Abstract
High hyperdiploidy is the single largest subtype of childhood acute lymphoblastic leukemia (ALL) and is defined by the presence of 51-68 chromosomes in a karyotype. The 5 or more extra chromosomes characterizing this subtype are known to occur in a single mitotic event, prenatally. We screened for RAS mutations among 517 acute childhood leukemias (including 437 lymphocytic, of which 393 were B-cell subtypes) and found mutations in 30% of high hyperdiploids compared to only 10% of leukemias of other subtypes (P < 0.0001). We assessed whether KRAS mutations occurred before birth using a PCR-restriction enzyme-mediated Taqman quantitative PCR reaction, and found no evidence for prenatal KRAS mutations in 14 patients tested. While RAS mutations were previously associated with prior chemical exposures in childhood and adult leukemias, in this study RAS-mutated cases were not significantly associated with parental smoking when compared to study controls. IGH rearrangements were backtracked in three RAS-positive patients (which were negative for KRAS mutation at birth) and found to be evident before birth, confirming a prenatal origin for the leukemia clone. We posit a natural history for hyperdiploid leukemia in which prenatal mitotic catastrophe is followed by a postnatal RAS mutation to produce the leukemic cell phenotype.
Keywords: childhood leukemia, high hyperdiploidy, NRAS KRAS, epidemiology
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer in children. Within ALL, the B-cell phenotype predominates, among which the high hyperdiploid subgroup (defined by a karyotype with 51-68 chromosomes) is the most common cytogenetic group representing 25-40% of ALL. High hyperdiploid leukemia is an archetypal subtype of “common” ALL leukemia (CD10+, CD19+ in young children), the leukemia subtype which accounts for most of the 30% increase in leukemia rates since World War II in Western countries [1]. This leukemia subtype has excellent prognosis, with cure rates exceeding 90%; however, most children cured of leukemia have chronic health problems, a quarter of which are life-threatening [2]. Understanding the etiology of pediatric leukemia may aid prevention and early detection efforts. Here we explore the role and timing of additional mutations in childhood leukemia with high hyperdiploidy, and the relationship between RAS mutation and demographic factors.
High hyperdiploidy has a typical complement of extra chromosomes including chromosomes 21, X, 4, 10, 17, and 18. Studies using microsatellite markers support a mechanism whereby all the chromosomes were gained in a single abnormal mitosis [3; 4]. High hyperdiploidy was directly shown to be prenatal in a study which backtracked three IGH rearrangements in Guthrie cards [5] and in another study which noted the presence of a hyperdiploid clone in cord blood of a neonate who later contracted leukemia [6]. Further indirect evidence for prenatal origin was demonstrated by presence of single IGH rearrangements in the Guthrie cards of most hyperdiploid children [7; 8]. Leukemia IGH sequences were found in Guthries more often in lower birthweight children and nearly all hyperdiploid patients in a recent study [9].
We previously reported that leukemias with higher rates of RAS mutation were associated with young maternal age when the child was born, and high hyperdiploidy was inversely associated with paternal pre-pregnancy smoking [10]. Because most of these associations were of borderline significance we have repeated the analysis with a larger sample size and more robust mutation screening methods. In addition, we have explored the timing of origin for both IGH rearrangements and RAS mutations as well as an exploratory assessment of RAS mutation status in relationship to parental smoking.
Materials and Methods
The patient population consisted of 517 consecutive leukemia patients enrolled in the 9 hospitals participating in Northern California Childhood Leukemia Study during the years 1995 to 2002. Intensive cytopathological review [11], parental interviews [12; 13] and biologic and environmental sampling [14] were instituted. A detailed description of this approximately population-based study design can be found elsewhere [15]. Parental demographic characteristics and smoking information was provided by the case mother (97.5%) or father (2.5%) through in-person interviews in the home of the parents. For comparisons with healthy children, controls which were individually matched by birthdate, gender, and ethnicity to cases, were utilized.
Characterization of high hyperdiploidy
Many of the high hyperdiploid childhood ALL cases were identified through cytogenetic reports obtained from clinical laboratories where classical banding methods are generally applied. However, there were many cases without any cytogenetic reports or a “normal” karyotype. Therefore, we employed fluorescence in situ hybridization (FISH) to further assess status of two major but cryptic cytogenetic subtypes of childhood ALL, TELAML1 translocation and cryptic high hyperdiploidy, using combined gene-loci specific FISH probes for chromosomes 12 (TEL) and 21 (AML1), as well as centromere probe for chromosome X. Because >96% of high hyperdiploid patients have extra copies of both chromosomes 21 and X in the same cell [16; 17], cases with both extra X and 21 were classified as high hyperdiploidy in the current study, as we have previously described [11].
RAS mutation characterization
Genomic DNA was extracted from patient bone marrow (Qiagen QIAamp DNA Mini Kit). Amplification of 20 ng of genomic DNA was performed in a 50 μl reaction containing 1X reaction buffer, 1.5 mM MgSO4, 200 μM each of dNTPs, 0.4 μM of each forward and reverse primer, and 1.0 μl of Optimase polymerase (Transgenomics). KRAS and NRAS mutations in exon 1 were detected with the following sets of oligonucleotide primers, respectively: KRAS2F and KRAS2R, and NRAS1F and NRAS1R (Table 1). Reactions were denatured for 5 minutes at 95 °C, followed by amplification for 40 cycles of 94 °C, annealing for the indicated temperature (Table 1), and extension for 1 minute at 72 °C, and then a final extension for 5 minute at 72 °C. Samples that amplified poorly were subjected to a nested-PCR approach, which involved a first round of amplification with the following primers: KRAS2NF and KRAS2NR, and NRAS1NF and NRAS1NR (Table 1). DNA hybridization was performed to form hetero- and homoduplexes prior to denaturing high-performance liquid chromatography (DHPLC) analysis: denaturing at 95 °C for 2 minutes, followed by a gradual re-annealing at a rate of -2 °C per second to 85 °C, and then from 85 °C to 25 °C at a rate of -0.1 °C per second. Each PCR product was analyzed using the WAVE DHPLC system (Varian, Inc) at column temperature of 55.5 °C and 60.1 °C, respectively, for KRAS and NRAS samples with a flow rate of 0.9 mL/min and 5 μl injection volume. Wild type samples were run concurrently as negative controls. Samples with chromatograms containing multiple or abnormal peaks, compared to the wild type samples, were scored as positive. Bidirectional sequencing was performed on all samples in parallel to confirm the presence of mutations.
Table 1.
Primers used for RAS screening and backtracking (all 5' to 3').
| Primer | Sequence | Amplicon size (bp) | TM (°C) | DHPLC TM (°C) |
|---|---|---|---|---|
| NRAS1NF | AGGCCGATATTAATCCGGTG | 481 | 60 | n/a |
| NRAS1NR | GGACAGGTTTTAGAAACTTCAGC | |||
| NRAS1F | GGTTTCCAACAGGTTCTTGC | 191 | 55 | 60.1 |
| NRAS1R | TCCGACAAGTGAGAGACAGG | |||
| KRAS2NF | ATCCTTTGAGAGCCTTTAGCC | 670 | 60 | n/a |
| KRAS2NR | CAGAGAGTGAACATCATGGAC | |||
| KRAS2F | CATTACGATACACGTCTGCAGTC | 507 | 60 | 55.5 |
| KRAS2R | GGACCCTGACATACTCCCAAG | |||
| 5BKIT | TATAAACTTGTGGTAGTTGGACCT | 82 | 60 | |
| 3K2 | CGTCCACAAAATGATTCTGA | |||
| 5BK36 | CTAGAACAGTAGACACAAACCA | 130 | 60 | |
| 3K37 | GATTTTGCAGAAAACAGATC | |||
| 5BK38 | GTACACATGAAGCCATCGTATA | 215 | 60 | |
| 3K39 | CCACTTGTACTAGTATGCCTTAAG | |||
| KRAS-probe | CTGTATCGTCAAGGCACTCTTGCCTAC | n/a | 60 | |
Statistical Analysis
The distribution of RAS mutation by childhood leukemia subtypes, child's demographics, and parental characteristics was evaluated using Pearson's Chi-Square tests or Fisher's Exact tests (when 25% of the cells had expected counts <5). Multivariable analysis was performed using log-linear regression to assess the associations between seven variables (presence of RAS mutation, presence of high hyperdiploidy, paternal smoking three months before pregnancy, maternal smoking during pregnancy, maternal age at child's birth, child's race/ethnicity, and household income) in a case-case analysis. Since the variable “income” has been found to be associated with leukemia risk in previous analyses, it was included in this model [12; 15]. We chose log-linear over logistic models because log-linear model does not require an outcome variable and is more appropriate for this analysis since it is not clear whether the high hyperdiploidy is an antecedent event to the RAS mutation. Whereas Pearson's Chi-Square test is used to assess pairwise association in a two-dimensional (row by column) contingency table, log-linear model allows for evaluation of associations in a multi-dimensional contingency table. A log-linear model consists of two parts, the additive part with the main effect terms and the part with the measures of association terms (interaction terms). The statistical significance of each interaction term was assessed by a p-value generated from the log-likelihood ratio test comparing the sub-model without the measure of association (interactive) term to the full model including the measures of association. This process identifies the degree of the pairwise association between the RAS mutation and each variable adjusted for the potential confounding by the other variables included in the analysis.
KRAS backtracking
KRAS backtracking was performed for 14 subjects. NRAS backtracking was not possible by REMS-PCR due to the lack of a thermostable restriction endonuclease which can cleave the wild-type NRAS codon 12 and 13 sites. Guthrie card DNA was isolated using the Qiagen QIAamp DNA Micro Kit. To increase the DNA yield, 1 ug of carrier RNA was added to the sample during the DNA extraction process. One eighth of a 1 cm archived neonatal blood spot (ANB or Guthrie card) yielded approximately 100-300 ng of DNA. The TaqMan and restriction endonuclease-mediated selective-PCR (REMS-PCR) were simultaneously used to backtrack mutations in codon 12 of KRAS to increase the sensitivity of mutation detection. This analysis was built on the methods of Fuery et al [18]. Three sets of primers were used in multiplex: (a) diagnostic primers, 5BKIT and 3K2, that amplify KRAS exon 1; (b) restriction endonuclease control primers, 5BK36 and 3K37, that amplify a region of KRAS exon 3 containing a BstNI site; and (c) PCR control primers, 5BK38 and 3K39, that amplify a region of KRAS exon 4b. Each 20 μl reactions contained 25 ng of DNA, 50 mM Tris, 100 mM NaCl, 4 mM MgCl2, 1 mM DDT; 1 μM of 5BKIT, 1 μM of 3K2, 40 nM of 5BK36, 40 nM of 3K37, 40 nM of 5BK38, 40 nM of 3K39, each dNTP at 50 μM, 300 nM of ROX Reference Dye, 250 nM of KRAS-probe, 20 U of BstNI; and water to 20 μl. TaqStart® antibody (Clontech Laboratories, Mountain View) was mixed with 2.5 U of AmpliTaq DNA polymerase (ABI, Foster City) at molar ratio of 1:5 and incubated 15 minutes at room temperature before adding to each mixture. Restriction endonuclease control reactions contained no BstNI. Fourteen samples positive for KRAS codon 12 mutations in their bone marrow samples were tested for the presence of those mutations in their corresponding Guthrie card DNA. Guthrie card DNA from nonmutant samples and cell line DNA were used as negative controls. For each run, reactions were performed in triplicate and run on an ABI 7900HT Sequence Detection System (ABI, Foster City) as follows: Reaction mix was warmed to 65°C for 30 minutes for BstN1 to cleave wild-type DNA prior to PCR. Reactions were denatured at 94 °C for 2 minutes, then 10 cycles of 58 °C for 2 minute followed by 92 °C for 20 seconds and finally 40 cycles of 58 °C for 1 minute followed by 92 °C for 20 seconds. A 10-fold dilution series ranging from 100 ng to 1 pg of genomic mutant cell line DNA from SW837 diluted into wild type cell line DNA was used to measure the sensitivity of the assay. The BstN1 enzyme cuts the wild-type KRAS sequence prior to the PCR, and again in the early rounds of the PCR reaction if any wild-type sequences are produced during these initial rounds, preventing further amplification. A key improvement of the assay to gain the requisite sensitivity was a 30 minute pre-incubation at 65°C, the enzyme's optimal temperature, prior to PCR. This resulted in an assay with a greater than requisite sensitivity of one mutant targets in 8,000 or more normal copies (25 ng of DNA), the quantity of Guthrie DNA available for PCR. This sensitivity was determined using limiting dilution experiments, and “mock” Guthrie cards constructed of SW837 cells (which harbor a KRAS mutation) mixed into blood, spotted onto cards, and used for mock backtracking experiments. Sensitivity was confirmed during each PCR run using limiting dilutions of patient leukemia DNA which harbored the mutation being assessed. Following amplification and analysis with the SDS 2.2.2 software (ABI, Foster City), 15 μl of each reaction was further visualized by electrophoresis on a 5% NuSieve GTG agarose gel. Electrophoresis allowed the detection of the restriction endonuclease and PCR control bands, for which the TaqMan® probe did not target.
Results
RAS mutations and clinical/demographic variables
Eighty-two cases (15.9%) had mutations in codons 12 or 13 of NRAS and KRAS (Table 2). This frequency was slightly higher in ALL cases (16.6%) than in AML (12.5%). Among ALL cases, mutations were found more often in B-lineage types (17.8%) than T-ALL (6.8%). Our current patient list (N = 517) includes the 191 patients that we previously reported [10] as well as 326 additional patients; however, we re-analyzed all patients (including the original 191) with a DHPLC technique and DNA sequencing. Four patients were reclassified from the previous report – two that were incorrectly classified as negative, and two as positive. Thirty-three KRAS and 51 NRAS mutations were discovered among 517 patients (2 had both). Sixty-nine mutations were transversions G->T, with one T->G; six transitions G->T, as well as six G->C. All point mutations were nonsynomymous and in codon 12 or 13, except for one in codon 19 (L19F in KRAS). RAS mutations were nearly twice as common in Hispanic (19.8%) as non-Hispanic white patients (10.3%) and more common in children of younger mothers (Table 2). Note that Hispanics have higher rate of high hyperdiploidy [11]. Among B-cell leukemia patients, RAS mutations were three-times more frequent in high hyperdiploid cases than the remainder of B-cell cases (29.6% and 10.0%, respectively, P < 0.0001). Within the hyperdiploid subgroup, white blood cell counts (WBC) were higher in the RAS-mutant group than the RAS-wild type group (median WBC = 13 × 103/ul, and 7 × 103/ul, respectively; P = 0.08 Wilcoxon rank-sum test). There was no relationship between RAS mutation and WBC in the nonhyperdiploid subgroup (P = 0.80).
Table 2.
Characteristics of the Northern California Childhood Leukemia Study patient series subjects by RAS mutations.
| RAS mutation All leukemias (n=517) | P-value | RAS Mutations B lineage, ALL only (n = 393) | P-value | |||
|---|---|---|---|---|---|---|
| Yes (%) | No (%) | Yes (%) | No (%) | |||
| Childhood leukemia | 82 (15.9) | 435 (84.1) | 70 (17.8) | 323 (82.2) | ||
| Childhood leukemia phenotype | ||||||
| ALL | 73 (16.6) | 368 (83.5) | 0.55 | |||
| AML | 8 (12.5) | 56 (87.5) | ||||
| Others | 1 (8.3) | 11 (91.7) | ||||
| ALL subtypes | ||||||
| B lineage | 70 (17.8) | 323 (82.2) | 0.06 | |||
| T lineage | 3 (6.8) | 41 (93.2) | ||||
| Race/ethnicity | ||||||
| Hispanic | 42 (19.8) | 170 (80.2) | 0.03 | 37 (21.8) | 133 (78.2) | 0.08 |
| Non-Hispanic White | 18 (10.3) | 156 (89.7) | 16 (12.0) | 117 (88.0) | ||
| Other | 15 (19.0) | 64 (81.0) | 11 (20.0) | 44 (80.0) | ||
| Father's age at child's birth | ||||||
| < 30 | 36 (16.7) | 179 (83.3) | 0.87 | 31 (18.5) | 137 (81.5) | 0.91 |
| ≥ 30 | 38 (16.2) | 197 (83.8) | 32 (18.0) | 146 (82.0) | ||
| Mother's age at child's birth | ||||||
| < 30 | 51 (18.2) | 229 (81.8) | 0.10 | 44 (20.4) | 172 (79.6) | 0.09 |
| ≥ 30 | 23 (12.5) | 161 (87.5) | 19 (13.5) | 122 (86.5) | ||
| Annual household income | ||||||
| < $30,000 | 24 (14.2) | 145 (85.8) | 0.14 | 22 (16.7) | 110 (83.3) | 0.16 |
| $30,000-$75,000 | 37 (20.2) | 146 (79.8) | 31 (22.3) | 108 (77.7) | ||
| >$75,000 | 14 (12.4) | 99 (87.6) | 11 (12.6) | 76 (87.4) | ||
| Non-high hyperdiploid B cell | 23 (10.0) | 207 (90.0) | < 0.0001 | |||
| High hyperdiploid B cell (Chromosome 51-68) | 39 (29.6) | 93 (70.4) | ||||
A log-linear analysis was performed to explore relationships of RAS mutation to other covariates in a case-case analysis. This is a multivariable analysis, which is able to detect relationships between variables while statistically “adjusting” for all other variables in the model. This analysis was applied previously in our leukemia epidemiology study to find relationships between maternal age and RAS, as well as high hyperdiploidy and RAS [10]. In the current better-powered analysis, RAS mutation was not associated with maternal age or ethnicity (Table 3). Instead, the dominant relationship was an extremely strong association between RAS and high hyperdiploidy. While our original hypothesis was to draw a link between RAS and a mutagenic exposure, namely, parental smoking, we did not find any association between RAS and parental smoking in this log-linear analysis (P = 0.81 for father's pre-pregnancy smoking, and P = 0.34 for mothers's pregnancy smoking) and only a borderline significant association of paternal smoking with high hyperdiploidy (Table 3).
Table 3.
Multivariable log-linear model and selected associations: B-cell acute lymphoblastic leukemia cases in the Northern California Childhood Leukemia Study.
| Measure of Association Terms | Coefficient Estimate | DF† | Chi-square value‡ | P-value§ |
|---|---|---|---|---|
| FSMpre* RAS | -0.09 | 1 | 0.06 | 0.81 |
| MSMpreg* RAS | -0.56 | 1 | 0.92 | 0.34 |
| FSMpre* Hyperdip | -0.61 | 1 | 4.21 | 0.04 |
| MSMpreg* Hyperdip | 0.11 | 1 | 0.06 | 0.81 |
| MomAge<30* RAS | 0.37 | 1 | 1.01 | 0.31 |
| Hyperdip* RAS | 1.42 | 1 | 19.82 | <0.0001 |
| RAS* Race | 2 | 1.50 | 0.47 | |
| RAS* Hispanic | 0.12 | |||
| RAS* Non-Hispanic White | -0.36 |
Details: FSMpre = paternal smoking during preconception (two categories, one parameter); MSMpreg = maternal smoking during pregnancy (two categories, one parameter); MomAge<30 = maternal age at birth < 30 yrs (two categories, one parameter); Hyperdip = high hyperdiploidy (two categories, one parameter); RAS = any RAS mutation (two categories, one parameter); Race = child's race/Ethnicity (three categories, two parameters)
DF = Degrees of freedom
Chi-square value derived from log-likelihood ratio test comparing the sub-model without the measure of the association term with the full model including the measure of association term.
P-value associated with the Chi-square value derived from log-likelihood ratio test.
Backtracking KRAS2
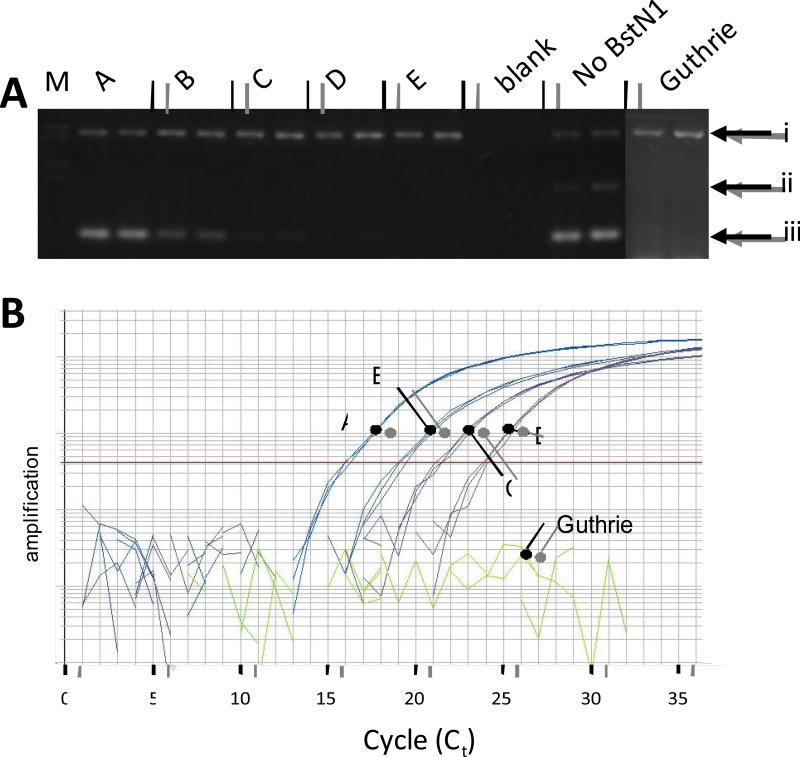
The strong link between hyperdiploid and RAS was investigated further by backtracking genetic markers to birth in neonatal blood spots. We backtracked IGH rearrangements as a marker of origin of the hyperdiploid clone, for four KRAS-mutant leukemias (which were also assessed for KRAS, see below). IGH rearrangements were backtracked positive in three of four patients tested, using IGH rearrangement screening primer sets and the same gel-based backtracking assay as Taub et al [7]. We discontinued this analysis with further information that IGH nearly always backtracks to birth [9], which was replicated here in three out of four patients tested. KRAS mutations were tested using a novel backtracking method that involves a real-time digestion of wild-type PCR product while concurrently amplifying the mutant. The quantitative PCR probe cleavage assay confirmed that no KRAS mutations in diagnostic DNA “backtracked” to birth. The two PCR reactions besides the diagnostic were necessary to monitor PCR and BstN1 digestion success. Southern blots of electrophoretic gels containing PCR products from this assay did not increase sensitivity over what is shown in the electrophoresis gels (data not shown), and the quantitative PCR detection limit was several logs better the ethidium bromide electrophoresis results. Visible 82 bp bands (Figure 1A) were cut and sequenced; these bands harbored 100% mutant DNA confirming the digestion of wild-type DNA. The KRAS diagnostic band was absent in 14 patients’ Guthrie samples (150 ng apiece), demonstrating no evidence for prenatal origin. The KRAS mutants tested all had mutations within the cleavage site of BstN1: a nt 34 G->T mutation (2 patients), nt 34 G->C mutation (2 patient), nt 35 G->A (9 patients), and nt 35 G->T (1 patient).
Figure 1. TaqMan + REMS-PCR Backtracking.
Electrophoresis analysis of REMS-PCR. SW837 DNA and various dilutions in REH DNA are shown, followed by two lanes with no template (blank) and two lanes of REH DNA (25 ng) run without BstN1 enzyme. All lanes with DNA include 25 ng; dilutions indicate the amount of SW837 within a background of REH DNA. Arrows indicate (i) – PCR control DNA, (ii) - BstN1 restriction enzyme control product, and (iii) – diagnostic mutant KRAS band. A, SW837 DNA, 4,166 copies (25 ng); B, 417 copies of SW837 DNA diluted in 25 ng WT DNA; C, 42 copies SW837 DNA diluted in 25 ng WT DNA; D, 4 copies of SW837 DNA in 25 ng of WT DNA; E, 25 ng REH cell line DNA; Blank, no DNA added to reaction; Guthrie shows an example of one Guthrie patient with positive control amplification band and negative bands for BstN1 control and diagnostic band. B. Amplification plot for testing Guthrie Card DNA. All reactions were performed in triplicate with 1 mM of each diagnostic primer, 40 nM of both the PCR control primer set and the RE control primer set. Each line represents the amplification of A to D as above; Guthrie, 25 ng of a test Guthrie DNA from a patient who had a KRAS mutation, which does not have detectable mutant KRAS. REH DNA (lane E) and “Blank” lanes also did not yield amplification, similar to Guthrie (data not shown).
Discussion
The largest subgroup among the childhood acute leukemias are the high hyperdiploids, representing about 25% of pediatric leukemias in clinical series involving conventional Giemsa-banding karyotype characterization. This subtype totals to larger proportions of case series screened when by flow cytometry or FISH. Our case series, derived from a consecutive sampling in a population base in Northern and Central California, has a 35% frequency of this subtype when screened by a FISH technique [11]. Strikingly, 30% of these are RAS-mutation positive with only 10% of non-hyperdiploids being RAS-positive (P < 0.0001).
Other investigators have identified additional RAS-pathway mutations to be frequent in this subtype, namely FLT3, with occasional BRAF and PTPN11 mutations [19; 20; 21; 22; 23], indicating a strong genetic selection for RAS-pathway mutations in hyperdiploids. The mutations are generally exclusive of one another [19], meaning that any mutation in a RAS pathway (receptor tyrosine kinase – RAS – MEK- ERK) may preclude the need for another. Our case series here have a similar overall rate of RAS mutations among ALL cases (16.6%, or 73 out of 441) as another large epidemiologic case series [15.2%, or 127 out of 870 cases [24]]. The rate of RAS mutations among high hyperdiploids in the current study at 30% (39/132) was far higher than another recent case series which demonstrated only 10% (8/78) RAS mutations [19]. The Paulsson, et al., study however also demonstrated a large number of FLT3 and PTPN11 mutations (7 each among 78 high hyperdiploids). Our series only had one functional mutation each for PTPN11 and BRAF out of 230 childhood leukemias screened (data not shown); we discontinued the analysis due to low yields. We also discovered 18 FLT3 mutations out of the same 517 cases screened for the current report, with an only 4% (5/132) FLT3 mutation frequency among high hyperdiploid ALL (MK and JLW, data not shown). The higher rate of RAS mutation in California high hyperdiploids and comparative absence of other RAS-pathway mutations [compared to Paulsson et al. [19]] is interesting since both studies resulted in an overall frequency of about one-third of high hyperdiploid patients harboring RAS-pathway mutations but a different mutation spectrum. The cause of this difference is unknown may be attributable to different etiologic risk factors or genetic background between the two populations. We did not yield any consistent support for our original hypothesis that parental smoking would be exhibited at a higher prevalence in families with RAS mutation-positive leukemia compared to families who had a RAS mutant-negative leukemia.
Our current study attempted to identify the timing of RAS mutation in the subtype most likely to display this mutation – high hyperdiploids. High hyperdiploidy is thought to arise from a single catastrophic mitosis, based on studies that demonstrated that the pattern of microsatellites and allelic ratios only compatible with one catastrophic event, rather than sequential gains of chromosomes [3; 4]. Prior to our current report, other investigators have “backtracked” high hyperdiploid patients using IGH sequences. First, Yagi et al (2000) and Taub and colleagues (2002) backtracked fourteen cases, and seven of these, all which were hyperdiploid, had IGH sequences present at birth [7; 8]. Panzer-Grumayer and colleagues demonstrated that three specific IGH rearrangements, from three chromosomes 14, all backtracked positive to birth providing definitive evidence that the chromosomal event had occurred before birth [5]. Further, in an interesting case with available cord blood, Maia and colleagues demonstrated that sorted precursor B cells harbored the hyperdiploid clone at the time of birth from a child who later contracted hyperdiploid leukemia [6]. In a recent report, 10 of 11 hyperdiploid patients had backtracked positive [9], and we determined 3 of 4 of our KRAS patients backtracked their IGH sequences to birth (data not shown). Clearly, the combined evidence places the high hyperdiploid mutation before birth with KRAS mutation afterwards. A recent report made use of paired diagnosis/relapse samples and confirmed this conclusion indirectly by noting that high hyperdiploidy pattern was retained in relapsed primary leukemias with high hyperdiploidy (the ancestral clone) while the RAS pathway mutations and structural changes were variable between diagnosis and relapse, indicating that they are secondary [25].
In the current study we demonstrate the strength of association between RAS and high hyperdiploidy in our Northern California population, and demonstrate that while the mitotic catastrophe event is prenatal, the RAS mutation is postnatal in most (possibly all) cases. High hyperdiploidy is likely to provide some clonal advantage, as evidenced by the ease at which these clones are found in neonatal blood spots using specific IGH sequence probes. However, the hyperdiploid genotype is not sufficient for leukemia as most children with this subtype get leukemia at the peak age range of cALL, ages 2-6. RAS mutations are a candidate leukemogenic essential secondary mutation. While RAS mutations are not important prognostic indicators in childhood leukemia overall [26], they may be re-evaluated in light of high hyperdiploidy as defined by our classification protocol which includes both conventional karyotyping and an additional FISH analysis which is not the clinical convention (see Methods). A suggestion that RAS mutations can influence white blood cell count in our cases provides a preliminary suggestion that RAS may be a prognostic marker in this subtype; we do not currently have long-term outcome information for the cases presented here.
We re-analyzed all samples that we screened previously [191 patients [10]] which changed four patient classifications, and have analyzed 326 new patients. The DHPLC technique is highly sensitive, but we excluded marginal sequence calls to ensure that all called RAS mutations were present in the dominant leukemia clone. We did not confirm our previous findings of independent effects of Hispanic status or maternal age on RAS mutation status; while these factors were related to RAS (Table 2), they are also strongly correlated with each other and do not independently associate with RAS (Table 3). We also did not find consistent associations of RAS mutation status to parental smoking, which was our original hypothesis. A suggestive increased risk with parental smoking was apparent in the high hyperdiploid subgroup, but no strong and consistent relationships with parental smoking and RAS were discovered.
In conclusion, we have found that RAS mutation is clearly a secondary mutation to high hyperdiploidy by employing a backtracking assay in children who contract high hyperdiploidy leukemia, and usually if not always occurs after birth. Future studies should focus on the postnatal period to identify causal mechanisms and exposures related to RAS mutation.
Acknowledgements
The authors wish to thank the families who participated in the NCCLS and the clinical collaborators from University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. ML and Katherine Matthay), Children's Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children's Hospital (Dr. Gary Dahl), Children's Hospital Oakland (Dr. James Feusner), Kaiser Permanente Sacramento (Dr. Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo and Alan Wong), Kaiser Permanente San Francisco (Dr. Kenneth Leung), and Kaiser Permanente Oakland (Dr. Stacy Month). JLW is a Scholar in Clinical Research of the Leukemia and Lymphoma Society of America. Supported by NIH grant R01-CA089032 (JLW), P42-ES04705 (MTS) and R01-ES09137 (PAB), P01-ES018172 (PAB, JLW) the Children with Leukaemia Fund, UK (JLW, PAB). JSC is also supported by the National Cancer Institute R25-CA112355.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greaves MF, Alexander FE. An infectious etiology for common acute lymphoblastic leukemia in childhood? Leukemia. 1993;7:349–60. [PubMed] [Google Scholar]
- 2.Oeffinger KC, Mertens AC, Sklar CA, Kawashima T, Hudson MM, Meadows AT, Friedman DL, Marina N, Hobbie W, Kadan-Lottick NS, Schwartz CL, Leisenring W, Robison LL. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355:1572–82. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 3.Onodera N, McCabe NR, Rubin CM. Formation of a hyperdiploid karyotype in childhood acute lymphoblastic leukemia. Blood. 1992;80:203–8. [PubMed] [Google Scholar]
- 4.Paulsson K, Morse H, Fioretos T, Behrendtz M, Strombeck B, Johansson B. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2005;44:113–22. doi: 10.1002/gcc.20222. [DOI] [PubMed] [Google Scholar]
- 5.Panzer-Grumayer ER, Fasching K, Panzer S, Hettinger K, Schmitt K, Stockler-Ipsiroglu S, Haas OA. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood. 2002;100:347–9. doi: 10.1182/blood-2002-01-0144. [DOI] [PubMed] [Google Scholar]
- 6.Maia AT, Tussiwand R, Cazzaniga G, Rebulla P, Colman S, Biondi A, Greaves M. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes Chromosomes Cancer. 2004;40:38–43. doi: 10.1002/gcc.20010. [DOI] [PubMed] [Google Scholar]
- 7.Taub JW, Konrad MA, Ge Y, Naber JM, Scott JS, Matherly LH, Ravindranath Y. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood. 2002;99:2992–6. doi: 10.1182/blood.v99.8.2992. [DOI] [PubMed] [Google Scholar]
- 8.Yagi T, Hibi S, Tabata Y, Kuriyama K, Teramura T, Hashida T, Shimizu Y, Takimoto T, Todo S, Sawada T, Imashuku S. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood. 2000;96:264–8. [PubMed] [Google Scholar]
- 9.Gruhn B, Taub JW, Ge Y, Beck JF, Zell R, Hafer R, Hermann FH, Debatin KM, Steinbach D. Prenatal origin of childhood acute lymphoblastic leukemia, association with birth weight and hyperdiploidy. Leukemia. 2008;22:1692–7. doi: 10.1038/leu.2008.152. [DOI] [PubMed] [Google Scholar]
- 10.Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L, Smith MT, Ma X, Selvin S, Buffler PA, Wiencke JK. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19:415–19. doi: 10.1038/sj.leu.2403641. [DOI] [PubMed] [Google Scholar]
- 11.Aldrich MC, Zhang L, Wiemels JL, Ma X, Loh ML, Metayer C, Selvin S, Feusner J, Smith MT, Buffler PA. Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiol Biomarkers Prev. 2006;15:578–81. doi: 10.1158/1055-9965.EPI-05-0833. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Buffler PA, Selvin S, Matthay KK, Wiencke JK, Wiemels JL, Reynolds P. Daycare attendance and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2002;86:1419–24. doi: 10.1038/sj.bjc.6600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JS, Selvin S, Metayer C, Crouse V, Golembesky A, Buffler PA. Parental smoking and the risk of childhood leukemia. Am J Epidemiol. 2006;163:1091–100. doi: 10.1093/aje/kwj143. [DOI] [PubMed] [Google Scholar]
- 14.Metayer C, Buffler PA. Residential Exposures to Pesticides and Childhood Leukaemia. Radiat Prot Dosimetry. 2008 doi: 10.1093/rpd/ncn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X, Buffler PA, Gunier RB, Dahl G, Smith MT, Reinier K, Reynolds P. Critical windows of exposure to household pesticides and risk of childhood leukemia. Environ Health Perspect. 2002;110:955–60. doi: 10.1289/ehp.02110955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorman AV, Clark R, Farrell DM, Hawkins JM, Martineau M, Secker-Walker LM. Probes for hidden hyperdiploidy in acute lymphoblastic leukaemia. Genes Chromosomes Cancer. 1996;16:40–5. doi: 10.1002/(SICI)1098-2264(199605)16:1<40::AID-GCC6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Raimondi SC, Pui CH, Hancock ML, Behm FG, Filatov L, Rivera GK. Heterogeneity of hyperdiploid (51-67) childhood acute lymphoblastic leukemia. Leukemia. 1996;10:213–24. [PubMed] [Google Scholar]
- 18.Fuery CJ, Impey HL, Roberts NJ, Applegate TL, Ward RL, Hawkins NJ, Sheehan CA, O'Grady R, Todd AV. Detection of rare mutant alleles by restriction endonuclease-mediated selective-PCR: assay design and optimization. Clin Chem. 2000;46:620–4. [PubMed] [Google Scholar]
- 19.Paulsson K, Horvat A, Strombeck B, Nilsson F, Heldrup J, Behrendtz M, Forestier E, Andersson A, Fioretos T, Johansson B. Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2008;47:26–33. doi: 10.1002/gcc.20502. [DOI] [PubMed] [Google Scholar]
- 20.Stam RW, den Boer ML, Schneider P, Meier M, Beverloo HB, Pieters R. D-HPLC analysis of the entire FLT3 gene in MLL rearranged and hyperdiploid acute lymphoblastic leukemia. Haematologica. 2007;92:1565–8. doi: 10.3324/haematol.11220. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto T, Isomura M, Xu Y, Liang J, Yagasaki H, Kamachi Y, Kudo K, Kiyoi H, Naoe T, Kojma S. PTPN11, RAS and FLT3 mutations in childhood acute lymphoblastic leukemia. Leuk Res. 2006;30:1085–9. doi: 10.1016/j.leukres.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Case M, Matheson E, Minto L, Hassan R, Harrison CJ, Bown N, Bailey S, Vormoor J, Hall AG, Irving JA. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res. 2008;68:6803–9. doi: 10.1158/0008-5472.CAN-08-0101. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, Sallan SE, Korsmeyer SJ. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103:3544–6. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 24.Shu XO, Perentesis JP, Wen W, Buckley JD, Boyle E, Ross JA, Robison LL. Parental Exposure to Medications and Hydrocarbons and ras Mutations in Children with Acute Lymphoblastic Leukemia: A Report from the Children's Oncology Group. Cancer Epidemiol Biomarkers Prev. 2004;13:1230–5. [PubMed] [Google Scholar]
- 25.Davidsson J, Paulsson K, Lindgren D, Lilljebjorn H, Chaplin T, Forestier E, Andersen MK, Nordgren A, Rosenquist R, Fioretos T, Young BD, Johansson B. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–31. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- 26.Perentesis JP, Bhatia S, Boyle E, Shao Y, Shu XO, Steinbuch M, Sather HN, Gaynon P, Kiffmeyer W, Envall-Fox J, Robison LL. RAS oncogene mutations and outcome of therapy for childhood acute lymphoblastic leukemia. Leukemia. 2004;18:685–92. doi: 10.1038/sj.leu.2403272. [DOI] [PubMed] [Google Scholar]