprene by terrestrial plant life. Importantly, the PcISPS structure promises to guide future protein engineering studies potentially leading to hydrocarbon fuels and products that do not rely on traditional petrochemical sources.
Keywords: protein crystallography, terpenoid biosynthesis, hemiterpene synthase, enzyme mechanism
Introduction
The worldwide atmospheric emission of the hydrocarbon isoprene from natural sources is ~100 Tg/year (100 billion kg/year).1 It is sometimes not fully appreciated that plant metabolism generates more atmospheric hydrocarbon than human activity.2–4 Isoprene emission by woody plants was first discovered by Sanadze,5 and since then myriad isoprene-emitting plant species have been identified.6 In the Blue Mountains of Australia and the Blue Ridge Mountains of the US, eponymous blue hazes result from Rayleigh or Tyndall scattering of biogenic isoprene and monoterpenes released into the atmosphere; interestingly, such blue hazes were also described in the Tuscan countryside in the notebooks of Leonardo Da Vinci.7
The reasons for plant isoprene generation are only partly understood, since at first glance it seems metabolically futile to return 2–5% of photosynthetically fixed carbon to the atmosphere.8 There is growing evidence that isoprene emission protects against environmental stresses, such as transiently high temperatures9–11 and oxidative damage.12, 13 Computational studies suggest that isoprene stabilizes lipid membranes against thermally induced phase transitions.14 Isoprene also influences direct15 and indirect16 defense mechanisms of herbivore-infested plants. However, regardless of its particular biological or ecological function, biogenic isoprene emission can lead to adverse consequences for atmospheric chemistry. For example, isoprene oxidation in the troposphere leads to increased concentrations of ozone, organic nitrates, organic acids, and secondary organic aerosol formation.17, 18
Plant isoprene biosynthesis represents the archetype of “green” chemistry in fuel science and materials science, given that this biosynthetic pathway enables atmospheric CO2 fixation for the generation of isoprenoid-based fuels otherwise derived from petrochemical refinement.19, 20 Isoprene is also a precursor in the generation of petrochemical-derived polymers, and plant isoprene biosynthetic pathways are currently being harnessed to address this industrial demand. For example, given that the global market for biotechnologically-based isoprene is expected to rise to ~109 kg/year, Genencor and Goodyear recently forged a collaboration to use fermentation technology for the generation of isoprene utilized in petroleum-based rubber and styrene-based elastomers.21
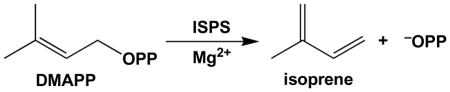
Isoprene synthase (ISPS) is the enzyme responsible for the worldwide generation of isoprene in nature and biotechnology.22–24 ISPS is a Mg2+-requiring terpenoid synthase that catalyzes the elimination of inorganic pyrophosphate from dimethylallyl diphosphate (DMAPP) to yield isoprene and inorganic pyrophosphate (−OPP):
The enzyme from the grey poplar hybrid Populus x canescens (syn. Populus tremula x P. alba) lacking the putative N-terminal plastidial targeting sequence has been cloned and expressed with an N-terminal histidine tag in Escherichia coli and is a 64 kD monomer exhibiting highest amino acid sequence identities (~40%) with plant monoterpene cyclases such as (4S)-limonene synthase (LMNS) and (+)-bornyl diphosphate synthase (BPPS).23, 25 Here, the crystal structure of native PcISPS has been determined at 2.7 Å resolution and the structure of its complex with the unreactive substrate analogue, dimethylallyl-S-thiolodiphosphate (DMASPP, in which the prenyl phosphoester oxygen atom is replaced with a sulfur atom) and three Mg2+ ions has been determined at 2.8 Å resolution.
Results and Discussion
PcISPS structure and metal binding motifs
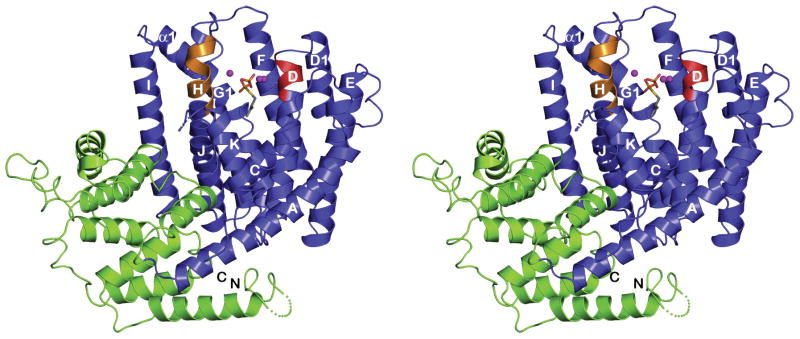
The PcISPS structure is comprised of two α-helical domains (Figure 1). The N-terminal domain adopts an α-barrel class II terpenoid synthase fold but has no known catalytic activity; the C-terminal domain contains the active site and adopts the α-helical class I terpenoid synthase fold first observed in farnesyl diphosphate synthase26 and subsequently observed in monoterpene and sesquiterpene cyclases.27–29 There is compelling biochemical evidence that this fold is also found in enzymes that catalyze all four fundamental isoprenoid coupling reactions of chain elongation, branching, cyclopropanation, and cyclobutanation.30 Thus, the class I terpenoid synthase fold is particularly adept in the evolution of templates for diverse isoprenoid biosynthetic reactions. Parenthetically, we note that a 3.05 Å resolution structure of native Populus tremuloides ISPS (98.5% amino acid sequence identity) has been recently described in a patent application31 and exhibits a homologous structure.
Figure 1.
The C-terminal catalytic domain of PcISPS adopts the α-helical class I terpenoid synthase fold (blue), and the noncatalytic N-terminal domain adopts an α-helical fold similar to that of a class II terpenoid synthase (green). In the catalytic domain, the D345DXXD and N489DXXSXXXE metal binding motifs are red and orange, respectively. Disordered polypeptide segments are indicated by dotted lines. The active site is indicated by the binding of DMASPP (stick figure) and three Mg2+ ions (magenta spheres).
In the mechanism of isoprene biosynthesis, the departure of the DMAPP diphosphate group is triggered by metal ions such as Mg2+ or Mn2+.24 The PcISPS structure reveals conservation of metal-binding motifs (the “aspartate-rich” motif D345DXXD and the “NSE/DTE” motif N489DXXSXXXE) characteristic of terpenoid cyclases from plants, bacteria, and fungi.27–29 Notably, the structure of PcISPS is the first to reveal the metal binding motifs of a terpenoid cyclase in a terpenoid synthase that is not a cyclase. For example, a terpenoid synthase such as farnesyl diphosphate synthase, which catalyzes an isoprenoid chain elongation reaction, typically contains two aspartate-rich metal binding motifs rather than one aspartate-rich motif and one NSE/DTE motif.26
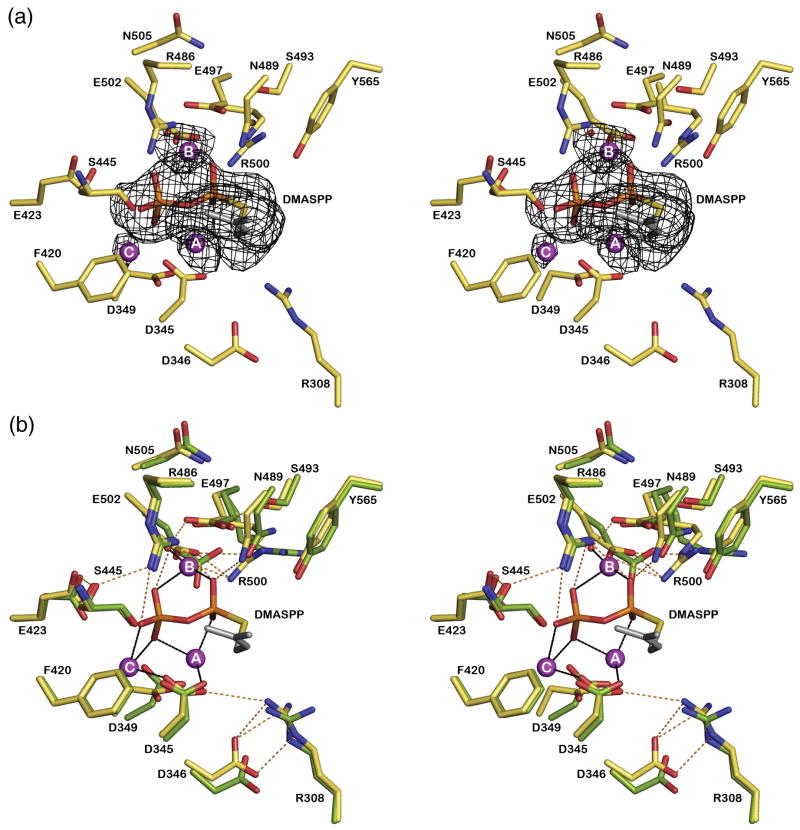
The metal-binding motifs of PcISPS interact with a trinuclear magnesium cluster in the complex with DMASPP (Figure 2a). However, while Mg2+A is bound with full occupancy, the occupancies of Mg2+B and Mg2+C are each estimated to be 0.75 (the occupancy of DMASPP is similarly estimated to be 0.75), and metal coordination geometries are less than ideal: the average metal-ligand coordination distance is somewhat long at 2.5 Å, instead of 2.0–2.2 Å, as more typically observed in higher resolution structures. Furthermore, Mg2+B interacts with only one residue of three (E497) in the “NSE/DTE” motif that typically chelates Mg2+B in terpenoid cyclase active sites,27–29 and this interaction occurs only through a bridging water molecule in monomer B. Such deviations from expected protein-metal interactions could be a consequence of the modest resolution of the crystal structure determination, but they could also be a consequence of incomplete active site closure. We speculate that full occupancy binding of Mg2+B and Mg2+C may accompany the transition to a closed active site conformation characterized by optimal metal coordination interactions, as recently observed for aristolochene synthase.32 Regardless, it is clear that catalysis by a trinuclear magnesium cluster is conserved in PcISPS as well as the greater family of class I terpenoid synthases, which include monoterpene, sesquiterpene, and diterpene cyclases.33
Figure 2.
(A) Simulated annealing omit maps of the PcISPS-DMASPP complex showing the binding of DMASPP (contoured at 4σ) and three Mg2+ ions (contoured at 4σ) in the active site of monomer B. Atoms are color-coded as follows: carbon = tan, nitrogen = blue, oxygen = red; Mg2+ ions appear as magenta spheres. (B) Superposition of the PcISPS-DMASPP complex with unliganded PcISPS reveals no significant conformational changes upon ligand binding; the view is the same as in (A). Selected active site residues are indicated; metal coordination and hydrogen bond interactions in the PcISPS-DMASPP complex are indicated by black and red dashed lines, respectively. Metal coordination interactions appear to be somewhat weak and range 2.2–2.8 Å (average = 2.5 Å). Atom color codes are as in (A), except that carbon = green for unliganded PcISPS. Active site solvent molecules in unliganded PcISPS are omitted for clarity.
No significant conformational changes are observed between the structures of native PcISPS and the PcISPS-DMASPP complex, and the average r.m.s. deviation is 0.49 Å for 514 Cα atoms (monomer A) and 0.38 Å for 516 Cα atoms (monomer B). A superposition of liganded and unliganded active sites is shown in Figure 2b. The DMASPP diphosphate group accepts hydrogen bonds from R486 and N489 in addition to its interactions with metal ions (R486 is conserved in the monoterpene cyclases BPPS and LMNS, whereas N489 is an aspartic acid in these cyclases). The Mg2+A ion is coordinated by D345 and one (monomer A) or two (monomer B) oxygen atoms of the DMASPP diphosphate group. The Mg2+B ion is coordinated by one (monomer A) or two (monomer B) oxygen atoms of the DMASPP diphosphate group, and additionally by a water molecule in monomer B (which in turn donates a hydrogen bond to E497). Finally, the Mg2+C ion is coordinated by D345 and one oxygen atom of the DMASPP diphosphate group.
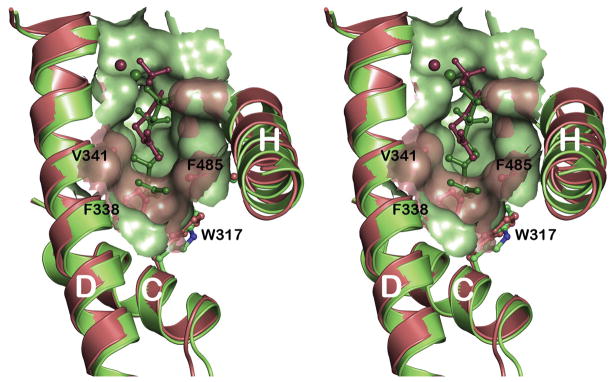
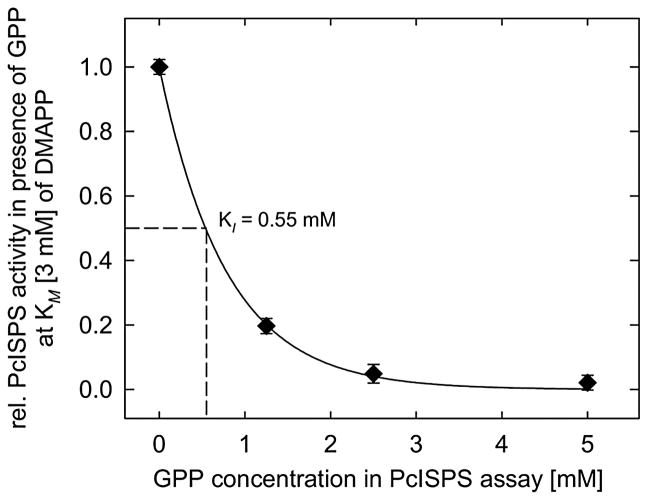
The active site pocket of PcISPS is hydrophobic in nature but shallower in comparison with the active site pockets of monoterpene cyclases,34, 35 consistent with the binding of a 5-carbon substrate rather than a 10-carbon substrate (Figure 3). The isoprenoid moiety of DMASPP makes van der Waals interactions with F338, V341, and F485 in the active site pocket. While in its first description PcISPS was reported to exhibit very weak limonene synthase activity,23 suggesting that the 10-carbon isoprenoid substrate geranyl diphosphate (GPP) can be accommodated in the shallow PcISPS active site, gas chromatography-mass spectrometry (GC-MS) analyses of enzyme assays with GPP give no indication that limonene or any other monoterpene is generated by the PcISPS construct utilized for the crystal structure determination. However, incubation of PcISPS with GPP under a non-saturating concentration of DMAPP (KM = 3 mM) results in competitive inhibition of isoprene formation with Ki = 0.55 mM for GPP (Figure 4). This indicates that the linear monoterpene GPP can bind in the PcISPS active site and block the binding of the hemiterpene substrate DMAPP.
Figure 3.
Cut-away view of superimposed active site contours of PcISPS (salmon; DMASPP is shown as a dark salmon stick figure) and the monoterpene cyclase, bornyl diphosphate synthase (BPPS, light green, PDB ID: 1N20; 3-azaGPP, a geranyl diphosphate analogue, is shown as a dark green stick figure). The active site contour of PcISPS is shallower mainly due to the bulky aromatic side chains of F338 and F485 at the base of the cleft.
Figure 4.
Competitive inhibition of isoprene biosynthesis by geranyl diphosphate (GPP) at non-saturating concentration (3 mM, corresponding to the Michaelis constant KM) of DMAPP by PcISPS. The assays were incubated at 40°C for 60 min. Isoprene and volatile terpenes were trapped continuously during the assay from the aqueous matrix by stir bar sorptive extraction (SBSE) and analyzed by GC-MS. PcISPS activity was normalized to the initial activity with no GPP added (n = 3 ± sd).
Structural inferences on catalysis
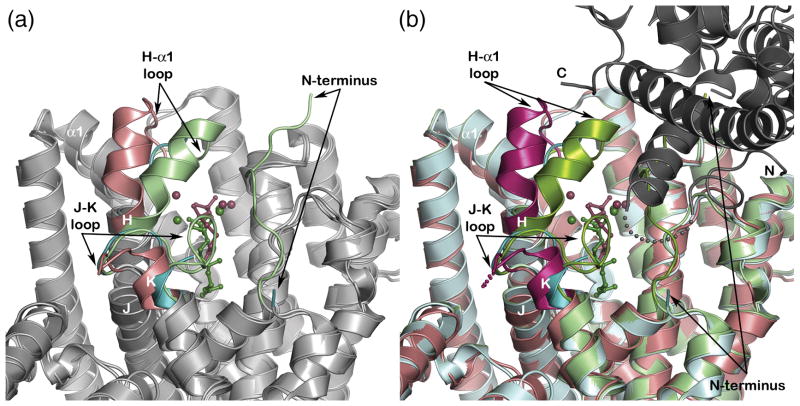
In general, class I terpenoid synthases undergo a significant structural transition from an open to a closed active site conformation upon the binding of 3 metal ions and the substrate diphosphate group, and this conformational transition is presumed to shield reactive carbocation intermediates in catalysis from premature quenching by bulk solvent.27–29 No significant structural differences are evident between native PcISPS and the PcISPS-Mg2+3-DMASPP complex; furthermore, comparisons of the latter structure with open and closed active site conformations of BPPS confirm that the active site of PcISPS remains in the open conformation in the DMASPP complex (Figure 5a). The active site closure mechanism in BPPS involves conformational changes of helix H-α1 and its flanking loop segments, the J-K loop, and the N-terminal polypeptide that serve to cap the active site. By analogy with BPPS, we hypothesize that corresponding conformational changes occur in PcISPS to achieve a fully closed active site conformation. However, some of these structural changes, particularly those involving the N-terminal polypeptide, may be blocked by the packing of a symmetry-related PcISPS molecule in the crystal lattice (Figure 5b). While cocrystallization of the PcISPS-DMASPP complex instead of soaking DMASPP into pre-formed crystals of unliganded PcISPS could conceivably yield a structure in which substrate-induced conformational changes of the N-terminus are not hindered, we were unable to prepare suitable crystals of the PcISPS-DMASPP complex by cocrystallization.
Figure 5.
(A) Superposition of the PcISPS-Mg2+3-DMASPP complex (salmon) with unliganded BPPS (blue, PDB ID: 1N1B) and its Mg2+3-3-azaGPP complex (light green, PDB ID: 1N20). The active site of PcISPS remains in an open conformation like that of unliganded BPPS. By analogy with BPPS, complete active site closure in PcISPS is expected to involve conformational changes of helix H-α1 and its flanking loops, the J-K loop, and the N-terminus (these polypeptide segments are colored with darker shades). (B) Comparison of unliganded bornyl diphosphate synthase (BPPS) (blue, PDB ID: 1N1B) and its Mg2+3-3-azaGPP complex (light green, PDB ID: 1N20) shows that conformational changes or ordering of the H-α1 loop, the J-K loop, and the N-terminus accompany ligand binding and complete active site closure. The structure of the PcISPS-DMASPP complex (salmon) reveals a conformation closer to that of unliganded BPPS. If full active site closure in PcISPS requires conformational changes similar to those observed in BPPS, these conformational changes (especially active site “capping” by the N-terminus) may be hindered by crystal lattice interactions with the first helix in the N-terminal domain of a symmetry-related PcISPS molecule (grey; the N- and C-termini of the symmetry-related molecule are labeled “N” and “C”, respectively).
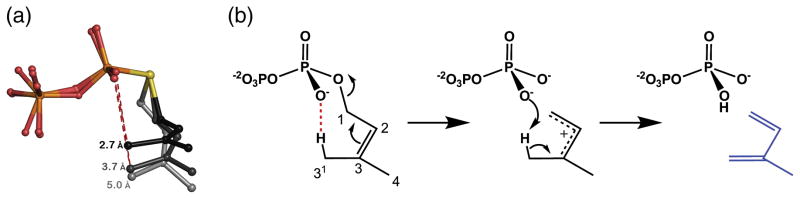
By analogy with other terpenoid synthases, it is reasonable to hypothesize that the simple elimination reaction catalyzed by PcISPS proceeds via an allylic carbocation intermediate, by analogy with the initiation step in the most closely related plant monoterpene synthases as well as the greater family of class I terpenoid synthases. However, the simple elimination reaction catalyzed by PcISPS could also proceed in concerted fashion. In either case, the reaction would require the assistance of a suitable general base, but inspection of the active site does not reveal any such residue. Intriguingly, analysis of the DMASPP conformation suggests that the diphosphate leaving group itself could serve as the general base. Specifically, the favorable chair-like binding conformation of DMASPP suggests a syn-periplanar elimination reaction with full or partial development of an allylic carbocation intermediate and proton abstraction from the C31 methyl group by the diphosphate leaving group (Figure 6). Thus, the structure of the PcISPS-DMASPP complex strongly implicates substrate-assisted catalysis in the mechanism of isoprene generation.
Figure 6.
Proposed catalytic mechanism for ISPS. (A) DMASPP binds to monomer B (grey) and monomer A (darker grey) with a favorable 7-membered ring chair-like conformation that can become more optimal with relatively small bond rotations (black). This conformation would support the proposed elimination mechanism in DMAPP by bringing the O2 atom of the αphosphate group closer to the C31 atom (dark grey). (B) The generation of isoprene (blue) likely occurs in two steps via an allylic carbocation intermediate, but it could also occur in concerted (synchronous or asynchronous) fashion with substrate-assisted catalysis. The favorable chair-like conformation of DMASPP is highlighted by the red dashed line and is consistent with syn-periplanar elimination geometry. Deprotonation at C31 appears likely if the diphosphate group serves as the general base.
Structural basis of cooperativity
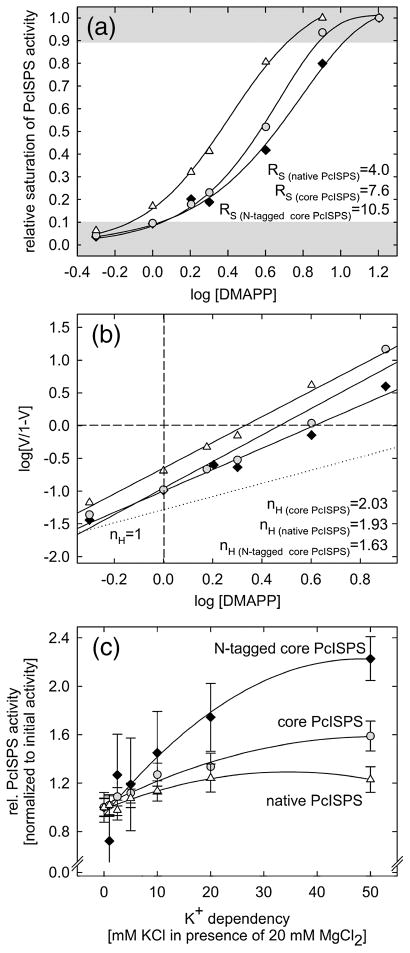
Although PcISPS was initially found to be monomeric by gel filtration chromatography and non-denaturing polyacrylamide gel electrophoresis,25 catalytic activity exhibits positive cooperativity with RS < 81 and the Hill coefficient36 nH >1 (Figures 7b, c); enzyme activity is enhanced by the addition of K+ in the presence of Mg2+ (Figure 7c). While the molecular weight of ISPS from kudzu (Pueraria montana) has not been determined, this enzyme too exhibits positive cooperativity with nH =4.1.37 It would be unusual, although not unprecedented, for a monomeric enzyme to exhibit positive cooperativity.38 However, the crystal structure of PcISPS provides compelling evidence that cooperativity is the consequence of dimeric quaternary structure (Figure 8a): PcISPS forms an isologous dimer through significant interactions between C-terminal catalytic domains that bury 1056 Å2 surface area per monomer at the dimer interface (4% of the total surface area per monomer). Consistent with these crystal structure data, our chemical crosslinking studies with PcISPS in solution at concentrations as low as 8 μM indicate PcISPS dimerization (data not shown).
Figure 7.
Cooperativity and enhancement of activity by K+ for native full-length PcISPS, recombinant core PcISPS lacking the putative N-terminal plastidial targeting sequence, and recombinant PcISPS with the N-terminal His-tag used for crystallization. The DMAPP dependence of PcISPS activity in semilogarithmic projection (A) and linearized as a Hill plot (B) demonstrates the cooperativity of PcISPS activity. The RS values < 81 (ratio of ligand (DMAPP) concentration at 90% and 10% saturation of PcISPS activity) and Hill coefficients (nH; slope of regression line) >1 indicate positive cooperativity. Each curve represents the mean of 4 independent experiments (see also reference 25). The dotted line in (B) shows the theoretical slope of a substrate dependence with no cooperativity (nH = 1). (C) The addition of K+ ions into the enzyme assay solution (with 20 mM Mg2+) enhances activity of native PcISPS (△), recombinant core PcISPS ( ), and recombinant core PcISPS with the N-terminal His-tag (◆) (n = 3 ± sd).
), and recombinant core PcISPS with the N-terminal His-tag (◆) (n = 3 ± sd).
Figure 8.
Dimeric quaternary structures are essentially identical for (A) PcISPS, (B) bornyl diphosphate synthase (BPPS) (PDB ID: 1N1B), and (C) limonene synthase (LMNS) (PDB ID: 2ONG). The catalytic C-terminal domain comprises the dimerization interface with buried surface areas of 1218, 1178, and 1049 Å2/monomer, respectively.
Slight structural changes at the PcISPS dimer interface are observed upon the binding of DMASPP, in that the buried surface area increases to 1218 Å2/monomer (5% of the total surface area per monomer) and two additional hydrogen bonds and four additional salt bridges are formed between monomers. Strikingly, the PcISPS dimer interface is essentially identical to that of the monoterpene cyclases BPPS (Figure 8b, 1178 Å2/monomer buried surface area,34 confirmed to be a dimer in solution by gel filtration chromatography39) and LMNS (Figure 8c, 1049 Å2/monomer buried surface area,35 found to be monomeric by gel filtration chromatography and coupled non-denaturing PAGE/SDS-PAGE analysis40). Comparison with BPPS is particularly informative since this enzyme structure was determined at high resolution (2.0 Å) and it is confirmed to be a dimer in solution and in the crystal. Several conserved or partially conserved residues are located at the dimer interface (Table 1), and the amino acid sequence identity of 45% for residues at the dimer interface is greater than the overall sequence identity of 40% between PcISPS and BPPS, although the interactions of conserved residues are not always identical. Such differences could account for the intersubunit communication implied by the positive cooperativity measured for isoprene generation by dimeric PcISPS.
Table 1.
Interactions at the Dimer Interfaces of ISPS and BPPS
| ISPS |
BPPS |
||||
|---|---|---|---|---|---|
| Residuea | Buried?b | Interacting Residuec | Residuea | Buried?b | Interacting Residuec |
| R296 | - | K399* (HB) | S302 | - | - |
| R297 | Yes | D396 (SB), K399* (HB) | R303 | Yes | D398, D402 (SB) |
| V298 | Yes | Y387* (HB) | L304 | Yes | Y401* (HB) |
| G299 | Yes | - | C305 | Yes | - |
| L300 | Yes | - | F306 | Yes | - |
| K303 | Yes | K399* (HB) | K309 | Yes | - |
| R366 | - | N370* (HB) | R372 | - | - |
| W367 | Yes | N370* (HB) | W373 | Yes | - |
| V369 | Yes | - | T375 | Yes | R416 (HB) |
| N370 | Yes | R366*, W367* (HB) | E376 | - | - |
| I372 | Yes | - | I378 | Yes | - |
| N373 | - | N404 (HB) | T379 | - | - |
| D377 | - | - | Y383 | Yes | L404*, K405* (HB) |
| K380 | Yes | - | Q386 | Yes | - |
| L381 | Yes | - | L387 | Yes | - |
| L384 | Yes | - | W390 | Yes | S397* (HB) |
| A385 | Yes | - | G391 | Yes | - |
| Y387 | Yes | Y387 (HB) | H393 | Yes | - |
| N388 | Yes | N391* (HB) | N394 | Yes | S397*, D398 (HB) |
| N391 | Yes | N388* (HB) | S397 | Yes | W390*, N394* (HB) |
| E392 | Yes | R297 (SB) | D398 | Yes | R303 (SB), N394 (HB) |
| A394 | Yes | - | A400 | Yes | - |
| Y395 | Yes | V298* (HB) | Y401 | Yes | L304* (HB) |
| D396 | Yes | R297 (SB) | D402 | Yes | R303 (SB) |
| L398 | Yes | - | L404 | Yes | Y383* (HB) |
| K399 | Yes | R297*, K303* (HB) | K405 | Yes | Y383* (HB) |
| N404 | Yes | N373 (HB) | F410 | Yes | - |
| L406 | Yes | - | L412 | Yes | - |
| T410 | - | - | R416 | - | T375 (HB) |
Only the residues oriented toward the dimer interface are indicated. Residues in the same row align in both sequence and structure; conserved residues are in boldface.
Residue is considered buried if 40% of its surface area is buried upon dimer formation.
Residue in the other monomer; HB: hydrogen bond, SB: salt bridge. Interactions labeled with an asterisk involve ]a backbone oxygen of one of the residues.
Concluding Remarks
New structural knowledge regarding the mechanism of isoprene formation by PcISPS sets the stage for future experiments aimed at developing non-petrochemical based carbon fuels in bioreactors. For example, it is possible that hydroxylated isoprenoid products could be obtained by site-directed mutagenesis experiments that facilitate water access to the allylic carbocation intermediate. It is notable, too, that the chemistry of the elimination step yielding isoprene is identical to that yielding farnesene from farnesyl diphosphate. Farnesene is currently being explored as a diesel fuel substitute,20 and farnesene is readily generated by site specific mutants of sesquiterpene cyclases; for example, F96A epi-izozizane synthase generates 70% farnesene.41 Additionally, isoprene itself is a precursor for the generation of rubber and plastic products, and biogenic isoprene is of course a “greener” source of carbon for such applications than petrochemically-derived isoprene. Future studies of PcISPS in such settings promise to yield new pathways for the structure-based engineering of terpenoid synthase function to elaborate and exploit the roots of isoprenoid biosynthetic diversity in myriad biotechnological applications.
Materials and Methods
Crystallization of isoprene synthase
Isoprene synthase from grey poplar hybrid Populus x canescens (syn. Populus tremula x P. alba) (PcISPS) with the 52-residue N-terminal plastidial targeting sequence replaced by a 12-residue hexahistidine tag and linker and containing the mutations N59D, K308R, and C533W (ISPS-pQE30) was expressed in E. coli TG1 cells and purified as previously described.23, 25 PcISPS was crystallized by the sitting drop vapor diffusion method. Typically, a 1 μL drop of protein solution [10 mg/mL PcISPS, 50 mM Tris-HCl (pH 8.5), 20 mM MgCl2, 5% glycerol, 2 mM dithiothreitol] was added to a 1 μL drop of precipitant solution [100 mM Bis-Tris (pH 6.5), 25% polyethylene glycol 3350, 200 mM (NH4)2SO4] and equilibrated against a 1 mL well reservoir of precipitant solution. Rod-like crystals appeared within 2–3 days and grew to maximal dimensions of 35 μm × 35 μm × 500 μm. Crystals were flash-cooled after transfer to a cryoprotectant solution consisting of the mother liquor augmented with 10% glycerol. For the preparation of the enzyme-inhibitor complex, crystals were soaked in a cryoprotectant solution containing 0.2 M NaCl, 2.5 mM MgCl2, and 2.5 mM dimethylallyl-S-thiolodiphosphate (DMASPP; Echelon Biosciences, Inc.) for 12 h at 15°C prior to flash-cooling.
X-ray diffraction data collection and processing
Crystals of the native enzyme and the enzyme-inhibitor complex diffracted X-rays to 2.7 Å and 2.8 Å resolution, respectively, at the National Synchrotron Light Source (NSLS, beamlines X25 and X29, respectively). PcISPS crystals belong to space group P43212 with unit cell parameters a = b = 155.42 Å, c = 142.03 Å, α = β = γ = 90°; with two monomers in the asymmetric unit, the Matthews coefficient VM = 3.41 Å2 / Da, corresponding to a solvent content of 63.7%. Diffraction data were processed with HKL2000.42
Structure refinement
For the structure determination of native ISPS, the program suite CNS43 was used for phasing by molecular replacement using the structure of (+)-bornyl diphosphate synthase (40% amino acid sequence identity; PDB ID: 1N1B)34 as the search model for rotation and translation function calculations. A clear solution was calculated with rotation function scores of 0.0320 and 0.0297 and translation search scores of 0.182 and 0.155 (twice the next best values). After the complete registration of the PcISPS amino acid sequence, iterative cycles of refinement and manual model rebuilding were performed using CNS43 and O44 or COOT,45 respectively. Noncrystallographic symmetry restraints were applied in the initial stages of refinement and later relaxed due to structural differences between monomers. Individual atomic B-factors were utilized during refinement. Water molecules were included in later cycles of refinement. A total of 531 of 555 residues are present in the final model of the native enzyme; disordered segments not included in the final model are N-terminal residues M41-R56 (M41 is the N-terminus of the construct) and loops S73-E77 and D569-H571. Ramachandran plot statistics were calculated with PROCHECK.48 Data reduction and refinement statistics are recorded in Table 2.
Table 2.
Data Collection and Refinement Statistics
| Native PcISPS | PcISPS-Mg2+3-DMASPP complex | |
|---|---|---|
| A. Data Collection | ||
| Incident wavelength (Å) | 0.979 | 1.075 |
| Resolution Range (Å) | 39.1–2.70 | 46.4–2.80 |
| No. of reflections (total/unique) | 392710/48217 | 344302/43719 |
| Completenessa (%) | 99.8 (100) | 100 (99.9) |
| I/σ | 14 (3.3) | 16 (3.3) |
| Rmergeb | 0.148 (0.654) | 0.135 (0.628) |
| B. Refinement | ||
| No. of reflections, work/test sets | 41244/4653 | 39758/2084 |
| Rwork/Rfreec | 0.198 / 0.245 | 0.199 / 0.249 |
| Protein atomsd | 8652 | 8633 |
| Solvent atomsd | 324 | 323 |
| Ligand atomsd | 0 | 34 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.007 | 0.007 |
| Angles (°) | 1.1 | 1.0 |
| Dihedral angles (°) | 18.8 | 18.8 |
| Improper dihedral angles (°) | 0.8 | 0.7 |
| Average B factors (Å2) | ||
| Main chain | 34 | 41 |
| Side chain | 35 | 43 |
| DMASPP-Mg2+3 | 49 | |
| Solvent | 30 | 32 |
| Ramachandran plot | ||
| Allowed (%) | 92.2 | 92.4 |
| Additionally allowed (%) | 7.8 | 7.5 |
| Generously allowed (%) | 0 | 0.1 |
| Disallowed (%) | 0 | 0 |
Number in parentheses refer to the outer 0.1 Å shell of data.
Rmerge = Σ|I-〈I〉|/ΣI, where I is the observed intensity and 〈I〉 is the average intensity calculated from replicate data.
Rwork = Σ||Fo|-|Fc||/Σ|Fo| for reflections contained in the working set, and Rfree = Σ||Fo|-|Fc||/Σ|Fo| for reflections contained in the test set held aside during refinement. | Fo | and | Fc | are the observed and calculated structure factor amplitudes, respectively.
Per asymmetric unit.
Structure determination of the enzyme-substrate analogue complex
For the structure determination of the PcISPS-Mg2+3-DMASPP complex, the difference Fourier technique was used since crystals were isomorphous with those of the native enzyme. Individual atomic B-factors were utilized during refinement. Water molecules, DMASPP, and Mg2+ ions were included in later cycles of refinement. A total of 521 (524 in monomer B) of 555 residues are present in the final model of the PcISPS-Mg2+3-DMASPP complex; disordered segments not included in the final model are N-terminal residues M41-P62, loops S73-E80 and D569-H571, and C-terminal residue R595. The final model includes a DMASPP molecule in each active site with some residual positive electron density in monomer B; possibly, this density corresponds to an alternative conformation of the DMASPP isoprenoid group or a trapped water molecule (e.g., as observed in the active site of the BPPS-product complex34), but it was not readily interpretable. The occupancy of Mg2+A was estimated to be 1.0, and the occupancies of Mg2+B, Mg2+C, and DMASPP were estimated to be 0.75, which yielded refined B factors for these ligands in line with average B factors for protein residues. Ramachandran plot statistics were calculated with PROCHECK.46 Data reduction and refinement statistics are recorded in Table 2. Structures were superposed with the SSM Superpose function of COOT45 for comparison. Protein structure figures were prepared with the graphics program PyMol (http://www.pymol.org) and labelled for publication using PhotoshopCS.
Analysis of ISPS activity
Protein extracts of native and heterologously-expressed PcISPS were obtained as described.25 Routine catalytic activity assays were performed as described.47 For the analysis of terpene formation from geranyl diphosphate (GPP) as a possible substrate and for the GPP inhibition studies, a new assay procedure was developed. Enzyme assays were performed in a total volume of 200 μL in 200 μL PCR tubes. Enzymatically formed isoprene and volatile terpenes were trapped during the assay from the aqueous matrix by stir bar sorptive extraction (SBSE) with polydimethylsiloxane (PDMS) coated stir bars (Twister, PDMS film thickness 0.5 mm, Gerstel, Mülheim, Germany). The enzyme assays were terminated by removing the stir bars. After rinsing in deionized H2O, the twisters were analyzed with a Thermo Desorption Unit (TDU, Gerstel, Mülheim, Germany) interfaced with a gas chromatograph-mass spectrometer (GC-MS) (Agilent GC 7890 with 5975C inert XL MSD interface, Agilent Technologies, Böblingen, Germany). The twisters were desorbed at 240°C for 2 min with a helium flow of 100 ml min−1. Volatiles were focused on a sorbent trap (Tenax GR, Supelco, Deisenhofen, Germany) at −100°C. For injection into the analytical column (J&W DB-1701, 30 m x 0.25 mm ID, 1.0 μm film thickness, Agilent Technologies, Böblingen, Germany), the cold trap was rapidly heated to 250°C with a split flow of 3 ml min−1. The temperature program started at 35°C (6-min hold) and rose with a rate of 6°C min−1 to 200°C, followed by a second temperature increase with 20°C min−1 to 240°C (2-min hold). The column effluent was ionized by electron impact ionization at 70 eV. Mass scanning was done from 25 to 230 m/z with a scan rate of 6.47 scans s−1. Compounds were identified by comparing the mass spectra with those of authentic standards or with NIST 05 and Wiley library spectra.
Acknowledgments
This work was supported by US National Institutes of Health grant GM56838 and the German Research Foundation grant DFG SCHN653/4. We thank the National Synchrotron Light Source, Brookhaven National Laboratory, for access to synchrotron data collection facilities at beamlines X25 and X29.
Abbreviations
- GC-MS
gas chromatography-mass spectrometry
- ISPS
isoprene synthase
- PcISPS
isoprene synthase from Populus x canescens
- BPPS
bornyl diphosphate synthase
- LMNS
limonene synthase
- DMAPP
dimethylallyl diphosphate
- PPi
inorganic pyrophosphate
- DMASPP
dimethylallyl-S-thiolodiphosphate
- NSLS
National Synchrotron Light Source
Footnotes
Protein Data Bank accession codes
The atomic coordinates and the crystallographic structure factors of isoprene synthase from Populus x canescens and of its complex with Mg2+3 and dimethylallyl-S-thiolodiphosphate have been deposited in the Protein Data Bank (www.rcsb.org) with accession codes 3N0F and 3N0G, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guenther A, Hewitt CN, Erickson D, Fall R, Geron C, Graedel T, Harley P, Klinger L, Lerdau M, Mckay WA, Pierce T, Scholes B, Steinbrecher R, Tallamraju R, Taylor J, Zimmerman P. A global model of natural volatile organic compound emissions. J Geophys Res. 1995;100(D5):8873–8892. [Google Scholar]
- 2.Wang KY, Shallcross DE. Modelling terrestrial biogenic isoprene fluxes and their potential impact on global chemical species using a coupled LSM–CTM model. Atmos Environ. 2000;34:2909–2925. [Google Scholar]
- 3.Monson RK, Holland EA. Biospheric trace gas fluxes and their control over tropospheric chemistry. Ann Rev Ecol System. 2001;32:547–576. [Google Scholar]
- 4.Purves DW, Capersen JP, Moorcroft PR, Hurtt GC, Pacala SW. Human-induced changes in US biogenic volatile organic compound emissions: evidence from long-term forest inventory data. Global Change Biol. 2004;10:1737–1755. [Google Scholar]
- 5.Sanadze GA. The nature of gaseous substances emitted by leaves of Robina pseudoacacia. Soobshch Akad Nauk Gruz SSR. 1957;19:83–86. [Google Scholar]
- 6.Kesselmeier J, Staudt M. Biogenic volatile organic compounds (VOC): an overview on emission, physiology and ecology. J Atmos Chem. 1999;33:23–88. [Google Scholar]
- 7.Went G. Blue Hazes in the Atmosphere. Nature. 1960;187:641–643. [Google Scholar]
- 8.Sharkey TD, Yeh S. Isoprene emission from plants. Ann Rev Plant Physiol Plant Mol Biol. 2001;52:407–436. doi: 10.1146/annurev.arplant.52.1.407. [DOI] [PubMed] [Google Scholar]
- 9.Sharkey TD, Singsass EL. Why plants emit isoprene? Nature. 1995;374:769. [Google Scholar]
- 10.Singsaas EL, Lerdau M, Winter K, Sharkey TD. Isoprene increases thermotolerance of isoprene-emitting species. Plant Physiol. 1997;115:1413–1420. doi: 10.1104/pp.115.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behnke K, Ehlting B, Teuber M, Bauerfeind M, Louis S, Hänsch R, Polle A, Bohlmann J, Schnitzler JP. Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J. 2007;51:485–499. doi: 10.1111/j.1365-313X.2007.03157.x. [DOI] [PubMed] [Google Scholar]
- 12.Loreto F, Velikova V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001;127:1781–1787. [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers CE, Possell M, Cojocariu CI, Velikova VB, Laothawornkitkul J, Ryan A, Mullineaux PM, Hewitt CN. Isoprene synthesis protects transgenic tobacco plants from oxidative stress. Plant Cell Environ. 2009;32:520–531. doi: 10.1111/j.1365-3040.2009.01946.x. [DOI] [PubMed] [Google Scholar]
- 14.Siwko ME, Marrink SJ, de Vries AH, Kozubek A, Schoot Uiterkamp AJM, Mark AE. Does isoprene protect plant membranes from thermal shock? A molecular dynamics study. Biochim Biophys Acta. 2007;1768:198–206. doi: 10.1016/j.bbamem.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Loivamäki M, Mumm R, Dicke M, Schnitzler JP. Isoprene interferes with the attraction of bodyguards by herbaceous plants. Proc Natl Acad Sci USA. 2008;105:17430–17435. doi: 10.1073/pnas.0804488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laothawornkitkul J, Paul ND, Vickers CE, Possell M, Taylor JE, Mullineaux PM, Hewitt CN. Isoprene emissions influence herbivore feeding decisions. Plant Cell Environ. 2008;31:1410–1415. doi: 10.1111/j.1365-3040.2008.01849.x. [DOI] [PubMed] [Google Scholar]
- 17.Paulot F, Crounse JD, Kjaergaard HG, Kürten A, St Clair JM, Seinfeld JH, Wennberg PO. Unexpected epoxide formation in the gas-phase photooxidation of isoprene. Science. 2009;325:730–733. doi: 10.1126/science.1172910. [DOI] [PubMed] [Google Scholar]
- 18.Kiendler-Scharr A, Wildt J, Dal Maso M, Hohaus T, Kleist E, Mentel TF, Tillmann R, Uerlings R, Schurr U, Wahner A. New particle formation in forests inhibited by isoprene emissions. Nature. 2009;461:381–384. doi: 10.1038/nature08292. [DOI] [PubMed] [Google Scholar]
- 19.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54:656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 20.Rude MA, Schirmer A. New microbial fuels: a biotech perspective. Curr Op Microbiol. 2009;12:274–281. doi: 10.1016/j.mib.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Ondrey G. Bio-based isoprene. Chem Eng. 2008;115:14. [Google Scholar]
- 22.Silver GM, Fall R. Enzymatic synthesis of isoprene from dimethylallyl diphosphate in aspen leaf extracts. Plant Physiol. 1991;97:1588–1591. doi: 10.1104/pp.97.4.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller B, Oschinski C, Zimmer W. First isolation of an isoprene synthase gene from poplar and successful expression of the gene in Escherichia coli. Planta. 2001;213:483–487. doi: 10.1007/s004250100557. [DOI] [PubMed] [Google Scholar]
- 24.Silver GM, Fall R. Characterization of aspen isoprene synthase, an enzyme responsible for leaf isoprene emission to the atmosphere. J Biol Chem. 1995;270:13010–13016. doi: 10.1074/jbc.270.22.13010. [DOI] [PubMed] [Google Scholar]
- 25.Schnitzler JP, Zimmer I, Bachl A, Arend M, Fromm J, Fischbach RJ. Biochemical properties of isoprene synthase in poplar (Populus × canescens) Planta. 2005;222:777–786. doi: 10.1007/s00425-005-0022-1. [DOI] [PubMed] [Google Scholar]
- 26.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-Å resolution. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 27.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 28.Christianson DW. Unearthing the roots of the terpenome. Curr Op Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesburg CA, Caruthers JM, Paschall CM, Christianson DW. Managing and manipulating carbocations in biology: terpenoid cyclase structure and mechanism. Curr Op Struct Biol. 1998;8:695–703. doi: 10.1016/s0959-440x(98)80088-2. [DOI] [PubMed] [Google Scholar]
- 30.Thulasiram HV, Erickson HK, Poulter CD. Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science. 2007;316:73–73. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 31.Cervin MA, Whited GM, Miasnikov A, Peres CM, Weyler W, Wells DH, Bott RR. Isoprene synthase variants for improved microbial production of isoprene. 20100003716 US Patent Application. 2010
- 32.Shishova EY, Yu F, Miller DJ, Faraldos JA, Zhao Y, Coates RM, Allemann RK, Cane DE, Christianson DW. X-ray crystallographic studies of substrate binding to aristolochene synthase suggest a metal ion binding sequence for catalysis. J Biol Chem. 2008;283:15431–15439. doi: 10.1074/jbc.M800659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aaron JA, Christianson DW. Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl Chem. 2010 doi: 10.1351/PAC-CON-09-09-37. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittington DA, Wise ML, Urbansky M, Coates RM, Croteau RB, Christianson DW. Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc Natl Acad Sci USA. 2002;99:15375–15380. doi: 10.1073/pnas.232591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyatt DC, Youn B, Zhao Y, Santhamma B, Coates RM, Croteau RB, Kang CH. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc Natl Acad Sci USA. 2007;104:5360–5365. doi: 10.1073/pnas.0700915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taketa K, Pogell BN. Allosteric inhibition of rat liver fructose 1,6-diphosphatase by adenosine 5’-monophosphate. J Biol Chem. 1965;240:651–652. [PubMed] [Google Scholar]
- 37.Sharkey TD, Yeh S, Wiberley AE, Falbel TG, Gong D, Fernandez DE. Evolution of the isoprene biosynthetic pathway in Kudzu. Plant Physiol. 2005;137:700–712. doi: 10.1104/pp.104.054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ainslie GR, Jr, Shill JP, Neet KE. Transients and cooperativity. A slow transition model for relating transients and cooperative kinetics of enzymes. J Biol Chem. 1972;247:7088–7096. [PubMed] [Google Scholar]
- 39.Wise ML, Savage TJ, Katahira E, Croteau R. Monoterpene synthases from common sage (Salvia officinalis) J Biol Chem. 1998;273:14891–14899. doi: 10.1074/jbc.273.24.14891. [DOI] [PubMed] [Google Scholar]
- 40.Alonso WR, Rajaonarivony JIM, Gershenzon J, Croteau R. Purification of 4S-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha x piperita) and spearmint (Mentha spicatu) J Biol Chem. 1992;267:7582–7587. [PubMed] [Google Scholar]
- 41.Aaron JA, Lin X, Cane DE, Christianson DW. Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry. 2010;49:1787–1797. doi: 10.1021/bi902088z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Jones TA, Zou JY, Cowan SW, Kjeldgaaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 47.Mayrhofer S, Teuber M, Zimmer I, Louis S, Fischbach RJ, Schnitzler JP. Diurnal and seasonal variation of isoprene biosynthesis-related genes in grey poplar leaves. Plant Physiol. 2005;139:474–484. doi: 10.1104/pp.105.066373. [DOI] [PMC free article] [PubMed] [Google Scholar]