Abstract
Background
Cyclooxygenase-2 (COX-2) plays a key role in breast cancer progression and metastasis. Effective therapeutic targeting of COX-2 would require the knowledge of whether a tumor is addicted to COX-2, and if we can counter the potential resistance to anti-COX-2 therapy. Herein we tested the hypothesis that celecoxib-resistance involves selection of cancer cells that overexpress COX-2.
Materials and Methods
We selected celecoxib-resistant (CER) variants from two metastatic cell lines, SUM149 inflammatory breast cancer (IBC) cell line and MDA-MB-231-BSC60 cell line, by culturing them in the presence of celecoxib. We measured the relative levels of COX-2 protein and its network components Bcl-2, Bcl-xL, and Bax in the parental cell lines and their CER variants by western blotting. To determine whether celecoxib-resistance would increase tumorigenicity, we performed an in vitro clonogenicity assay. We determined the statistical significance of differences between the groups using the two-sample t-test.
Results
Both the celecoxib-resistant cell lines SUM149-CER and BSC60-CER produced significantly higher levels of COX-2 protein than their parental counterparts (p < 0.05). The CER variants produced a reduced level of pro-apoptosis protein Bax (both cell lines) and increased levels of anti-apoptosis proteins Bcl-2 (BSC60) or Bcl-xL (SUM149). Importantly, the CER variants had significantly higher clonogenicity than their parental cell lines (p < 0.05). The siRNA-mediated COX-2 knockdown in SUM149-CER cell line resulted in a significant decrease in clonogenicity and in Bcl-xL and Bcl-2 protein levels, thus supporting our hypothesis.
Conclusion
Celecoxib-resistant variant cells present in breast cancer cell lines overexpress COX-2, which is robustly linked with survival pathways and clonogenicity. Since COX-2 is important in the variant cancer cells of aggressive nature, it represents a good therapeutic target.
Keywords: Breast cancer, inflammatory breast cancer, tumor heterogeneity, COX-2, metastasis, apoptosis, targeted therapy, therapy resistance
INTRODUCTION
Cyclooxygenase-2 (COX-2), a mediator of inflammation, is expressed in premalignant and malignant breast epithelial cells, where it plays a key role in breast cancer progression and metastasis [1,2]. COX-2 blockade is effective for both cancer prevention and therapy [reviewed in 3]. Retrospective epidemiological studies indicate a correlation between the use of specific and non-specific COX-2 inhibitors and the reduced breast cancer incidence. As an example, a case-controlled study showed that exposure to selective COX-2 inhibitors celecoxib or rofecoxib produced a significant (71%) reduction in the risk of breast cancer, underscoring their strong potential for breast cancer chemoprevention [4]. An extensive meta-analysis (involving 38 studies) performed recently supports an inverse association between non-steroidal anti-inflammatory drugs (NSAID) use and risk of breast cancer [5].
Overexpression of COX-2 induces genomic instability in premalignant breast epithelial cells and in breast cancer cells [6,7]. Others have shown that COX-2 promotes the progression of ductal carcinoma in situ to invasive breast carcinoma [8], and that the COX-2 gene is a part of the gene expression signatures for metastases to lungs and brain [9,10]. We have investigated the role of COX-2 protein in metastasis of breast cancer to bone, which is the major site of metastasis seen in breast cancer. In a preclinical mouse model, we have shown that COX-2 is active in metastasis of breast cancer to bone via production of prostaglandin E2 (PGE2), interleukin-8 (IL-8), interleukin-11 (IL-11), and urokinase plasminogen activator (uPA) [11–14]. Furthermore, expression of COX-2 in stages I–III of breast cancer correlates with the presence of disseminated tumor cells (DTC) in the bone marrow [15], which are independent predictors of clinical metastases [16]. It is interesting that we also detected COX-2 protein in solitary DTCs present in the bone marrow of breast cancer patients, further supporting a role of COX-2 in bone marrow micrometastasis (BMM) [15].
COX-2 is highly expressed in inflammatory breast cancer (IBC), an aggressive subset of breast cancers [17]. We are particularly interested in IBC because of an urgent need for effective therapies against this disease. COX-2 may be a driver of IBC as indicated by the analysis of 60 NF-κB targets in non-IBC versus IBC, which revealed that COX-2 was one of the only two genes, (the other one being CXCL1), that were also expressed in metastases in addition to primary tumors [17]. Effective therapeutic targeting of COX-2 would require the knowledge of whether a tumor is addicted to COX-2 and whether the potential resistance to anti-COX-2 therapy can be countered. A clearer understanding of the mechanisms of celecoxib resistance should identify new models for COX-2 inhibition as a therapeutic approach for IBC and other aggressive forms of breast cancer that overexpress COX-2.
MATERIALS AND METHODS
Cell lines and culture
The SUM149 inflammatory breast cancer cell line, originally obtained from Dr Stephen Ethier (Barbara Ann Karmanos Cancer Institute, Detroit, MI, USA), was grown in Ham’s F-12 medium supplemented with 5% fetal bovine serum (FBS), 5 µg/ml of insulin, 1 µg/mL of hydrocortisone, 100 U/ml of penicillin, and 100 µg/ml of streptomycin in a humidified 5% CO2 atmosphere. The MDA-MB-231-BSC60 (abbreviated to BSC60) cell line, a metastatic variant of MDA-MB-231 isolated in our laboratory after two passages in female nude mice by cardiac ventricle inoculation followed by cell culture from bone metastases [12,14], was grown in RPMI 1640 medium supplemented with 10% FBS.
Selection and culture of celecoxib-resistant variants
We added celecoxib (10 µM final concentration; LKT Laboratories, Inc., St. Paul, MN) to the culture medium soon after the SUM149 cells (passage 16) were trypsinized and plated. The medium was changed with new medium containing celecoxib two times to remove dead floating cells. After 15 days, we pooled the colonies of surviving cells by trypsinization, and plated into the medium with 20 µM celecoxib for 16 days followed by 10µM celecoxib for 11 days. After a selection with celecoxib for 3 rounds, the variants were considered celecoxib-resistant (named SUM149-CER passage 0), and were maintained in the presence of 10 µM celecoxib unless specified otherwise. All experiments were carried out within 6 passages after the initial selection on celecoxib.
The MDA231-BSC60-CER variant was established with 5 rounds of selection with celecoxib. The BSC60 cells (passage 16) were treated with 20 µM for 17 days, 40 µM for 14 days, and 3 passages with 10 µM for 10, 11,and 10 days respectively. After selection, these cells were considered celecoxib-resistant, named BSC60-CER passage 0, and were maintained in the presence of 10 µM celecoxib unless specified otherwise. All experiments were carried out within 11 passages after the initial selection on celecoxib.
Western immunoblotting
Expression of COX-2 protein was detected by western blot analysis, as described previously [6]. Samples with equal amounts of protein were separated on an 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a 0.45-µm nitrocellulose membrane. The COX-2 protein was detected with a monoclonal antibody (Cayman Chemical, Ann Arbor, MI) and with the ECL Advance western blot detection reagents (Amersham Biosciences). To analyze Bcl-2, Bcl-xL, and Bax protein by western blotting, they were resolved on a 12% gel, transferred to a 0.2-µm nitrocellulose membrane, and detected with monoclonal antibodies (all from Cell Signaling Technology, Danvers, MA). The filters were stripped by incubating the membrane in 0.5% Triton X-100 and were re-probed with a monoclonal β-actin antibody (Sigma-Aldrich, St. Louis, MO) to serve as gel-loading controls. We performed each western blot at least 3 times. We quantified the COX-2, Bcl-2, Bcl-xL, Bax, and beta-actin protein bands on X-ray films by the ImageJ image processing program (National Institutes of Health).
Clonogenicity assay
To determine the tumorigenic potential of cancer cells, we performed clonogenicity assays as described previously [7] with some modifications. Subconfluent cultures of SUM149 and SUM149-CER were dissociated with trypsin-EDTA and plated at 7,500 cells/10 cm dish in Ham’s F-12 medium supplemented with 0.5% fetal bovine serum, 0.5 µg/ml of insulin, and 0.1 µg/mL of hydrocortisone. These cells were grown for 25 days and then were stained for 15–30 minutes in 0.1% crystal violet (w/v) plus 6% glutaraldehyde fixative (v/v) dissolved in water [18]. The plates were rinsed in water 5–10 times, until no more dye was detected in the rinse. After air drying the colonies, we counted the colonies with the Bio-Rad Gel Documentation System XR equipped with Quantity One version 4.6.1 software and photographed them with a digital camera.
We performed the clonogenicity assays with BSC60 and BSC60-CER cells as described above, but in RPMI 1640 complete media with 10% FBS (7,500 cells/10 cm dish plated, and colonies stained after 13 days). In the second assay involving low-serum, the cells were plated at 30,000 cells/10 cm dish in RPMI 1640 medium supplemented with 3% FBS, and colonies were stained after 15 days.
COX-2 knockdown with siRNA
We performed COX-2 knockdown in SUM149-CER cell line using COX-2 specific Silencer Select siRNAs s11472 and s11473, and a Silencer Select negative control #1 siRNA (all from Applied Biosystems, Foster City, CA). Soon after trypsinization to dissociate cells on a confluent culture dish, we transfected 120,000 cells plated in 15 ml low-serum medium (see clonogenicity assay) in a 10 cm culture dish with 20 nM siRNAs (final concentration) that were pre-incubated with 30 µL siPORT NeoFX transfection agent (Applied Biosystems) in 1.2 ml OPTI-MEM serum-free medium (Invitrogen, Carlsbad, CA), according to manufacturer’s instructions. After 15 days, colonies were stained with crystal violet. For the western blot analysis of COX-2 and other proteins after siRNA transfection, 240,000 SUM149-CER cells were transfected as above, in parallel to the clonogenicity assay. We prepared cell lysates at 16 days after transfection, and subjected them to western blotting analysis.
Statistical analysis
We performed statistical analysis of western blots and clonogenicity assays using the two-sample t-test. For western blots, we normalized the relative protein levels by dividing with β-actin signal. We used the normalized values from three bots to determine average ± standard deviation, and to determine p-value. P ≤ 0.05 was considered significant.
RESULTS
Selection of celecoxib-resistant variants
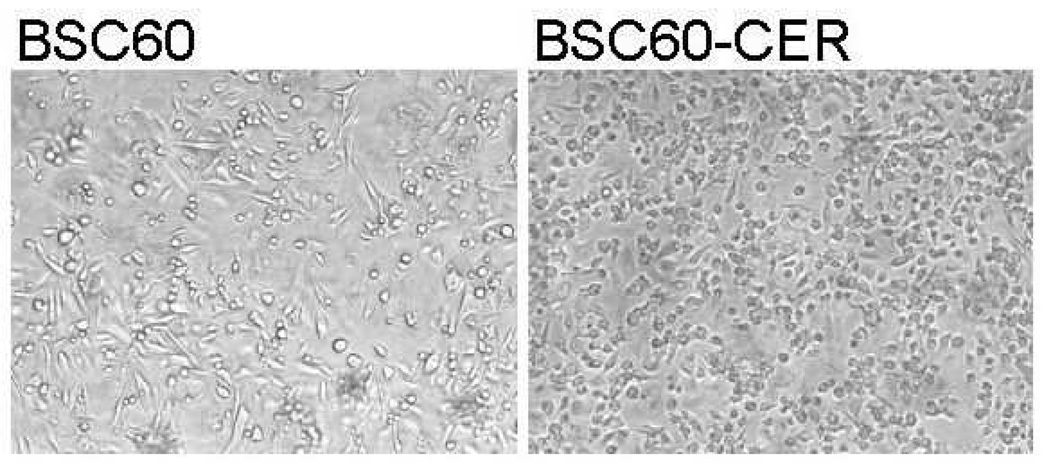
To gain an understanding of how resistance to therapy may develop in breast cancer when COX-2 is inhibited with celecoxib, we chose two appropriate cell line models: 1) the SUM149 IBC cell line since COX-2 has a dominant role in IBC, and 2) the BSC60 metastatic variant of MDA-MB-231 that has a functional COX-2 network [11–14]. We treated both the cell lines with celecoxib as described in Materials and Methods and selected the surviving variants that grew and gave rise to stable cell lines that grow continuously in the presence of 10 µM celecoxib. One significant morphological difference we noted was between BSC60-CER and BSC60; when cells were grown to confluency, the CER variants grew to a significantly higher cell density than the parental cells (compare the images in Fig. 1). This difference was due to the fact that the BSC60 cells stopped dividing at a lower cell density than the BSC60-CER did. If left on the dish for a long period after confluency, the parental BSC60 would come off the dish, while the CER variants continued to pile up on each other, thus reaching a higher cell density. As an example, the average cell count per 10 cm dish after 17 days of culture, beyond the confluency stage, was more than double in BSC60-CER than BSC60 (5.7 million cells versus 2.3 million cells, respectively).
FIG. 1.
Morphology of cells. BSC60 (passage 9) and BSC60-CER (passage 7) cultures were allowed to grow beyond confluency for a total 17 days, and were examined under a Nikon microscope. Representative images of both the cultures photographed at 100X magnification with a digital camera are shown. The CER variant reached a significantly higher cell density than the parental cell line, which was confirmed by counting cells on day 18 (5.7 million cells/10 cm dish versus 2.3 million cells/10 cm dish respectively).
Increased level of COX-2 protein in CER variants
To investigate the mechanism behind the celecoxib-resistance, first we analyzed the target of celecoxib, COX-2. One obvious mechanism of resistance would be that the CER variants overexpress COX-2 thus escaping the cytotoxic effects of celecoxib. Our past investigations involving dose-dependent inhibition with celecoxib indicated that a small percentage of the COX-2 protein would be resistant (B Singh and A Lucci, unpublished data); therefore, CER variants overexpressing COX-2 may be expected to contain a higher overall pool of celecoxib-resistant COX-2 than the parental cell line. To test our prediction, we compared the level of COX-2 protein in CER variants and their parental cell lines by western blotting. Both SUM149-CER and BSC60-CER produced significantly higher level of COX-2 protein than their parental counterparts (Figs. 2 and 3). Although the basal level of COX-2 expression was superior in SUM149 cell line than BSC60, the relative increase was higher in BSC60-CER (3.9-fold as determined by densitometry of bands, 0.2819 ± 0.0439 versus 0.0732 ± 0.0139, p < 0.05) than SUM149-CER (2.9-fold, 0.4494 ± 0.0344 versus 0.1566 ± 0.0246, p < 0.05) as compared to their parental cell lines.
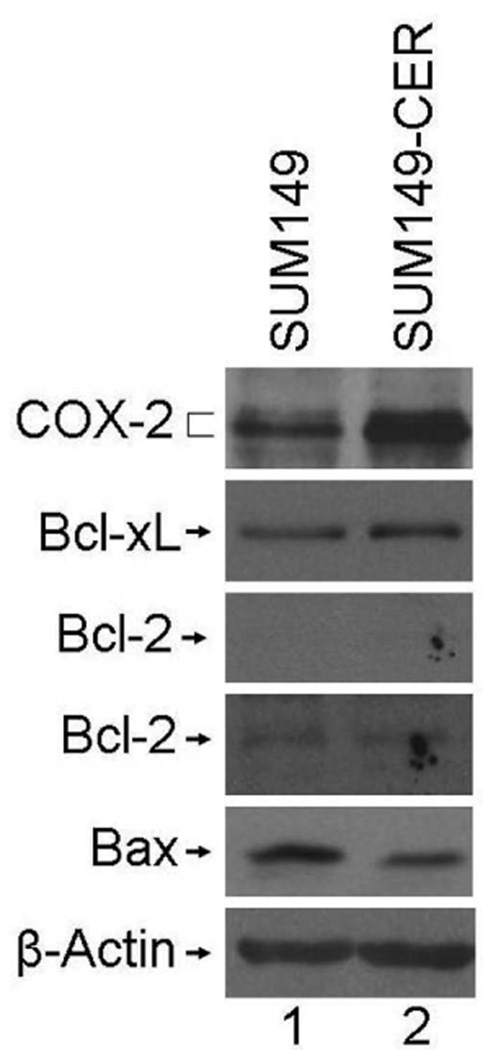
FIG. 2.
Increased COX-2, increased Bcl-xL, and decreased Bax levels in SUM149-CER variants. The cell lysates prepared from both the cell lines in normal growth conditions were subjected to western blot analysis as described in Materials and Methods. Two exposures of the Bcl-2 blot are shown- a light exposure (upper panel) identical to the Bcl-2 blot for BSC60 (Fig. 3A), and a longer exposure to visualize the weak Bcl-2 band (lower panel). SUM149, passage 24; SUM149-CER, passage 6.
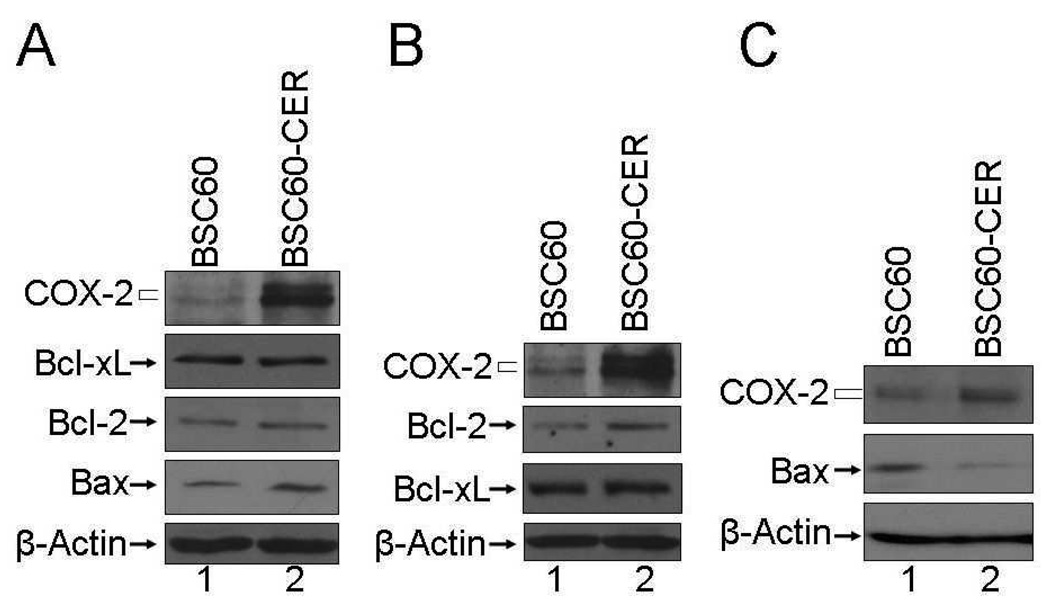
FIG. 3.
Increased COX-2, increased Bcl-2, and decreased Bax levels in BSC60-CER variants. The cell lysates prepared from both the cell lines were subjected to western blot analysis as described in Materials and Methods. A. Early passage cells grown in normal serum (10% FBS). BSC60, passage 24; BSC60-CER, passage 4. B. Early passage cells (same as in A) were grown in normal serum until subconfluency and then were shifted to low-serum (0.5% FBS) for 24 hours before preparing cell lysates for a western blot. C. Late passage cells grown in normal serum (10% FBS). BSC60, passage 13; BSC60-CER, passage 11.
Evidence of increased survival signaling in CER variants
Apart from supporting our hypothesis regarding the mechanism of celecoxib resistance, our CER variants may provide an opportunity to investigate the COX-2 networks that are functional in the context of aggressive and metastatic disease. In this regard, we focused on some known members of anti-apoptosis/apoptosis machinery that appear to be linked with COX-2 in breast cancer, namely Bcl-2, Bcl-xL, and Bax [19]. This linkage is based on findings from tumors in MMTV-LTR-COX-2 transgenic mice and from COX-2 transfected MCF7 breast cancer cell line [7]. Another reason we chose to analyze these proteins is for their well-known involvement in pan-resistance to anti-cancer therapy [20].
We found by western blot analysis that the level of anti-apoptosis protein Bcl-xL increased (2.2-fold increase in a representative blot) and the level of pro-apoptosis protein Bax decreased (45% decrease in a representative blot) in SUM149-CER as compared to SUM149 (Fig. 2). The Bcl-2 protein is produced at a relatively low level in SUM149 cell line, which did not change significantly in the CER variant (Fig. 2). A similar analysis performed with BSC60-CER showed that the level of Bcl-2 anti-apoptosis protein is increased (63% increase in a representative blot) and the level of Bax protein is decreased (71% decrease in a representative blot) in BSC60-CER as compared to BSC60 (Fig. 3B and 3C). The Bcl-xL protein is produced at a comparable level in BSC60 and BSC60-CER. Overall these results indicate that the increased level of COX-2 protein produced in CER variants is functional and linked with the components of survival network in way which would be consistent with increased survival.
There is a noteworthy point regarding the Bcl-2 protein increase in BSC60-CER. This increase as compared to the parental cell line was not observed in normal growth medium with 10% FBS, but was observed when the cells were shifted to the low-serum medium for 24 h (Fig. 3B). This result implies that serum-mediated signaling may mask COX-2 specific signaling under these growth conditions. Another important point pertains to the Bax protein decrease in BSC60-CER. The decrease in Bax protein was not observed in an early passage (passage 4) even though the COX-2 protein level was high at this stage (Fig. 3A); however, it was observed in late passage (passage 11; Fig. 3C). The simple explanation of this result would be that continued COX-2 expression may trigger some epigenetic alterations that are responsible for a subsequent Bax decrease. Alternatively, COX-2 overexpression may trigger pro-apoptotic effects in some subpopulations during early passage, thus causing an increase in Bax at this stage, which would mask a decrease in true stable CER variants.
Stability of COX-2 protein overexpression in the CER variants
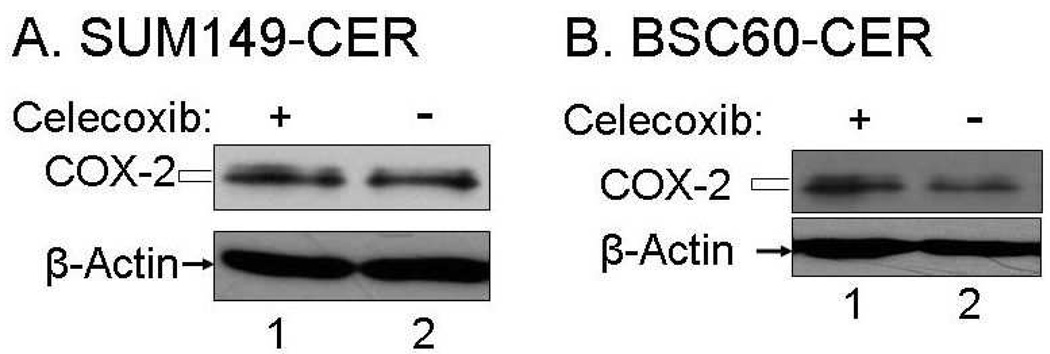
One important question that would be important in the clinical setting is whether the increased COX-2 expression observed in the CER variants persists after celecoxib is not added to the culture medium. Interestingly, we observed that lack of celecoxib for 8 days did not result in any significant decrease in COX-2 protein level in SUM149-CER (Fig. 4A). In contrast, COX-2 overexpression in BSC60-CER cells was highly dependent on the continued presence of celecoxib as a withdrawal of celecoxib for 5 days caused a dramatic decrease in the COX-2 protein level (Fig. 4B). One reason for this difference between the cell lines may be due to the difference in the maximum amount of COX-2 tolerated by the cell lines.
FIG. 4.
A decrease in COX-2 protein level upon celecoxib withdrawal from BSC60-CER. A. The BSC60-CER cell line at passage 11 was cultured in parallel with or without 10 µM celecoxib for one passage (5 days) before preparing the cell lysates for the western blot. B. The SUM149-CER cell line at passage 6 was cultured in parallel with or without 10 µM celecoxib for one passage (8 days) before preparing the cell lysates for the western blot.
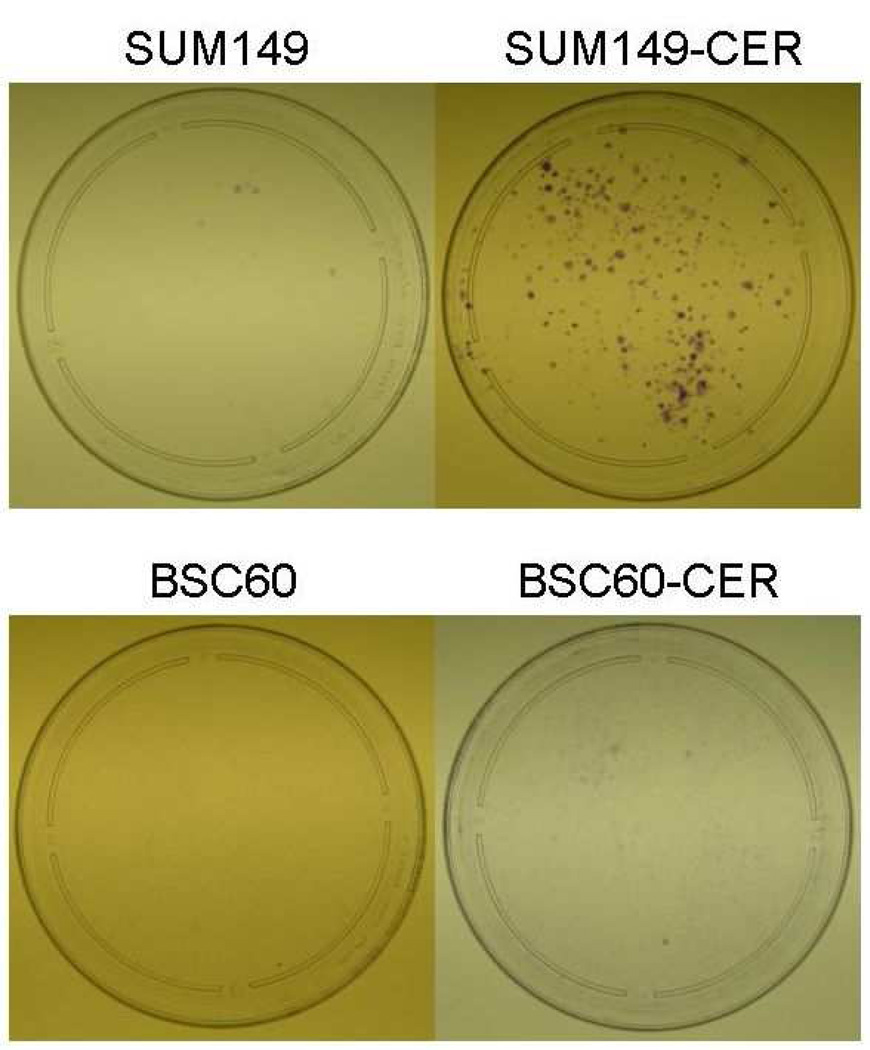
Increased clonogenicity in the CER variants
Next, we investigated whether an increase in the COX-2 protein and associated increase in Bcl-2/Bcl-xL, and decrease in Bax, would translate into increased clonogenicity of the CER variants. The results of this in vitro assay often correlate with tumorigenicity in vivo and also with pan-resistance to anti-cancer therapies. We performed these assays under low-serum conditions to increase the likelihood that the effects of survival pathways altered in CER variants are observed and that the serum-mediated effects are minimized. We observed a significant increase in the clonogenicity of both SUM149-CER and BSC60-CER as compared to their parental counterparts (Fig. 5). As a technical issue, although both the cell lines yielded similar results, this assay is better suited for SUM149-CER than for BSC60-CER. The BSC60-CER requires a high concentration of FBS (minimum 3%) in this assay, and therefore the effects of serum-mediated signaling are not minimized to the same degree as in case of SUM149-CER (which requires only 0.5% FBS).
FIG. 5.
Increased clonogenicity in the CER variants than their parental cell lines. We performed the assay in low-FBS medium as described in Materials and Methods. Comparison of the plates showed that the CER variants are more clonogenic than the parental cell lines, and 2) the CER variants are inhibited with celecoxib in this assay. Passage numbers: SUM149, 24 ; SUM149-CER, 4; BSC60, 9; BSC60-CER, 7.
Effect of COX-2 knockdown
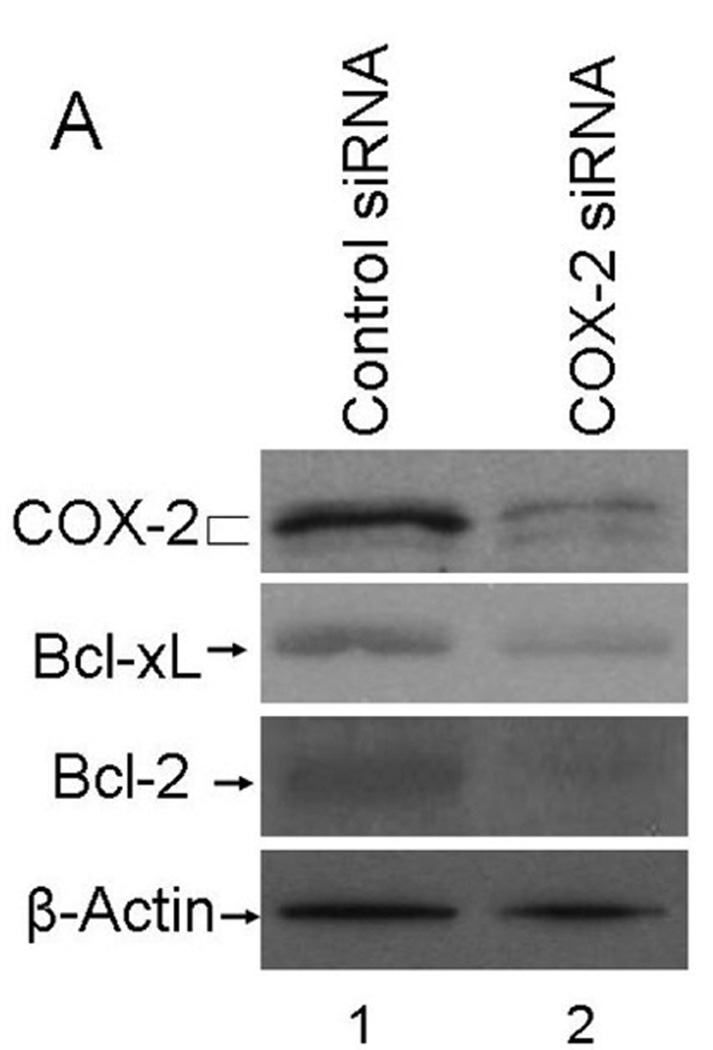
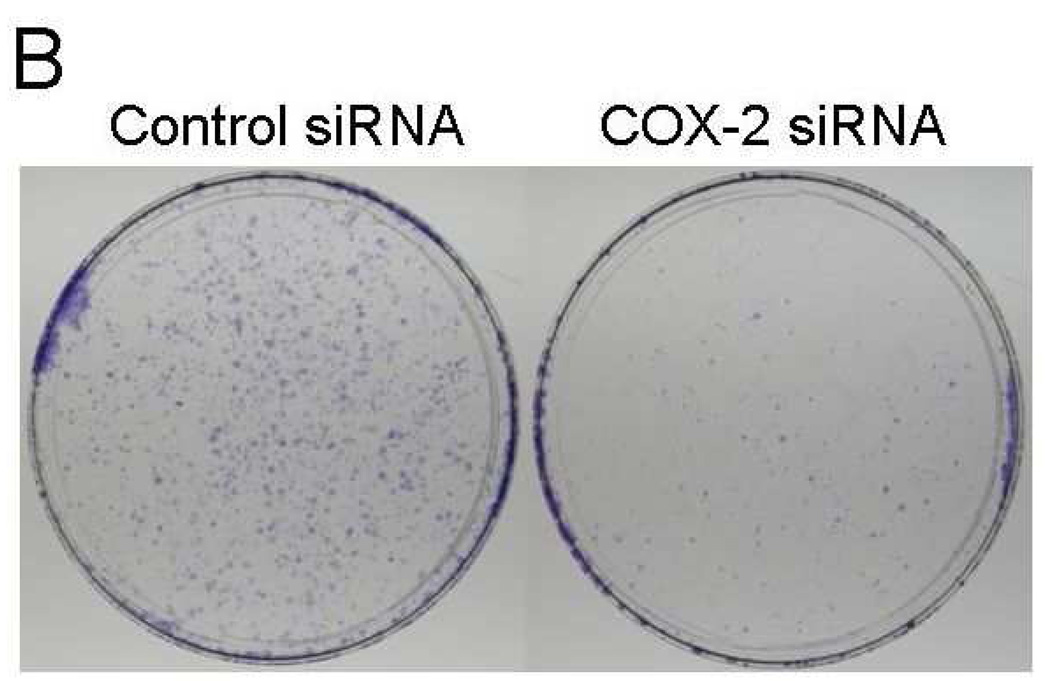
If increased COX-2 level is responsible for increased survival signaling in the CER variants, specific knockdown of COX-2 should reverse this effect. To test this, we performed the siRNA transfection experiment to knockdown COX-2. We performed these experiments with two different COX-2 specific siRNAs (Silencer Select siRNAs s11472 and s11473 from Applied Biosystems, Foster City, CA). The s11473 siRNA caused a superior knockdown of COX-2 than s11472 did (data not shown). The data presented in Figure 6 showed that s11473 siRNA specifically knocked down COX-2 (69% decrease in normalized COX-2 amount based on β-actin levels compared to the negative control siRNA transfected SUM19-CER cells in a representative blot) and there was a corresponding decrease in Bcl-xL and Bcl-2 protein levels (40 and 69% decrease in normalized amounts based on β-actin levels, respectively, in representative blots) (Fig. 6A). Of importance, COX-2 knockdown resulted in a dramatically reduced clonogenicity in SUM149-CER cell line (Fig. 6B). The decreased clonogenicity was observed both as reduced number of colonies (a 76% decrease from 301 to 70 colonies per 10 cm dish in a representative experiment) and as reduced size of colonies in COX-2 siRNA-transfected cells as compared to negative control siRNA-transfected cells. Transfection with COX-2 specific siRNA also caused COX-2 knockdown under normal growth conditions (in culture medium with 5% FBS; data not shown). We chose previously established clonogenicity assay conditions, involving growth on low-FBS medium for this experiment to minimize the effects of FBS.
FIG. 6.
Inhibition of survival signaling and clonogenicity in SUM19-CER cell line by COX-2 knockdown. A. SUM149-CER cell line at passage 6 was transfected with a negative control siRNA or COX-2 specific siRNA and subjected to western blot analysis as described in Materials and Methods. B. We performed a clonogenicity assay with SUM149-CER cells transfected with a negative control siRNA or COX-2 specific siRNA in parallel with the western blotting experiment shown in panel A. Representative western blotting and clonogenicity assay results obtained in two experiments are shown.
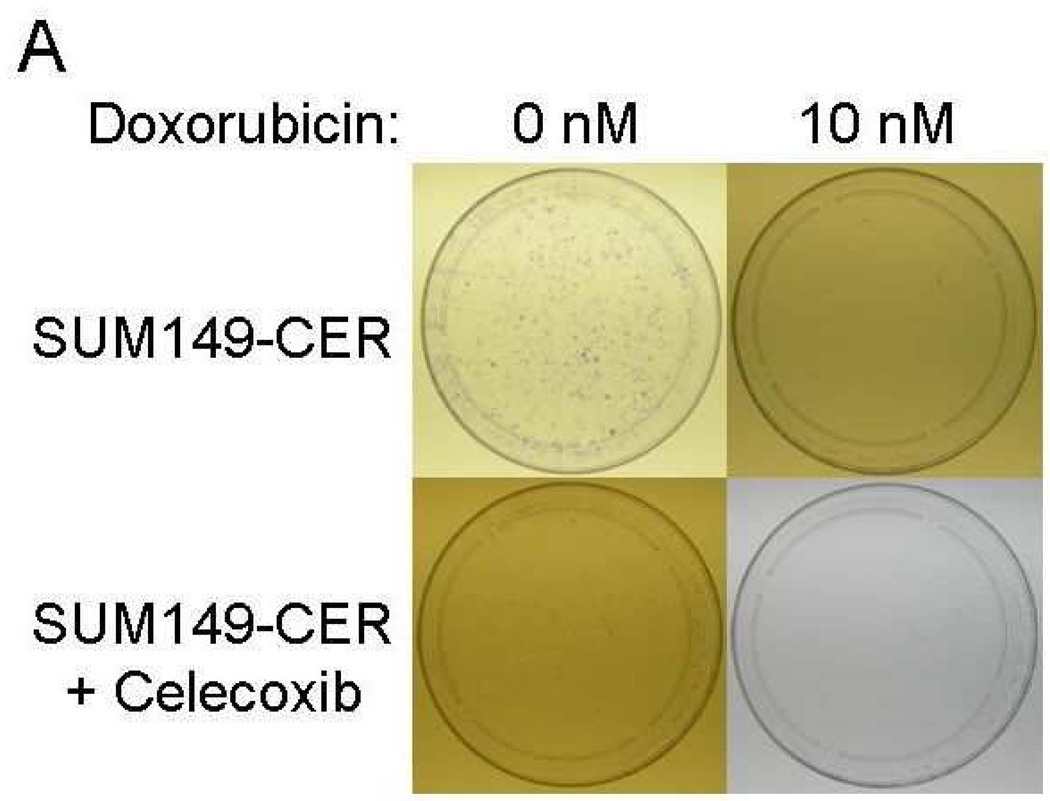
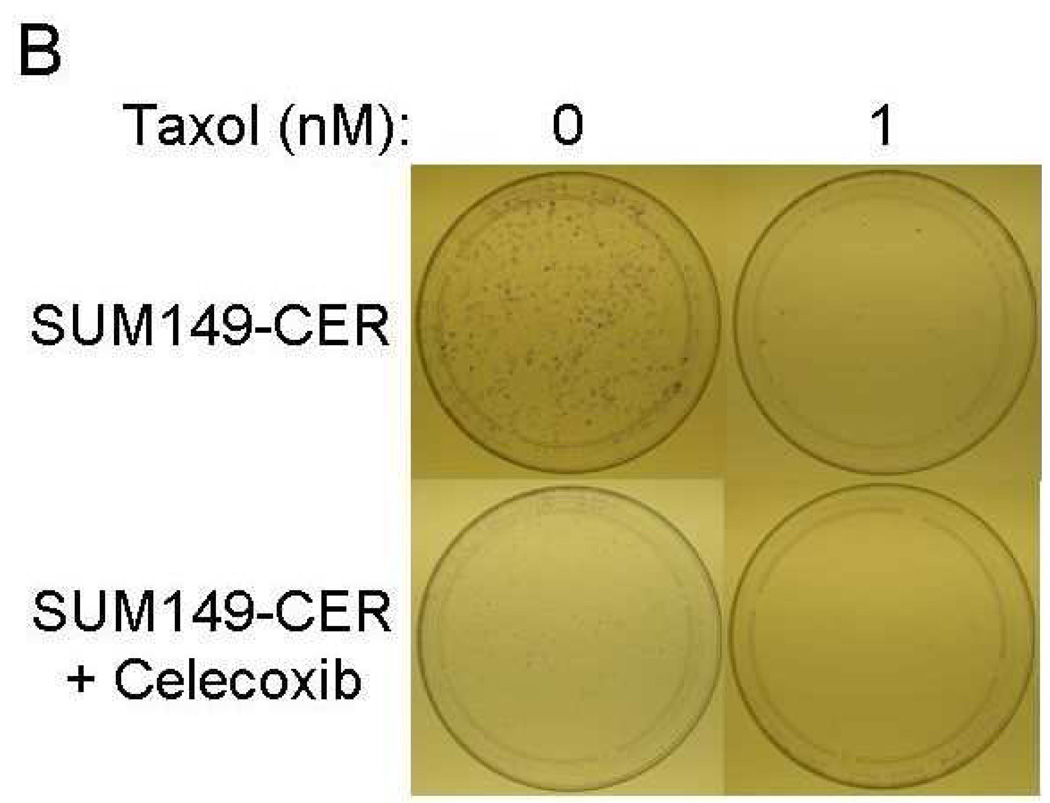
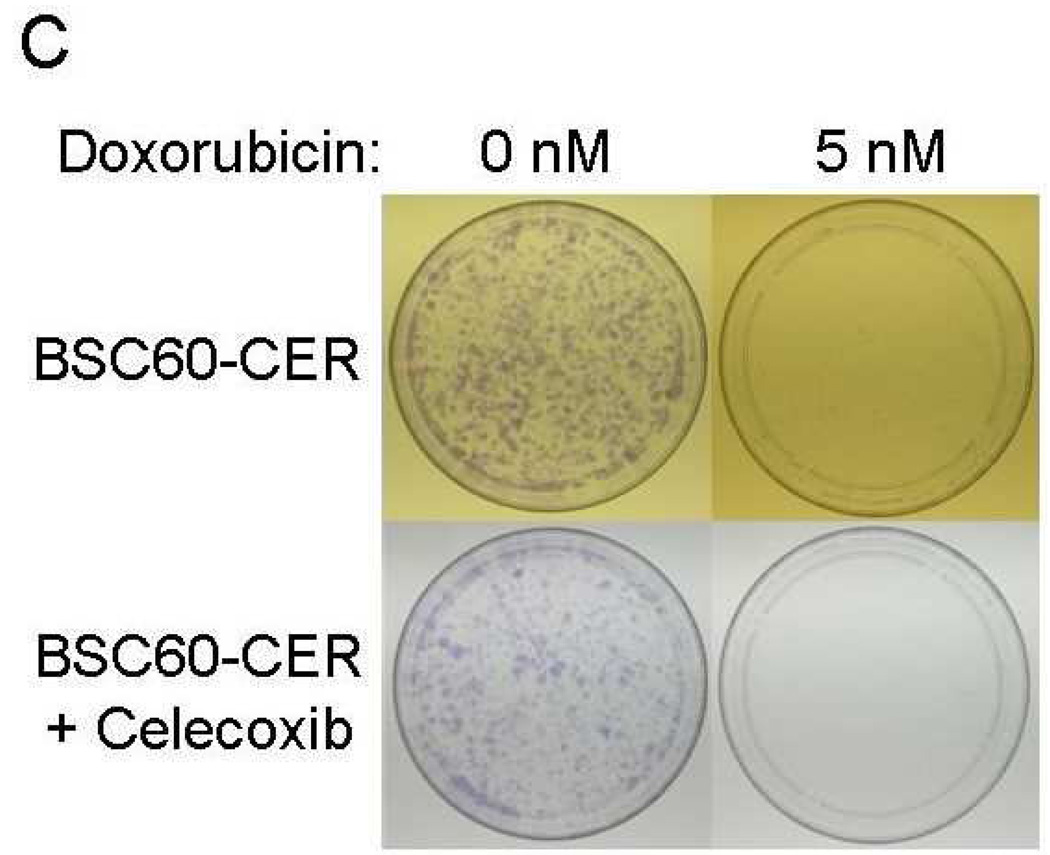
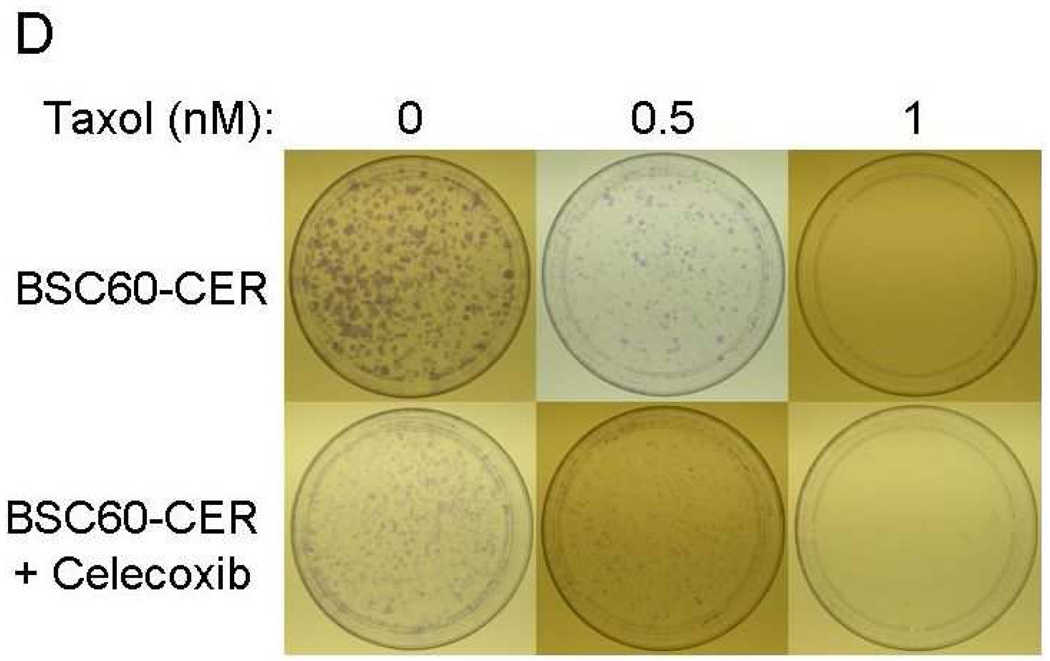
Inhibition of CER variants with celecoxib plus chemotherapeutic drugs
The important issue from the translation perspective is how to deal with the problem of celecoxib resistance. It seems logical that the problem may be easier to address sooner rather than later, i.e., incorporate celecoxib therapy as a combination therapy early in the treatment to minimize the expansion of CER variants. Our results indicate that since COX-2 (network) is highly functional in CER variants, celecoxib may be beneficial against them when combined with other therapy, e.g., chemotherapy. In this regard, a combination of celecoxib to either paclitaxel or doxorubicin was significantly more effective in inhibiting clonogenicity of the CER variants than the either chemotherapy drug alone (Fig. 7). We also noted that although SUM149-CER cells are resistant to celecoxib for growth in their medium with 5% FBS, celecoxib inhibited them significantly in the clonogenicity assay performed in medium with 0.5% FBS (Figs. 7A and 7B). We obtained a similar result with BSC60-CER, i.e., celecoxib inhibited them in a clonogenicity assay performed under low-FBS (Figs. 7C and 7D). However, this inhibition was less pronounced in BSC60-CER than that observed in SUM149-CER, which could be partly because of the confounding effect of FBS since it was not technically possible to reduce the FBS concentration below 3% for clonogenicity assays with BSC60. BSC60 cells were unable to survive in this assay with less than 3% FBS concentration.
FIG. 7.
Inhibition of clonogenicity of the CER variants with a combination treatment. We added Celecoxib (10 µM) or the DMSO solvent (as a control) along with different concentrations of doxorubicin or paclitaxel (taxol) at day 1. A and B, SUM149-CER cells (passage 5) were plated at 7,500 cells per 10 cm dish in a low FBS (0.5%) medium as described in Materials and Methods and stained at day 25. C and D, BSC60-CER cells (passage 4) were plated at 7,500 cells per 10 cm dish in medium with normal amount of FBS (10%) and stained at day 13. Comparison of the plates showed that 1) the CER variants are inhibited with celecoxib in this assay, and 2) celecoxib cooperates with the chemotherapy drugs to inhibit clonogenicity.
DISCUSSION
COX-2 in the context of a heterogeneous disease
Cancer is a heterogeneous disease at several levels, including heterogeneity at the level of tumor cell populations, a finding also observed in breast cancer cell lines. As a result, when any therapy (targeted therapy, chemotherapy, or radiation therapy) is applied, some pre-existing variants of the target cell population do not respond to therapy. This appears to be the major cause of “pan-resistance” often encountered in the clinic, although it is possible that therapy itself may modify the target cell population thus leading to resistance.
From the perspective of this study, cells are heterogeneous in a given breast cancer cell line with regards to COX-2 expression, and COX-2 may contribute to heterogeneity via its role in cancer stem-like phenotype [21]. Since COX-2 is involved in breast cancer metastasis, and relatively safe COX-2 inhibitors are available, we decided to investigate the mechanisms of potential resistance to COX-2 targeted therapy in the clinic. To increase the likelihood of translation, we chose cell lines for this study that have been selected for metastasis and that have robust COX-2 networks. Experimental models built on such cell lines would be ideal for evaluating promising new therapies.
Apart from learning more about the mechanism of celecoxib resistance, the CER variants may be valuable in evaluating new novel therapies since they have elevated level of functional COX-2 (and associated networks that control cell survival) in the relevant background of aggressive disease-causing ability. This is important because the COX-2 network can have very different functional output depending upon the cell background, ranging from growth arrest to cell survival and cell proliferation [e.g., see references 13, 22]. Our CER variants and their parental cell lines represent suitable isogenic pairs for investigating COX-2 signaling and its therapeutic targeting. As an important conceptual point, we view COX-2 as a component of networks wherein individual components of a network exhibit specific context-dependent relationships, e.g., cooperation with NF-κB pathway and antagonism with p53 signaling at an advanced stage of breast cancer [22]. The networks, rather than linear signaling better explain the role of COX-2 in cancer.
In most models of cancer wherein COX-2 has a role, including mouse models of breast cancer, COX-2 inhibition results in partial inhibition of tumor progression [e.g., see 14,23,24]. Similar to our results in this study, two recent studies with mouse models of breast cancer, involving C3 (1)-SV40 tumor antigen transgenic mice and 4T1 xenografts in mammary fat pad of BALB/c and COX-2 knockout mice, provided evidence that an escape from COX-2 specific inhibitors involves COX-2 overexpression in tumor cells [23,24]. Pertaining to the origin of CER variants in our study and possibly the similar variants that drive escape from COX-2 inhibition in mouse models [23,24], we favor a model wherein the variants and/or their precursors preexist prior to celecoxib therapy. There is a significant cellular heterogeneity within cell lines with regards to COX-2 protein level [21]. A high level of COX-2 protein may contribute to cellular heterogeneity through several mechanisms, including genomic instability [6,7]. The cells with a high level of COX-2 protein may also be endowed with a robust pro-survival network [19, this study]. In addition, the results obtained after COX-2 knockdown (Fig. 6) support the interpretation that a high level of COX-2 protein is linked with the anti-apoptosis apparatus in SUM149-CER cell line. Such cells may survive both anti-COX-2 therapy and other therapies, particularly if they are applied one at a time.
COX-2 signaling in IBC versus non-IBC
To briefly discuss similarities and differences between the IBC and non-IBC cell lines in their response to celecoxib, both cell lines respond in a fundamentally similar manner, but with some differences. Both cell lines yield CER variants with a high COX-2 protein level; however, the elevated level of COX-2 protein was more stable in SUM149-CER cell line upon celecoxib withdrawal than in BSC60-CER cell line (Fig. 4). One reason for this difference between the cell lines may be due to the difference in the maximum amount of COX-2 tolerated by the cell lines. Our limited analysis of pro- and anti-apoptosis proteins showed that COX-2 is linked with increased pro-survival signaling in both the backgrounds. This is not surprising given the similarity of IBC gene expression signature with the aggressive non-IBC gene expression signatures [25]. The difference between the CER variants was in the specific members of the Bcl-2 family; BSC60-CER had an elevated level of Bcl-2 while SUM149-CER had an elevated level of Bcl-xL (Figs. 2 and 3).
Both the CER variants had a decrease in pro-apoptosis protein Bax; this decrease occurred sooner (early passage) in SUM149-CER than in BSC60-CER, the latter depending possibly on an “evolution” that was forced by a high COX-2 expression in this background (Fig. 3A and 3C). One simple explanation for this result is that continued COX-2 expression may trigger some epigenetic alterations that are responsible for a subsequent Bax decrease. Alternatively, COX-2 overexpression may trigger pro-apoptotic effects in some subpopulations during early passage, thus causing an increase in Bax at this stage, which would mask a decrease in true stable CER variants. It is noteworthy that MMTV-LTR-COX-2 transgene-driven breast tumors have elevated Bcl-2 and reduced level of Bax as observed in the BSC60-CER variants in our study [19]. However, the tumors in the transgenic mice have reduced level of Bcl-xL as opposed to an increase in SUM149-CER. The explanation of this difference may lie in the epigenetics of specific subpopulations of cancer cells. Finally, we noted that the BSC60-CER variants grew to a significantly higher cell density on the dish than the parental cells. Such a property of the CER variants may be indicative of their relative aggressiveness.
COX-2 addiction in aggressive breast cancer
There were several reasons to pursue our current study. The key issue is whether the breast cancer cells that express COX-2 protein are addicted to it. The answer to this question may produce different results depending upon the nature of COX-2 networks that could be influenced by genetic and epigenetic changes within specific subpopulations of breast cancer cells. Therefore, we thought it was important to analyze this issue in important disease contexts, e.g., a human breast cancer cell line selected to metastasize (BSC60), and a human inflammatory breast cancer cell line (SUM149). The previous cited studies were performed with a transgenic mouse model and with a xenograft model in mice inoculated with a mouse-derived breast cancer cell line. Furthermore, unlike our study analyzing COX-2 protein, the COX-2 protein was not analyzed in any of those studies; it was inferred from the changes in COX-2 RNA [23] or from circulating levels of prostaglandin F1α [24]. Thus our study is a significant contribution to address the important issue of COX-2 addiction, and suggests that COX-2 addiction may be a general phenomenon in the context of aggressive disease.
We have shown recently that a subpopulation of cells present within a cell line overexpress COX-2, and that they are relatively resistant to low concentrations of celecoxib [21]. Importantly, COX-2high variants appear to behave like cancer stem cells that give rise to COX-2low cells [21]. To explain the data presented here, we propose that the celecoxib-resistant (CER) variants represent expansion of preexisting COX-2high variants, whereas majority of cells are killed by celecoxib. Another possibility, which is not mutually exclusive, is that the cancer cells are dependent on COX-2 activity for survival, and they must respond by overproducing COX-2 protein to counter the inhibition of COX-2 enzyme activity by celecoxib. The important point is that as long as cancer cells “depend” on COX-2 for survival, it is a therapeutic target. The most important issue that we face is how to kill the CER variants.
Our data in Figure 7 is informative in this regard. First, we showed that the celecoxib-resistant variants selected in complete medium with 5% serum are inhibited dramatically with celecoxib in clonogenicity assay which is performed under low serum (0.5%), indicating that resistance involves cooperation with serum-mediated signaling. More importantly, we also showed cooperation between celecoxib and chemotherapeutic drugs doxorubicin and paclitaxel. These results could be more clearly demonstrated in SUM149-CER (which tolerates lower level of serum than BSC60-CER for survival in this assay) than BSC60-CER (Fig. 7). Some other studies have also provided evidence of cooperation between celecoxib and chemotherapeutic drugs [e.g., 26–28]. We are currently pursuing several approaches to identify additional targeted therapies to overcome celecoxib resistance.
ACKNOWLEDGMENTS
These studies were supported in part by grants R21 DK067682 (AL) and CA16672 (Core) from the National Institutes of Health, DAMD17-03-1-0669 and W81XWH-09-1-0590 from the United States Army Medical Research and Material Command (AL), HostGator (AL), Breast Cancer Micrometastasis Research Program at M.D. Anderson Cancer Center (AL), Morgan Welch Inflammatory Breast Cancer Multi-Disciplinary Research Program at M.D. Anderson Cancer Center, and a Clinical Investigator Award from the Society of Surgical Oncology (AL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Howe LR. Inflammation and breast cancer. Cyclooxygenase/prostaglandin signaling and breast cancer. Breast Cancer Res. 2007;9:210. doi: 10.1186/bcr1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh-Ranger G, Salhab M, Mokbel K. The role of cyclooxygenase-2 in breast cancer: review. Breast Cancer Res Treat. 2008;109:189. doi: 10.1007/s10549-007-9641-5. [DOI] [PubMed] [Google Scholar]
- 3.Harris RE. Cyclooxygenase-2 (cox-2) and the inflammogenesis of cancer. Subcell Biochem. 2007;42:93. doi: 10.1007/1-4020-5688-5_4. [DOI] [PubMed] [Google Scholar]
- 4.Harris RE, Beebe-Donk J, Alshafie GA. Reduction in the risk of human breast cancer by selective cyclooxygenase-2 (COX-2) inhibitors. BMC Cancer. 2006;6:27. doi: 10.1186/1471-2407-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takkouche B, Regueira-Méndez C, Etminan M. Breast cancer and use of nonsteroidal anti-inflammatory drugs: a meta-analysis. J Natl Cancer Inst. 2008;100:1439. doi: 10.1093/jnci/djn324. [DOI] [PubMed] [Google Scholar]
- 6.Singh B, Vincent L, Berry JA, Multani AS, Lucci A. Cyclooxygenase-2 expression induces genomic instability in MCF10A breast epithelial cells. J Surg Res. 2007;140:220. doi: 10.1016/j.jss.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 7.Singh B, Cook KR, Vincent L, Hall CS, Berry JA, Multani AS, Lucci A. Cyclooxygenase-2 induces genomic instability, BLC2 expression, doxorubicin resistance, and altered cancer-inhibiting cell phenotype in MCF7 breast cancer cells. J Surg Res. 2008;147:240. doi: 10.1016/j.jss.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci U S A. 2009;106:3372. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh B, Berry JA, Shoher A, Ramakrishnan V, Lucci A. COX-2 overexpression increases motility and invasion of breast cancer cells. Int J Oncol. 2005;26:1393. [PubMed] [Google Scholar]
- 12.Singh B, Berry JA, Vincent LE, Lucci A. Involvement of IL-8 in COX-2-mediated bone metastases from breast cancer. J Surg Res. 2006;134:44. doi: 10.1016/j.jss.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Singh B, Berry JA, Shoher A, Lucci A. COX-2 induces IL-11 production in human breast cancer cells. J Surg Res. 2006;131:267. doi: 10.1016/j.jss.2005.11.582. [DOI] [PubMed] [Google Scholar]
- 14.Singh B, Berry JA, Shoher A, Ayers GD, Wei C, Lucci A. COX-2 involvement in breast cancer metastasis to bone. Oncogene. 2007;26:3789. doi: 10.1038/sj.onc.1210154. [DOI] [PubMed] [Google Scholar]
- 15.Lucci A, Krishnamurthy S, Singh B, Bedrosian I, Meric-Bernstam F, Reuben J, Broglio K, Mosalpuria K, Lodhi A, Vincent L, Cristofanilli M. Cylooxygenase-2 expression in primary breast cancers predicts dissemination of cancer cells to the bone marrow. Breast Cancer Res Treat. 2009;117:61. doi: 10.1007/s10549-008-0135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 17.Lerebours F, Vacher S, Andrieu C, Espie M, Marty M, Lidereau R, Bieche I. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 19.Liu CH, Chang SH, Narko K, Trifan OC, Wu MT, Smith E, Haudenschild C, Lane TF, Hla T. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. J Biol Chem. 2001;276:18563. doi: 10.1074/jbc.M010787200. [DOI] [PubMed] [Google Scholar]
- 20.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 21.Singh B, Cook KR, Vincent L, Martin C, Lucci A. Role of COX-2 in tumorospheres derived from a breast cancer cell line. J Surg Res. 2010 March 26; doi: 10.1016/j.jss.2010.03.003. doi 10.1016/j.jss.2010.03.003, Uncorrected Proof available Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fordyce C, Fessenden T, Pickering C, Jung J, Singla V, Berman H, Tlsty T. DNA damage drives an Activin A-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev Res (Phila Pa) 2009 Dec 22; doi: 10.1158/1940-6207.CAPR-09-0229. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mustafa A, Kruger WD. Suppression of tumor formation by a cyclooxygenase-2 inhibitor and a peroxisome proliferator-activated receptor gamma agonist in an in vivo mouse model of spontaneous breast cancer. Clin Cancer Res. 2008;14:4935. doi: 10.1158/1078-0432.CCR-08-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barry M, Cahill RA, Roche-Nagle G, Neilan TG, Treumann A, Harmey JH, Bouchier-Hayes DJ. Neoplasms escape selective COX-2 inhibition in an animal model of breast cancer. Ir J Med Sci. 2009;178:201. doi: 10.1007/s11845-009-0335-3. [DOI] [PubMed] [Google Scholar]
- 25.Van Laere S, Beissbarth T, Van der Auwera I, Van den Eynden G, Trinh XB, Elst H, Van Hummelen P, van Dam P, Van Marck E, Vermeulen P, Dirix L. Relapse-free survival in breast cancer patients is associated with a gene expression signature characteristic for inflammatory breast cancer. Clin Cancer Res. 2008;14:7452. doi: 10.1158/1078-0432.CCR-08-1077. [DOI] [PubMed] [Google Scholar]
- 26.Hashitani S, Urade M, Nishimura N, Maeda T, Takaoka K, Noguchi K, Sakurai K. Apoptosis induction and enhancement of cytotoxicity of anticancer drugs by celecoxib, a selective cyclooxygenase-2 inhibitor, in human head and neck carcinoma cell lines. Int J Oncol. 2003;23:665. [PubMed] [Google Scholar]
- 27.Ponthan F, Wickström M, Gleissman H, Fuskevåg OM, Segerström L, Sveinbjörnsson B, Redfern CP, Eksborg S, Kogner P, Johnsen JI. Celecoxib prevents neuroblastoma tumor development and potentiates the effect of chemotherapeutic drugs in vitro and in vivo. Clin Cancer Res. 2007;13:1036. doi: 10.1158/1078-0432.CCR-06-1908. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Shen HL, Yang J, Chen QY, Xu WL. Preventing chemoresistance of human breast cancer cell line, MCF-7 with celecoxib. J Cancer Res Clin Oncol. 2010 Mar 14; doi: 10.1007/s00432-010-0854-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]