Abstract
Vertebrates employ V(D)J recombination to generate diversity for an adaptive immune response. Born of a transposon, V(D)J recombination could conceivably cause more trouble than it's worth. However, of the two steps required for transposon mobility (excision and integration) this particular transposon's integration step appears mostly blocked in cells. The employment of a transposon as raw material to develop adaptive immunity was thus a less-risky choice than it might have been…but is it completely risk-free?
Keywords: transposon, RAG1, RAG2, V(D)J recombination, G4 DNA
1. Introduction
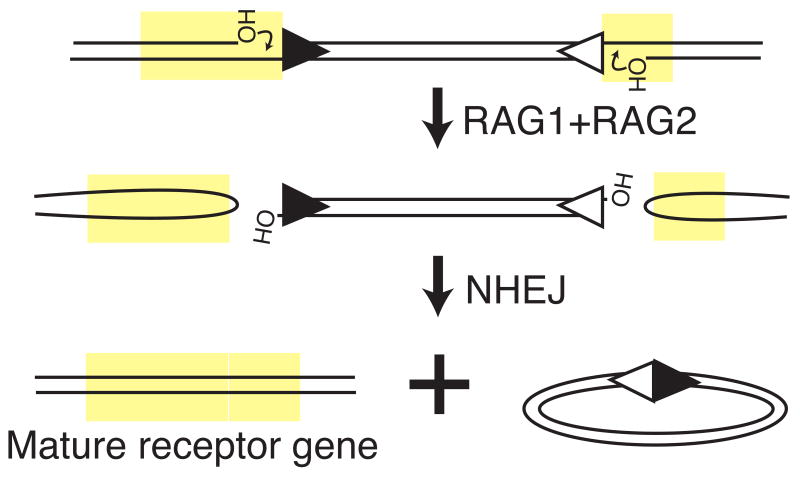
V(D)J recombination assembles the immune system's antigen specific receptors (immunoglobulins and T cell receptors)(see e.g. [1] for a general review). In the first step, lymphocyte specific recombination activating genes 1 and 2 (RAG1 and RAG2) introduce a pair of chromosome breaks between recombination targeting signals (RS) and receptor coding DNA. In typical RS arrangements this excises a fragment from the receptor locus with RS ends (signal ends) (Figure 1). “Host” repair machinery (nonhomologous end joining, or NHEJ), then resolves these breaks. Still poorly understood interactions between RAG1/RAG2 and NHEJ help ensure pair-wise resolution of receptor coding ends results in assembly of a mature receptor gene.
Figure 1.
V(D)J recombination. RAG1 and RAG2 direct chromosomal breakage at receptor gene loci. Nonhomologous end joining (NHEJ) resolves chromosome breaks to assemble a mature receptor gene (immunoglobulin or T cell receptor) and deleted extrachromosomal circle. Yellow rectangles denote receptor-coding DNA, and triangles denote recombination signals (RS).
In vitro studies have shown V(D)J recombination has clear origins in transposition. Most importantly, RAG1- RAG2 cleave and join DNA strands by direct (or one-step) transesterification [2]. A donor DNA strand 3′OH terminus is used as a nucleophile to attack a phosphodiester bond in a target DNA strand, breaking the bond 5′ of the phosphate attacked in the target strand and replacing it with a new bond, between the donor strand's 3′ end and the targeted phosphate. Initially, RAG-mediated one-step transesterification is critical for making the chromosome breaks that flank the RS pair. A nick adjacent to an RS generates a donor 3′OH, and this 3′OH attacks the immediately opposite phosphodiester bond. This transesterification generates a double strand break between the RS and receptor coding DNA, where the RS end is blunt ended and the coding end possesses a characteristic hairpin terminus. Cleavage requires recognition of a pair of RS and both RS are cleaved at the same time, resulting in excision of a linear fragment with RS ends (signal end fragment). In sum, initial breakage of DNA (e.g. at receptor loci) is analogous to transposon excision; indeed, the same end structures (blunt RS ends, hairpin terminated flanks) are generated when the hAT family transposons excise from their host genome [3].
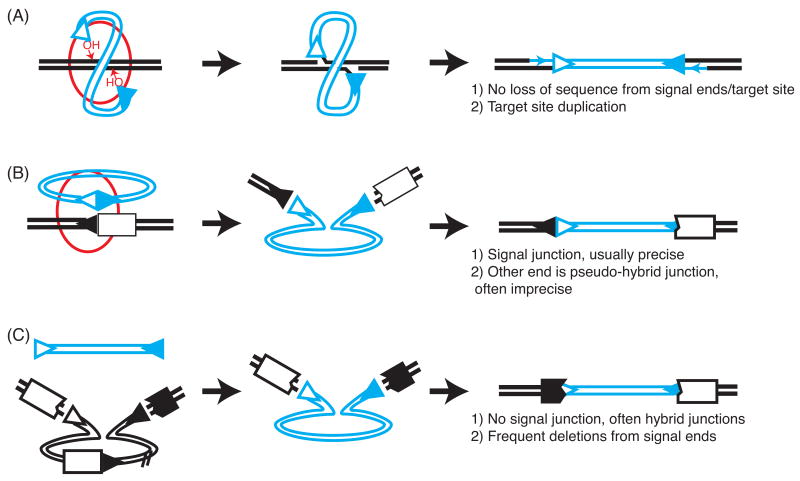
RAG proteins can remain bound to the excised signal end fragment in vitro and continue to perform direct transesterification, but now using the signal end 3′OH as a donor. The next transesterification step can target the recently generated hairpin-coding end, essentially reversing the initial cleavage reaction [4]. The products of these reactions are termed open-and-shut or hybrid junctions. Alternatively, RAG1/RAG2 bound signal ends can direct strand transfer of the excised element into a “foreign” DNA target in a process equivalent to the integration step in transposition (Figure 2a) [5, 6]. Strand transfers are often concerted such that both RS ends attack the same target duplex, but with each of the RS ends attacking bonds in opposite strands of the target 4-5 bp apart. As a result the excised “transposon” is integrated into the target and flanked by 4-5 nucleotide gaps. Gap repair generates a 4-5 direct repeat of flanking target DNA, or a target site duplication, that is considered a signature of transposition integrations.
Figure 2.
Mechanisms for integration of the excised signal end fragment/putative transposon. Blue and black lines distinguish the putative transposon from its target site, and triangles denote recombination signals (RS). RAG-bound signal end complexes are noted by red oval. The features that distinguish the products of each mechanism from the others are noted at right. (A) Transposition. Gap repair to generate the flanking target site duplication is noted by arrows. (B) Intermolecular recombination (recombination in trans). (C) End donation (capture by an independently generated double strand break).
At least in vitro, therefore, RAG1 and RAG 2 mediate the excision and integration of a RS bounded DNA fragment through direct transesterifications. V(D)J recombination is a modified transposon: it was born to be wild. However, while excision of the V(D)J recombination associated transposon is efficient in lymphoid cells and required for receptor assembly, this transposon appears tamed in cells by blocking the integration step. A number of mechanisms for blocking integration in cells have been discussed in other reviews [7-10], most significantly a C-terminal domain of RAG2 that suppresses transposition in vitro [11-13] and possibly related aberrant signal end resolutions in cells [14, 15].
Nevertheless…
2. It happens: Identification of V(D)J recombination associated transposition events in cells
Multiple mechanisms direct integration of signal end fragments into chromosomes in cells (see section 3.1 and 3.2, below). For the purpose of this review we term integrations as transpositions only if a strong case can be made that the structure of the integration is consistent with RAG-protein dependent transesterification. Specifically, integrations should be conservative (no lost sequence or non-templated insertion) both for the donor signal end fragment ends and target (chromosomal) site, and the target site should be flanked by a 4-5 bp target site duplication.
A screen for HPRT mutations in a human T cell line identified a fragment of the T cell receptor locus that was integrated within an intron of the HPRT gene [16]. T cell receptor RS ends were precisely inserted, with no lost sequence relative to the excised fragment (presumptive transposon). The HPRT intronic DNA flanking the integration site similarly possessed no deleted sequence or non-templated additions; instead, the insertion site was flanked by a 4 bp direct repeat of target DNA (TSD). V(D)J recombination associated transposition was subsequently observed in a reconstituted V(D)J recombination model in yeast [17], between donor and target episomes in mammalian cells [18], and between an integrated donor recombination substrate and genome target in mammalian cells [19]. All four reports identified a preference for GC rich targets (see also section 4.2), consistent with previous analyses of RAG mediated integrations in vitro [5, 6, 20]. Two of these reports identified transpositions in cells using exclusively full length RAG1 and RAG2 [16, 19]. The remaining two reports identified transpositions using both full length and core RAG constructs, and, together with the other studies, suggest suppression of transposition by RAG2's C-terminal domain is at best incomplete [17, 18].
Transposition is thus associated with V(D)J recombination in cells. Is it a significant threat to genome stability - How often does it occur in cells, and when it happens, where does it go?
3. How often?
3.1 How often?
As already noted, excision of the signal end fragment (the putative transposon) is obligatory for V(D)J recombination; integration is apparently the limiting step. Not all integrations occur by transposition, though. Assessment of the frequency of V(D)J recombination-associated transposition thus requires first determining the frequency of integration into host chromosomal DNA, followed by characterization of integrations as “transposition” (integration by transesterification) or not based on the criteria described above. Finally, transposition has only loosely defined target site preferences (see section 4) [5, 6, 20], thus the most useful estimations of transposition frequency require the ability to recovery integration events regardless of target site (i.e. a genome-wide screen).
Our group employed an integrated artificial substrate as the source of the putative transposon (signal end fragment) and a virally transformed pre-B cell line as host to address this issue [19]. We induced V(D)J recombination to release the signal end fragment (excision) and determined it integrates back into the genome once every 1.3×104 excisions (V(D)J recombinations). Notably, subsequent studies from both the Nadel [21] and Schlissel [14] groups also scored chromosomal integrations of signal end fragments excised from the T cell receptor locus during normal mouse development, a more biologically relevant model. Consistent with our study, both groups determined this event was surprisingly frequent: for example, the Schlissel group assessed its frequency as between 10-3 to 10-5.
Our group then characterized the structure of signal end fragment integrations to determine if they occurred by transposition. Our initial report isolated and characterized 21 integrations; we summarize here an expanded data set that includes an additional 23 integrations isolated and characterized using the same experimental model (Table 1). We observed transposition in 25% (11/44; 95% C.I. ∼15%-40%) of integrations (Table 2). Taking into account the frequency of all signal end fragment integrations (discussed above), we observed a transposition about once every 50,000 V(D)J recombinations (95% confidence interval; ∼ 1/30,000 to 1/90,000). This frequency predicts the V(D)J recombinations required for the daily lymphocyte output of an adult human would be associated with about 10,000 transposon integrations [19].
Table 1.
| Chr1 | 12RS flank2 | Δ 12/233 | 23RS flank2 | Notes4 |
|---|---|---|---|---|
| 5 | 3344948 | 0/0 | 3344952 | Tnp, cdk6, [10] |
| 4 | 149422359 | 0/0 | 149,422,356 | Tnp, [10] |
| 18 | 35990339 | 0/0 | 35990343 | Tnp, cxxc5, G4, [10] |
| 16 | 32914397 | 0/0 | 32,914,394 | Tnp, G4, [10] |
| 12 | 114663356 | 0/0 | 114,663,359 | Tnp, Sμ, G4, [10] |
| 12 | 1146621245 | 0/0 | 1146621245 | Tnp, Sμ, G4, [10] |
| 5 | 66248447 | 0/0 | 66248455 | Tnp, inv. rep., [10] |
| 11 | 102157220 | 0/0 | 102157216 | Tnp, atxn7L3, G4, |
| 18 | 35989974 | 0/0 | 35989971 | Tnp, cxxc5, G4 |
| 8 | 87230852 | 0/0 | 87230848 | Tnp, nfix, G4 |
| 15 | 96473384 | 0/0 | 96473380 | Tnp, G4 |
| 6 | Vk4-69 CE (2) | 0/0 | Vk4-69 SE (0) | IR, Igκ, [10] |
| 12 | VH7183.33 SE (0) | 0/0 | VH7183.22 CE (1) | IR, IgH, [10] |
| 12 | VH15B SE (0) | 0/6 | JH3 CE (2) | IR, 2°, IgH, [10] |
| 4 | 44719680 SE (0) | 0/5 | 44719677 CE (2) | IR, pax5 |
| 18 | 5378614 SE (0) | 0/17 | 5378609 CE (4) | IR |
| 6 | Vκ ai4 SE (0) | 0/0 | Vκ ai4 CE (1) | IR, Igκ |
| 6 | Vκ at4 SE (0) | 0/0 | Vκ kj4 SE (0) | IR, 2°, Igκ |
| 6 | Vκ af4 CE (4) | 0/0 | Vκ af4 SE (0) | IR, Igκ |
| 6 | Vκ am4 CE (2) | 0/0 | Vκ am4 SE (0) | IR, Igκ |
| 6 | Jκ1 SE (0) | 0/7 | Jκ1 CE (4) | IR, lgκ |
| 6 | Vκ gj38c CE (4) | 5/0 | Vκ gj38c SE (0) | IR, Igκ |
| 6 | Vκ 4-51 CE (2) | 2/0 | Vκ 4-51 SE (0) | IR, Igκ |
| 6 | Vκ gj38c CE (1) | 0/0 | Vκ cf9 SE (0) | IR, 2°, lgκ |
| 6 | Vκ fl12 CE (0) | 0/4 | Vκ fl12 SE (0) | IR, Igκ |
| 6 | Vκ 12-44 CE (3) | 5/0 | Vκ 12-44 SE (0) | IR, Igκ |
| 7 | 87352466 | 0/0 | (MMLV) 5 | ED, sema4b, [10] |
| 19 | 41086374 | 2/0 | (MMLV)5 | ED, [10] |
| 11 | 87565207 | 0/4 | 87565197 | ED, [10] |
| 19 | 4155742 | 25/16 | 4155825 | ED, [10] |
| 6 | Vκ 19-25 CE (4) | 0/0 | Jκ2 CE (5) | ED, Igκ, [10] |
| 6 | Vκ 4-50 CE (5) | 0/2 | Jκ5 CE (3) | ED, Igκ, [10] |
| 6 | 69581317 CE (1) | 0/0 | Vκ 4-51 CE (10) | ED, Igκ, [10] |
| 6 | Jκ1 CE (4) | 0/0 | Vκ 8-27 CE (4) | ED, Igκ, [10] |
| 6 | Vκ 4-73 CE (15) | 1/0 | Vκ 4-73 SE (1.3κb) | ED, Igκ, [10] |
| 6 | Jκ4 CE (6) | 47/59 | Vκ 4-70 CE (3) | ED, Igκ, [10] |
| 6 | Vκ 19-32 SE (38) | 24/76 | Vκ 8-27 CE (55) | ED, Igκ |
| 6 | Vκ aj4 CE (4) | 0/2 | Vκ ah4 CE (38) | ED, Igκ |
| 6 | Vκ by9 CE (1) | 0/0 | Vκ gj38c CE (3) | ED, Igκ |
| 6 | Vκ cq1 CE (3) | 9/31 | Vκ gj38c (83) | ED, Igκ |
| 6 | Jκ1 CE (4) | 0/0 | Vκ af4 CE (3) | ED, Igκ |
| 6 | 70673884 | 0/5 | Vκ 8-27 CE (4) | ED, Igκ |
| 16 | Vλ1 CE (6) | 0/24 | No flank | ED, Igλ |
| 6 | No flank | 68/0 | Vκ ko4 CE (1) | ED, Igκ |
Chromosome where integration occurred.
Genomic DNA flanking integrating element is noted relative to Ig gene segment (CE, coding end; SE, signal end; deleted nucleotides from end in parentheses) or UCSC genome browser position (mm9 assembly).
Deletion(s) from 12RS or 23 RS ends of integrating fragment.
Integrations classed as transposition (Tnp), intermolecular recombination (IR), or end donation (ED) as in Figure 2. 2°, consistent with secondary recombination after integration. Genes at target sites are noted in italics. G4; integration within G4 tract or site with high G4 forming potential as determined using http://bioinformatics.ramapo.edu/QGRS/index.php [26]. Previously characterized integrations described in [10].
Flank could not be unambiguously assigned.
Table 2.
Integrations from Table 1 classified by integration mechanism. Total; number for each class of integration, with % of the total integrations in parenthesis. Near Ig RS; the total number of integrations for each class near (typically within 100 bp) of an immunoglobulin locus recombination signal, with the % of near Ig/total integrations for that class in parenthesis.
| Total (%) | Near Ig RS (%) | |
|---|---|---|
| Transposition | 11 (25) | 0 (0) |
| Intermolecular recombinations | 15 (34) | 12 (80) |
| End donations | 18 (41) | 14 (78) |
3.2 Transposition vs. other integration mechanisms
Integrations of the candidate transposon are generally consistent with one of three mechanisms: transposition, intermolecular recombination, or end donation (Figure 2).
The primary pathway for integration of the candidate transposon is probably not transposition, but rather intermolecular recombination [14, 21] (reviewed in [22, 23]). Intermolecular recombinations possess a characteristic signal junction, where one of the signal ends of the integrating fragment is precisely fused to a genomic RS. The other RS is fused to DNA immediately flanking the genomic RS (pseudo-hybrid junction) [19, 21, 24, 25] (Figure 2b, Table 1). As characterized in detail by the Nadel group [21], such events are probably mediated by intermolecular recombination (or recombination in trans): a V(D)J recombination between the extrachromosomal RS in the signal end fragment and the genomic RS. Association of intermolecular recombinations with lymphoid malignancy has been well established [24, 26]; in this regard, we note a non-Ig locus intermolecular recombination in our data set targeted the first intron of Pax5 (Table 1), a site rearranged in a large fraction of in acute lymphoblastic leukemias [27].
Other integrations (classified as End donations) are characterized primarily by a lack of the previously described distinguishing features (target site duplications and signal junctions; Table 1, Figure 2c). In these events the RAG-bound signal end fragment did not actively participate in breaking the chromosomal target site. Instead, the integrating fragment simply “donated an end” to participate in NHEJ to an independently generated DSB. Consistent with this model integration structures more commonly have deletions of signal ends (Table 2). Nevertheless, these events can often still be linked to V(D)J recombination. In our study, at least one and often both signal ends were usually (14/18) fused to DNA that previously flanked a genomic RS (coding end) to generate a hybrid junction (Figure 2c). Hybrid junctions can be mediated by either NHEJ or RAG mediated transesterification [4] (see also section 1, section 3.2). An interesting (though unlikely) [15, 21, 28] possibility is that at least a portion of these events (those without deletions from signal ends) could be mechanistically equivalent to integration by RAG-directed transposition.
In cultured mammalian cells integrations clearly consistent with transposition accounts for 10% [18] to 25% [19](Table 2) of integrations, but this frequency may be significantly less in vivo. A genome wide-assessment of signal end fragment integrations [14] that occur during T cell development identified many events consistent with the other two mechanisms described above, but no obvious transpositions. About half of the events recovered (22/43) integrated near a genomic RS and formed either a signal junction or a hybrid junction; in only two cases could signal end integration be plausibly attributed to transposition (no deletion of signal end sequence, and no non-templated additions or nearby RS). Consistent with transposition, both examples (EL 2-4 and b108; [14]) integrated into “good” transposition targets (GC rich DNA, with one adjacent to a G tract; see section 3.2 below). However, the presence of target site duplications could not be verified (the other end of the integrating fragments were not recovered). Regardless, integration by transposition is significantly less frequent for T-cell receptor loci donors, in vivo, when compared to (for example) a transgene donor, in a transformed pre-B cell line – 0-2/43 [14], relative to 11/44 (Table 2) (p<0.01, Fisher's exact test).
There are many possible explanations for this difference. Defective DNA damage responses in cultured cells may allow for abnormally long persistence of signal end intermediates [29], and consequently more frequent transposition. RAG protein-signal end complexes assembled at endogenous loci may be assembled differently [30], and this could make them less prone to transposition, relative to RAG protein-signal end complexes assembled at artificial recombination substrates. The different cell types studied (B cells/fibroblasts in cell culture, vs. T cells in vivo) may even make unanticipated contributions. For example, TdT is active in thymocytes in vivo but not in pre-B cells active in Igκ recombination or in fibroblasts [31]: TdT could conceivably suppress transposition by modifying the substrate (donor 3′ termini), or by competing with the RAGs for donor 3′ termini. An additional not-trivial issue may be that transposition targets low complexity sequence (see 4.2, below), and these events are more difficult to recover. In vivo studies don't allow for a cell-based cloning step, making it likely that difficult to recover integration sites are under-represented. Finally, it must be emphasized that the sample size of these data sets are small. The difference in frequencies, while significant, may thus not be very large.
3.3 The lymphoid genome – a victim of hit-and-run transposition?
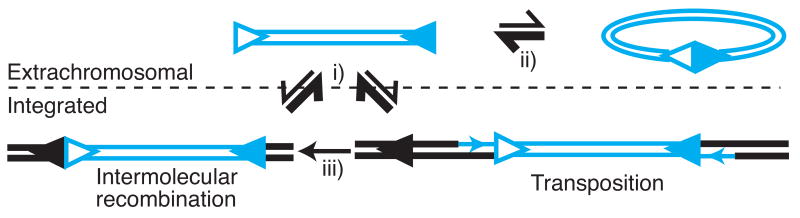
The transposon associated with V(D)J recombination accumulates in cells extra-chromosomally. It is generally assumed that this is because the RAGS perform the integration step in cells very badly. Another possible explanation, though, is that they are much better at excision: integration might be frequent, or at least more frequent than current estimates suggest, but efficient re-excision/secondary recombination of the integrated transposon results in accumulation of the extrachromosomal form. The transposon could effectively be in equilibrium between extrachromosomal and integrated forms, with the extrachromosomal form favored (Figure 3, i) (at least as long as RAG expression is sustained and the signals remain intact). Excision of the recently integrated transposon can occur through a transesterification reaction within branched integration intermediates termed disintegration [32], or after gap repair by the more standard cleavage reaction (Figure 1).
Figure 3.
Hit-and-run Transposition. High RAG-mediated cleavage activity may keep the transposon mostly extrachromosomal (i), consistent with accumulation of linear fragment at the expense of circle (ii) and observed secondary recombinations after integration (iii).
Several arguments make “hit and run” transposition tenable as a significant mechanism. The eventual fate of the extrachromosomal transposon fragment is usually its resolution by NHEJ into an extrachromosomal circle. However, as long as RAG expression is sustained, the linear form is often favored at the expense of the circle [33] primarily due to re-cleavage of the signal junction by the “nick-nick” mechanism [34] (Figure 3, ii). Moreover, the integrated element (however it integrates) not infrequently undergoes secondary recombination with a nearby RS ([24, 35])(Table 1). Such secondary recombinations are evident only when one of the signal end fragment's RS pairs with a nearby genomic RS (e.g. 2° in table 1, Figure 3, iii) - if both RS of the original integrating pair directs the recombination, as might be preferred, the signal end fragment excises and erases the evidence of the initial integration (“hit and run” integration; Figure 3, i), as noted above.
A presumption of efficient (>90%) excision or secondary recombination means that hit-and-run transpositions could be a major “hidden” source of near-random (see section 4.2) double strand breaks in cells active in V(D)J recombination. Cancer-causing chromosome abnormalities might be partly the consequence of aberrant repair of these breaks, in the same way that hit and run transposition is effectively employed as a tool for genome mutagenesis in fruit flies (e.g. [36]).
4 Where do they go?
A significant factor in determining the danger of a transposon to its host is its selection of target site. In vitro, RAGs directs transposition with only loose target site preference, and this is also apparent in cellular transpositions.
4.1 Transposition vs. other integration mechanisms
The candidate transposon can integrate by transposition, intermolecular recombination, or end donation. A comparison of where these integrations occur according to class is sufficient to clearly emphasize transpositions' loosely defined target site preference. Intermolecular recombinations and end donations both show an equivalent and strong preference for integration near a RS within an active receptor locus (∼80% in our study; Table 2, Figure 4). Preferential targeting to receptor loci RS is also observed in the Schlissel group's study [14], though to a lesser degree, and is consistent with very high frequencies of intermolecular recombination targeted to specific T cell receptor RS during T cell development [21]. Notably, aberrant resolutions of coding ends show a similar strong targeting preference (∼80%) for actively recombining receptor loci [37].
Figure 4.
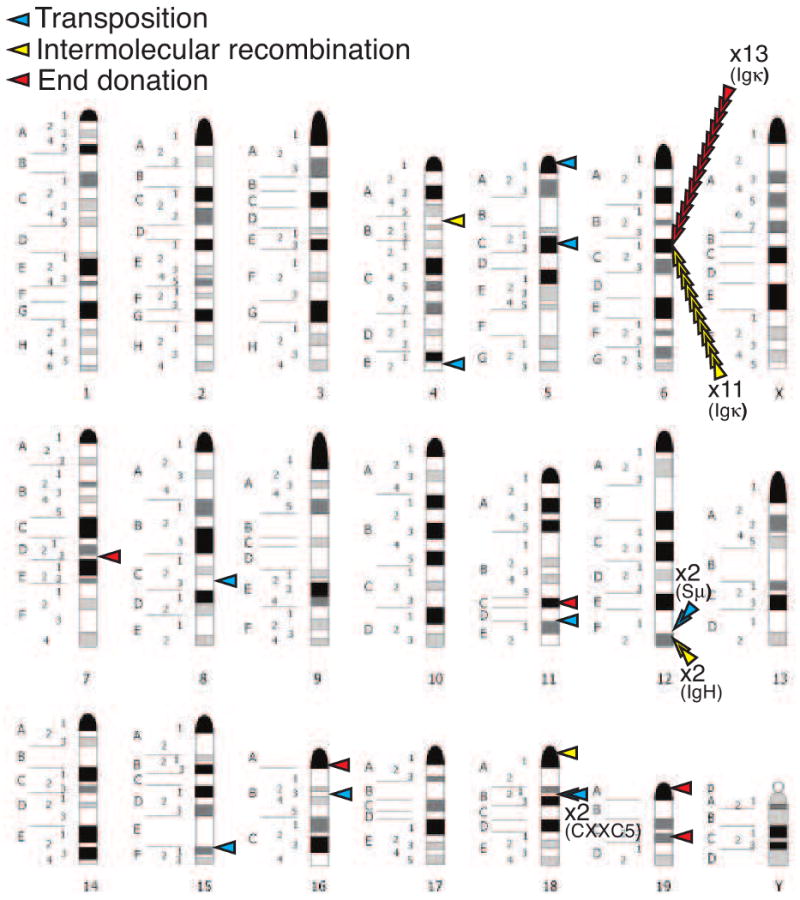
Location of integrations mapped onto a mouse chromosome idiogram (http://www.pathology.washington.edu/research/cytopages/ideograms/mouse). Integrations are classified as in Figure 3 and Tables 1, 2.
By comparison, target sites for transpositions were significantly more widely distributed; for example, the 11 transpositions we recovered were distributed over 8 different chromosomes [19] (Table 1, Figure 4). Relative to the other mechanisms, therefore, transposition is far more likely to integrate outside DNA already active for V(D)J recombination.
4.2 Sites of predicted secondary structure are hot spots
Target sites of V(D)J recombination associated transposition in cells are generally widely distributed, or a least more widely distributed than other reintegration mechanisms. Nevertheless, there were two regions that might be considered “hot-spots” – both sites had two independent transposon integrations occur near each other (Table 1, Figure 4). Two integrations were observed 1-2kb apart in the highly repetitive Sμ region of the IgH locus. Strikingly, two other events integrated 370 bp apart in low complexity DNA (an intron of the CXXC5 gene) in chromosome 18. Together with an additional four events (8/11 total), these integrations occur in regions with high potential [38] for forming a class of secondary structure termed G4 DNA (also termed G quartet, G quadruplex; reviewed in [39]); for example, 5/8 are within a tract of 4 or more Gs. This is a higher frequency than expected by chance, even after taking into account the known preference for targeting of transposition to GC rich DNA [5, 6, 20]. RAG-mediated transposition is generally directed towards secondary structures in vitro, including hairpin tips [40, 41] and bubbles [20], and targeting to G4 DNA is consistent with these preferences. In this regard, we note we previously characterized an integration into the center of a chromosome 5 inverted repeat as consistent with a transposition targeted to a hairpin tip [19].
5 Concluding remarks
As yet no V(D)J recombination transposition event has been definitively linked to a lymphoid malignancy. However, more sensitive and systematic analyses of cancer genomes (e.g. using array-based comparative genome hybridization; aCGH) suggests the full extent of genome instability in lymphoid malignancy is only now being appreciated [42]. It should be noted, though, that transposition does not vary copy number and may involve relatively small-scale (1-100 kbp) aberrations, thus may not be easily detected by either aCGH or spectral karyotyping. Moreover, ongoing recombination (including secondary recombinations [24, 35] and excisions, or hit-and-ran transpositions - section 3.3) may obscure the contributions of early and cancer-causing transpositions.
What is clear is that DNA elements mobilized entirely by the RAGs have disrupted a variety of genes (HPRT, CDK6, CXXC5 - twice, ATXN7L3, and NF I/X) in cells active in V(D)J recombination [16, 19](Table 1). Evidently, transposition associated with V(D)J recombination is a threat to the stability of lymphocyte genomes and is a potential contributor to lymphoid malignancy. Just how much of a threat? With regard to frequency, the putative transposon integrates into lymphocyte genomes often enough – on the order of one integration in 10,000 excisions [14, 19, 21]. However, less than 1/3rd (and possibly much less than 1/3rd; see 3.2 above) of these integrations can be clearly attributed to transposition [14, 18, 19]. The current data sets have small sample sizes, though, making it important for future studies to more accurately assess this frequency, particularly in in vivo settings (e.g. as in [14]). Additionally, for a variety of reasons noted above (section 3) transposon integrations may be under-reported. Very “loose” target site preferences, at least relative to other integration mechanisms (section 4.1), also means transposition is more likely to cause a problem when it does occur. For example, in our unbiased study of 44 aberrant signal end integrations only 7 disrupted a non-Ig gene, and 5 of these 7 were transpositions (Table 1).
The V(D)J recombination associated-transposon: born to be wild, and not yet fully tamed.
Acknowledgments
Work described above from the Ramsden lab was supported by the Searle Scholar foundation, the Leukemia and Lymphoma Society, the Department of Homeland security, and PHS grants CA 97096 and CA 84442.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gellert M, et al. V(D)J recombination: links to transposition and double-strand break repair. Cold Spring Harb Symp Quant Biol. 1999;64:161–7. doi: 10.1101/sqb.1999.64.161. [DOI] [PubMed] [Google Scholar]
- 2.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271(5255):1592–4. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432(7020):995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 4.Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280(5361):301–3. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 5.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94(4):463–70. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394(6695):744–51. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 7.Brandt VL, Roth DB. V(D)J recombination: how to tame a transposase. Immunol Rev. 2004;200:249–60. doi: 10.1111/j.0105-2896.2004.00161.x. [DOI] [PubMed] [Google Scholar]
- 8.Chatterji M, Tsai CL, Schatz DG. New concepts in the regulation of an ancient reaction: transposition by RAG1/RAG2. Immunol Rev. 2004;200:261–71. doi: 10.1111/j.0105-2896.2004.00167.x. [DOI] [PubMed] [Google Scholar]
- 9.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–48. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 10.Matthews AG, Oettinger MA. Regulation of RAG transposition. Adv Exp Med Biol. 2009;650:16–31. doi: 10.1007/978-1-4419-0296-2_2. [DOI] [PubMed] [Google Scholar]
- 11.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 2003;22(8):1931–8. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J Biol Chem. 2004;279(6):4034–44. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 13.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22(8):1922–30. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curry JD, et al. Chromosomal reinsertion of broken RSS ends during T cell development. J Exp Med. 2007;204(10):2293–303. doi: 10.1084/jem.20070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol Cell. 2001;8(6):1383–90. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 16.Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22(6):1381–8. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clatworthy AE, Valencia MA, Haber JE, Oettinger MA. V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell. 2003;12(2):489–99. doi: 10.1016/s1097-2765(03)00305-8. [DOI] [PubMed] [Google Scholar]
- 18.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26(4):1558–68. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20(12):1575–82. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai CL, Chatterji M, Schatz DG. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 2003;31(21):6180–90. doi: 10.1093/nar/gkg819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanura K, et al. In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability. PLoS Biol. 2007;5(3):e43. doi: 10.1371/journal.pbio.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnal SM, Roth DB. Excised V(D)J recombination byproducts threaten genomic integrity. Trends Immunol. 2007;28(7):289–92. doi: 10.1016/j.it.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Marculescu R, et al. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair (Amst) 2006;5(9-10):1246–58. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Marculescu R, Vanura K, Le T, Simon P, Jager U, Nadel B. Distinct t(7;9)(q34;q32) breakpoints in healthy individuals and individuals with T-ALL. Nat Genet. 2003;33(3):342–4. doi: 10.1038/ng1092. [DOI] [PubMed] [Google Scholar]
- 25.Messier TL, O'Neill JP, Finette BA. V(D)J recombinase mediated inter-chromosomal HPRT alterations at cryptic recombination signal sequences in peripheral human T cells. Hum Mutat. 2006;27(8):829. doi: 10.1002/humu.9440. [DOI] [PubMed] [Google Scholar]
- 26.Tycko B, Sklar J. Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells. 1990;2(1):1–8. [PubMed] [Google Scholar]
- 27.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 28.Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7(1):37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 29.Dujka ME, Puebla-Osorio N, Tavana O, Sang M, Zhu C. ATM and p53 are essential in the cell-cycle containment of DNA breaks during V(D)J recombination in vivo. Oncogene. 2010;29(7):957–65. doi: 10.1038/onc.2009.394. [DOI] [PubMed] [Google Scholar]
- 30.Hempel WM, Stanhope-Baker P, Mathieu N, Huang F, Schlissel MS, Ferrier P. Enhancer control of V(D)J recombination at the TCRbeta locus: differential effects on DNA cleavage and joining. Genes Dev. 1998;12(15):2305–17. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–7. [PubMed] [Google Scholar]
- 32.Melek M, Gellert M. RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell. 2000;101(6):625–33. doi: 10.1016/s0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 33.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9(19):2409–20. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 34.Neiditch MB, Lee GS, Huye LE, Brandt VL, Roth DB. The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol Cell. 2002;9(4):871–8. doi: 10.1016/s1097-2765(02)00494-x. [DOI] [PubMed] [Google Scholar]
- 35.Lewis S, Gifford A, Baltimore D. DNA elements are asymmetrically joined during the site-specific recombination of kappa immunoglobulin genes. Science. 1985;228(4700):677–85. doi: 10.1126/science.3158075. [DOI] [PubMed] [Google Scholar]
- 36.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118(3):461–70. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahowald GK, et al. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc Natl Acad Sci U S A. 2009;106(43):18339–44. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34(Web Server issue):W676–82. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13(12):1055–9. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 40.Lee GS, Neiditch MB, Sinden RR, Roth DB. Targeted transposition by the V(D)J recombinase. Mol Cell Biol. 2002;22(7):2068–77. doi: 10.1128/MCB.22.7.2068-2077.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posey JE, Pytlos MJ, Sinden RR, Roth DB. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4(11):e350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullighan CG, Downing JR. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009;23(7):1209–18. doi: 10.1038/leu.2009.18. [DOI] [PubMed] [Google Scholar]