Abstract
We here report the genetic basis for susceptibility and resistance to carcinogen-mediated (7,12-Dimethylbenz[a]anthracene (DMBA)) mammary tumorigenesis using the full panel of SS/BN consomic rat strains, in which substitutions of individual chromosomes from the resistant BN strain onto the genomic background of the susceptible SS strain were made. Analysis of 252 consomic females identified rat mammary Quantitative Trait Loci (QTLs) affecting tumor incidence on chromosomes 3 and 5, latency on chromosomes 3, 9, 14 and 19, and multiplicity on chromosomes 13, 16 and 19. In addition, we unexpectedly identified a novel QTL on chromosome 6 controlling a lethal toxic phenotype in response to DMBA. Upon further investigation with chromosomes 6 and 13 congenic lines, in which an additional 114 rats were investigated, we mapped 1) a novel mammary tumor QTL to a region of 27.1 Mbp in the distal part of RNO6, a region that is entirely separated from the toxicity phenotype, and 2) a novel and powerful mammary tumor susceptibility locus of 4.5 Mbp that mapped to the proximal q-arm of RNO13. Comparison of genetic strain differences using existing rat genome databases enabled us to further construct priority lists containing single breast cancer candidate genes within the defined QTLs, serving as potential functional variants for future testing.
Keywords: mammary cancer, consomic strains, DMBA, SS and BN rat
INTRODUCTION
Identification of novel breast cancer susceptibility and resistance genes in genetically diverse human populations is challenging. The use of inbred animal model systems in which the environmental and genetic conditions are controlled can facilitate the discovery of such genes, and should be translated rapidly to human populations with the use of comparative genomics. Samuelsson et al. (2007) recently illustrated the utility of the rat as a model organism to identify novel genetic polymorphisms associated with the etiology of breast cancer in humans. Rat and human mammary carcinomas share similar developmental and histopathological features and, unlike mouse carcinomas, show hormonal responsiveness and do not have a known viral etiology (Russo and Russo, 1987, 2000; Gould et al., 1989; Russo et al., 1990). The rat is therefore a model of choice for the recapitulation of human breast cancer etiology and gene discovery.
The current approach used to identify rat mammary cancer quantitative trait loci (QTLs) is to localize them in genetic crosses, then to generate congenic strains carrying individual QTLs, and finally to clone the gene. Even with the complete genomic DNA sequence of the laboratory rat now available (Gibbs et al., 2004), positional cloning remains a time-consuming and expensive process. An alternative to conventional mapping approaches for QTL mapping is to use chromosome substitution (consomic) rat strains, in which one chromosome at a time from a donor strain substitutes for the corresponding chromosome in the host strain. The method has the advantage of rapid generation of congenic strains from consomic animals (Cowley et al., 2004), and has a greater statistical power to detect linkage over traditional F2 crosses (Nadeau et al., 2000).
We recently reported the first use of a novel pair of rat strains and chromosome substitution techniques (consomic strains) for localization of QTLs important in 7,12-Dimethylbenz[a]anthracene (DMBA; a prototypical polycyclic aromatic hydrocarbon)-induced mammary cancer susceptibility or resistance (Adamovic et al., 2008). Polycyclic aromatic hydrocarbons are an important class of environmental chemical carcinogens and have been implicated in human breast carcinogenesis (Li et al., 1999; Steck et al., 2007). Moreover, the ability of DMBA to induce mammary cancer in rats has been known and studied for over 40 years (Huggins et al., 1961), although the molecular mechanisms by which the carcinogen mediates the transformation of cells in mammary tissue remains largely undefined. Thus, to define mechanisms and to identify genes that determine breast cancer risk, we used consomic rat strains developed from the SS (host) and BN (donor; the same strain used to sequence the rat genome (Gibbs et al., 2004)) parental inbred rat strains, that were identified as highly susceptible and completely resistant, respectively, to DMBA-induced mammary carcinogenesis (Adamovic et al., 2008). As described in detail in our previous study, the BN parental females developed no macroscopically detectable mammary carcinomas, whereas 93% of the SS parental females developed an average of 2.45 mammary adenocarcinomas per rat after DMBA-treatment, and a mean tumor latency time of 11.43 weeks. As a starting point in our pilot study, we chose to evaluate DMBA-induced mammary cancer phenotypes in SS-10BN and SS-12BN consomic females (in which the resistant BN chromosomes 10 and 12, respectively, are introgressed into the isogenic genetic background of the susceptible SS strain) because of the presence of the highly penetrant breast cancer susceptibility genes Brca1 and Tp53 on chromosome 10 and Brca2 on chromosome 12 (Adamovic et al., 2008). Studying just 2 out of 20 consomic SS/BN strains, we were able to rapidly identify and localize QTLs on the BN chromosome 10 that affect mammary cancer latency and multiplicity over a 15-week period. We further reported the identification of novel cancer target genes deregulated at the somatic level by using an appropriate control in tumor genomic profiling studies, demonstrating the unique resource of our physiologically relevant mammary tumor rat model (Adamovic et al., 2009).
The purpose of the present study was to complete the screening of the full SS/BN consomic rat panel (i.e., 18 additional consomic strains were investigated), in order to map genetic determinants of susceptibility to DMBA-induced mammary cancer. Using the chromosome substitution methodology, we systematically mapped mammary cancer QTLs to a limited number of rat chromosomes (Rattus norvegicus, RNO) controlling the tumor phenotypes of incidence, latency and/or multiplicity. Furthermore, we took advantage of a series of congenic lines generated directly from the significant consomic SS-6BN and SS-13BN strains, respectively, and isolated defined DMBA-induced QTLs, further increasing our understanding of carcinogen-mediated breast tumorigenesis.
Our study demonstrates the valuable resource of DMBA-induced consomic- and congenic strains, together with a powerful genetic toolkit available for the rat (Aitman et al., 2008) as an accelerated approach to positionally clone mammary cancer susceptibility genes.
MATERIALS AND METHODS
Animals, Treatment and Mammary Gland Samples
Eighteen consomic rat lines were derived from the SS/JrHsdMcwi and BN/NHsdMcwi parental rats, and 8 congenic lines were derived from the SS-6BN/Mcwi and SS-13BN/Mcwi consomic strains. The generation of consomic strains from the two parental strains, and derivation of congenic strains from the consomic strains have been described in detail previously (Cowley et al., 2004; Moreno et al., 2007) http://pga.mcw.edu/. All rats were received at weaning and were housed, and fed as described previously (Adamovic et al., 2008). All experimental procedures were conducted following approval of the Institutional Animal Care and Use Committee (IACUC). At the age of 47-57 days, SS-1BN/Mcwi (n=13), SS-2BN/Mcwi (n=14), SS-3BN/Mcwi (n=14), SS-4BN/Mcwi (n=9), SS-5BN/Mcwi (n=13), SS-6BN/Mcwi (n=20), SS-7BN/Mcwi (n=14), SS-8BN/Mcwi (n=15), SS-9BN/Mcwi (n=18), SS-11BN/Mcwi (n=11), SS-13BN/Mcwi (n=18), SS-14BN/Mcwi (n=15), SS-15BN/Mcwi (n=13), SS-16BN/Mcwi (n=13), SS-17BN/Mcwi (n=14), SS-18BN/Mcwi (n=10), SS-19BN/Mcwi (n=14), SS-20BN/Mcwi (n=14), SS.BN-(D6Rat149-D6RatArb3)/Mcwi (n=14), SS.BN-(D6Rat149-D6Rat18)/Mcwi (n=15), SS.BN-(D6Rat149-D6RatHF8)/Mcwi (n=15), SS.BN-(D6Rat119-D6RatArb3)/Mcwi (n=13), SS.BN-(D13Rat7-D13Rat60)/Mcwi (n=16), SS.BN-(D13Rat111-D13Got22)/Mcwi (n=12), SS.BN-(D13Rat123-D13Rat101)/Mcwi (n=14) and SS.BN-(D13Rat151-D13Rat197)/Mcwi (n=15) virgin females received a single 65 mg/kg DMBA, 200 mg DMBA (Acros Organics, NJ, USA) dissolved in 10 ml sesame oil (S3547, Sigma-Aldrich), dose by oral gavage. Controls were 2-4 SS, 2-4 consomic and congenic females, respectively, from each strain, age-matched, treated with an equivalent volume of sesame oil (without DMBA).
Beginning 3 weeks after inoculation, the rats were palpated weekly for mammary tumors until the 15th week. The location of each palpable tumor was noted. The time between the date of DMBA treatment and the date of detection of the first tumor was recorded and defined as latency. At week 15, the rats were sacrificed by CO2 asphyxiation and the number and location of the mammary tumors were recorded at necropsy. Portions of each tumor were fixed in 10% buffered formalin, processed and embedded. Paraffin sections, 5 μm thick, were stained with hematoxylineosin (H&E). Pathologic diagnoses of the mammary lesions were classified by a pathologist blind to strain identity, as previously described (Russo and Russo, 2000).
Statistical Analysis
Cancer phenotypes of tumor incidence (number of rats with tumors) and latency were compared between the susceptible parental strain, SS, and the 18 consomic- and 8 congenic-rat strains, respectively. For tumor incidence, we used classical chi-square tests and Fisher’s exact test when there are cell counts ≤5. The bootstrap method was adopted to adjust for the multiple testing. For tumor latency, we performed survival analysis using log-rank tests. Bonferroni adjustment was used to adjust for the multiple testing. Comparison of tumor multiplicity (number of tumors on average developed per rat) was also evaluated using the Bootstrap method. All presented p-values were adjusted ones. Adjusted p-values ≤0.05 were considered to be significant. All tests are 2-sided. For cumulative incidence plots, Kaplan-Meier estimators were used to compute the cumulative incidence of death, and the cumulative incidence probability for tumor development was estimated by treating death as a competing risk. The statistical analysis was implemented using the SAS software version 9.1 (SAS Institute, Cary, NC).
Search for Genes, Copy Number Variants (CNVs) and Single Nucleotide Polymorphisms (SNPs) in the Defined Mammary Tumor QTL-regions
The location, order and literature information of genes in the mapped QTL regions were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/genome/guide/rat/index.html). Haplotypes for the rat strains of interest were established using the software tool SNPlotyper from the Rat Genome Database (RGD: http://rgd.mcw.edu/). Mapped CNVs in the rat genomic regions of interest were obtained from our previously published data (Guryev et al., 2008).
Comparative Genomics
The rat (build 3.4) and human (build 37.1) genomes were compared within rat chromosomes of interest using the synteny view of the Ensembl genome browser (http://www.ensembl.org).
RESULTS
Virgin female rats, 47-57 days old, of 18 consomic rat strains were tested for their susceptibility to mammary adenocarcinoma development following DMBA treatment. The majority of the tumors (>90%) detected in the consomic rats were classified histologically as adenocarcinomas (ductal carcinomas in situ, papillary and cribriform carcinoma, or invasive carcinoma); the remaining tumors were classified as adenomas, as previously described for the SS-, SS-10BN- and SS-12BN-tumors (Adamovic et al., 2009). No difference in differentiation was found between the tumors developed in the SS females when compared to the tumors detected in the consomic females. Control SS- and consomic-rats treated with sesame oil alone developed no detectable tumors.
Evaluation of Tumor Incidence, Latency and Multiplicity in the Consomic Inbred Rat Strains
The phenotypes of tumor incidence, latency, and multiplicity were evaluated in the affected consomic inbred rat strains and were compared to the tumor phenotypes of the susceptible SS parental strain.
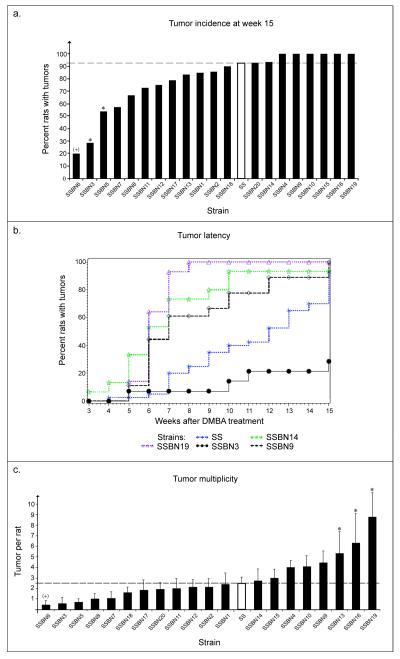
The average values of tumor incidence for control (SS) and 20 consomic strains at week 15 are illustrated in Figure 1a (data from SS-10BN and SS-12BN strains, previously investigated (Adamovic et al., 2008), are included in the figure). Female rats from the SS-3BN, SS-5BN and SS-6BN consomic strains had a tumor incidence of 29% (4/14), 54% (7/13) and 20% (4/20) at week 15, showing a significant phenotypic difference compared with the SS strain (93%; 37/40), (SS vs. SS-3BN, P<0.0001; SS vs. SS-5BN, P=0.0317; SS vs. SS-6BN, P<0.0001, Fisher’s exact test using Bootstrap). Unexpectedly, the SS-6BN consomic females were found to have a death rate of 95% (19/20) by the end of week 15, and was the only consomic strain with a significantly different rate of death from that of the SS strain (SS vs. SS-6BN, P<0.0001, Fisher’s exact test using Bootstrap). Due to the high death incidence early after carcinogen treatment (see Fig. 3c), a true value of tumor incidence at week 15 was not possible to obtain for the SS-6BN strain. Nevertheless, our data suggests the presence of a locus on the BN chromosome 6 controlling a highly toxic phenotype, and possibly a tumor phenotype, as the few rats that survived, developed very aggressive (fast growing) tumors. Thus, substitution of the entire chromosome 3 and 5 from the BN rat in the SS-3BN and SS-5BN consomic rat greatly reduced the tumor incidence phenotype after carcinogen treatment.
Figure 1.
Illustration of phenotypes from 20 consomic strains. a) The tumor incidence at week 15 after carcinogen treatment was calculated to 29% (4/14) and 54% (7/13) in the SS-3BN and SS-5BN consomic rat, respectively, which is significantly reduced compared with the SS parental strain (93%; 37/40) (SS vs. SS-3BN, P<0.0001; SS vs. SS-5BN, P=0.0317). b) Comparison of tumor latency showed a significantly different value for the SS-3BN (black line, filled circles in the figure), d), SS-9BN (black dashed line, open diamonds), SS-14BN (green dotted line, open stars) and SS-19BN consomic strains (purple dotted line, open triangles) vs. the SS (blue dotted line, stars) (SS vs. SS-3BN, P=0.0043; SS vs. SS-9BN, P=0.0018; SS vs. SS-14BN, P<0.0001; SS vs. SS-19BN, P<0.0001). c) A significant difference in tumor multiplicity was observed in the SS-13BN, SS-16BN and SS-19BN rat compared to the SS (SS vs. SS-13BN, P=0.0024; SS vs. SS-16BN, P=0.0002; SS vs. SS-19BN, P<0.0001; standard deviation (s.d.) is given). Due to a high death incidence (+) of the SS-6BN consomic females early after carcinogen treatment, a true value of tumor incidence and multiplicity was not possible to obtain.
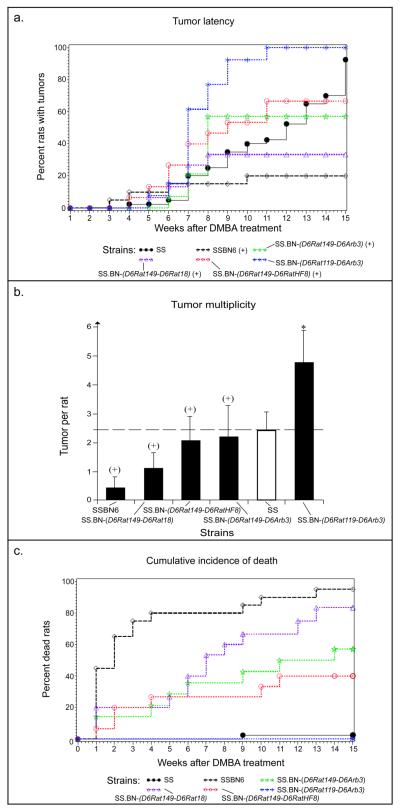
Figure 3.
Illustration of phenotypes from congenic lines derived from the SS-6BN consomic rat. a) Comparison of tumor latency showed a significantly different value for the SS.BN-(D6Rat119-D6RatArb3) strain compared to the SS (SS vs. SS.BN-(D6Rat119-D6RatArb3), P<0.0001). SS: black line, filled circles; SS.BN-(D6Rat149-D6Rat18): purple dotted line, open triangles; SS-6BN: dashed black line, open diamonds; SS.BN-(D6Rat149-D6RatHF8): red dotted line, open circles; SS.BN-(D6Rat149-D6Arb3): green dotted line, open stars; SS.BN-(D6Rat119-D6Arb3): blue dotted line, stars. b) Also a significant difference in tumor multiplicity was observed in the SS.BN-(D6Rat119-D6RatArb3) rat compared to the SS (SS vs. SS.BN-(D6Rat119-D6RatArb3), P=0.0003; s.d. is given). c) The death rates were evaluated to 57% (8/14) for the SS.BN-(D6Rat149-D6RatArb3), 80% (12/15) for the SS.BN-(D6Rat149-D6Rat18) and 40% (6/15) for the SS.BN-(D6Rat149-D6RatHF8) congenic by the end of week 15, all of which showed a significant phenotypic difference compared with the SS (SS vs. SS.BN-(D6Rat149-D6RatArb3), P<0.0001; SS vs. SS.BN-(D6Rat149-D6Rat18), P<0.0001; SS vs. SS.BN-(D6Rat149-D6RatHF8), P=0.0044). Due to a high death incidence (+) of the SS.BN-(D6Rat149-D6RatArb3), SS.BN-(D6Rat149-D6Rat18) and SS.BN-(D6Rat149-D6RatHF8) females (similarly to what was observed in the SS-6BN consomic females), a true value of the tumor phenotypes were not possible to obtain. SS: black line, filled circles; SS.BN-(D6Rat149-D6Rat18): purple dotted line, open triangles; SS-6BN, black dashed line, open diamonds; SS.BN-(D6Rat149-D6RatHF8): red dotted line, open circles; SS.BN-(D6Rat149-D6RatArb3): green dotted line, open stars; SS.BN-(D6Rat119-D6RatArb3): blue dotted line, stars.
We next evaluated the latency of tumor appearance in the consomic strains. We found the tumor latency time for the SS-3BN, SS-9BN, SS-14BN and SS-19BN consomic strains to be significantly different from that of the SS (SS vs. SS-3BN, P=0.0043; SS vs. SS-9BN, P=0.0018; SS vs. SS-14BN, P<0.0001; SS vs. SS-19BN, P<0.0001, log-rank tests and Kaplan-Meier survival estimators), see Figure 1b. The mean latency for the parental SS rats was 11.40±0.53 weeks, and in the SS-3BN, SS-9BN, SS-14BN and SS-19BN consomics was 13.64±0.89, 8.33±0.77, 6.67±0.58 and 6.29±0.22 weeks, thus, identifying a QTL located on RNO3, 9, 14 and 19 controlling the mammary tumor latency phenotype.
The number of tumors on average developed per SS-13BN, SS-16BN and SS-19BN rat was further significantly different from what was observed in the SS parental rat (SS vs. SS-13BN, P=0.0024; SS vs. SS-16BN, P=0.0002; SS vs. SS-19BN, P<0.0001, Bootstrap). The SS-13BN, SS-16BN and SS-19BN rat developed 5.33±4.38, 6.31±5.79 and 8.79±4.68 tumors on average, compared to 2.45±1.32 in the SS rat (see Fig. 1c). Accordingly, our data demonstrates that BN chromosomes 13, 16 and 19 increases the tumor multiplicity phenotype in the SS background.
Tumor Phenotype Evaluation of Congenic Lines Covering RNO6 and 13, Respectively
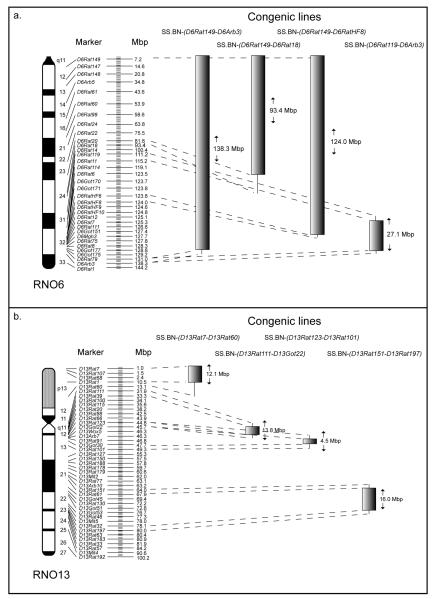
To further define the mapped genomic regions within RNO6 and RNO13, females from 4 congenic lines covering each chromosome were phenotyped after carcinogen treatment and compared to the SS parental strain. Figure 2a and 2b shows a schematic representation of the congenic lines SS.BN-(D6Rat149-D6RatArb3), SS.BN-(D6Rat149-D6Rat18), SS.BN-(D6Rat149-D6RatHF8) and SS.BN-(D6Rat119-D6RatArb3) generated from the SS-6BN-, and SS.BN-(D13Rat7-D13Rat60), SS.BN-(D13Rat111-D13Got22), SS.BN-(D13Rat123-D13Rat101) and SS.BN-(D13Rat151-D13Rat197) congenics generated from the SS-13BN-consomic rat (Moreno et al., 2007), respectively. The majority (>90%) of the mammary tumors developed in females from each congenic strain were histologically classified as adenocarcinomas (ductal carcinomas in situ, papillary and cribriform carcinoma, or invasive carcinoma). As before, no difference in differentiation was found between the tumors developed in the SS females when compared to the tumors detected in the congenic females. Control congenic rats treated with sesame oil alone developed no detectable tumors.
Figure 2.
Schematic representation of the congenic lines derived from the SS-6BN (a) and SS-13BN-consomic rat (b). The markers displayed on the right side of each chromosome were used to define the differential segments. Each markers cytogenetic and Mb position is indicated. The short vertical lines flanking the differential segments represent the intervals of recombination between the SS and BN parental genomes.
The size of the congenic regions ranged from 27.1 Mbp (SS.BN-(D6Rat119-D6RatArb3)) to 138.3 Mbp (SS.BN-(D6Rat149-D6RatArb3)) on RNO6, see Figure 2a. As described above, RNO6 substitution from the BN rat in the SS-6BN consomic rat caused death in 95% of the carcinogen-induced females by the end of week 15. Only four out of 20 (20%) SS-6BN females developed tumors, three of which had to be sacrificed before week 15 due to the large size of the fast growing tumors. The genetic make-up of the SS-6BN rat clearly created a background of susceptibility to the highly toxic phenotype in the presence of the carcinogen. Interestingly, substitution of the distal part of chromosome 6 from the BN rat in the SS.BN-(D6Rat119-D6RatArb3) congenic rat, completely eliminated the toxicity phenotype, and left only the presence of a strong tumor phenotype. Hence, all SS.BN-(D6Rat119-D6RatArb3) females survived carcinogen treatment and developed tumors, enabling statistical comparison of the tumor phenotypes between the SS parental and the SS.BN-(D6Rat119-D6RatArb3) congenic strain. We found the SS.BN-(D6Rat119-D6RatArb3) strain to develop tumors significantly earlier (7.54±0.42 weeks vs. 11.40±0.53, P<0.0001, see Fig. 3a) and had significantly more tumors developed (4.77±2.39 vs. 2.45±1.32, P=0.0003, see Fig. 3b). Consequently, we identified a 27.1 Mbp region in the SS.BN-(D6Rat119-D6RatArb3) congenic line controlling the tumor latency and multiplicity phenotypes, respectively. This region does not appear to contain the DMBA toxicity QTL.
The phenotype pattern was more complex for the SS.BN-(D6Rat149-D6RatArb3), SS.BN-(D6Rat149-D6Rat18) and SS.BN-(D6Rat149-D6RatHF8) lines. Both the toxicity- and the tumor phenotype were observed in each line (Figs. 3a, 3b, 3c). However, due to the high rate of death found in these lines, it was difficult to ascertain the presence of possible loci controlling tumor phenotype. The toxicity phenotype manifested with death rates of 57% (8/14) for the SS.BN-(D6Rat149-D6RatArb3), 80% (12/15) for the SS.BN-(D6Rat149-D6Rat18) and 40% (6/15) for the SS.BN-(D6Rat149-D6RatHF8) congenic by the end of week 15 (see Fig. 3c), all of which showed a significant phenotypic difference compared with the parental strain (SS vs. SS.BN-(D6Rat149-D6RatArb3), P<0.0001; SS vs. SS.BN-(D6Rat149-D6Rat18), P<0.0001; SS vs. SS.BN-(D6Rat149-D6RatHF8), P=0.0044). When these results are considered collectively, the BN chromosome region within (D6Rat149-D6Rat11) appears to contain a QTL of 115.2 Mbp controlling the toxicity phenotype.
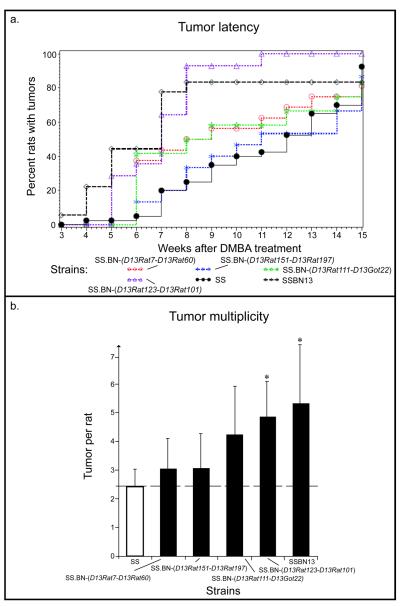
In contrast to the RNO6 congenic lines, the four congenics covering regions of RNO13 were discrete and non-overlapping, ranging in size from 4.5 Mbp (SS.BN-(D13Rat123-D13Rat101)) to 16.0 Mbp (SS.BN-(D13Rat151-D13Rat197)), see Figure 2b. The consomic SS-13BN strain had significantly increased tumor multiplicity when compared with the parental SS strain. In the congenics, significant difference in tumor latency was found between the SS.BN-(D13Rat123-D13Rat101) and the parental SS strain (see Fig. 4a; SS vs. SS.BN-(D13Rat123-D13Rat101), 6.93±0.45 vs 11.43±0.54, P<0.0001). Tumor multiplicity was also significantly different in the SS.BN-(D13Rat123-D13Rat101) congenic (see Fig. 4b; 4.86±2.63 vs 2.45±1.32; SS vs. SS.BN-(D13Rat123-D13Rat101), P=0.0343). No significant difference was observed between the SS and the four congenics with regard to tumor incidence. In summary, a defined QTL of 4.5 Mbp in the SS.BN-(D13Rat123-D13Rat101) congenic line was identified controlling both tumor multiplicity and latency.
Figure 4.
Illustration of phenotypes from congenic lines derived from the SS-13BN consomic rat. a) Comparison of tumor latency showed a significantly different value for the SS.BN-(D13Rat123-D13Rat101) strain compared to the SS (SS vs. SS.BN-(D13Rat123-D13Rat101), P<0.0001). SS.BN-(D13Rat7-D13Rat60): red dotted line, open circles; SS.BN-(D13Rat123-D13Rat101): purple dotted line, open triangle; SS.BN-(D13Rat151-D13Rat197): blue dotted line, stars; SS: black line, filled circles; SS.BN-(D13Rat111-D13Got22): green dotted line, open stars; SS-13BN: black dashed line, open diamonds. b) Also a significant difference in tumor multiplicity was observed in the SS.BN-(D13Rat123-D13Rat101) rat compared to the SS (SS vs. SS.BN-(D13Rat123-D13Rat101), P=0.0343; s.d. is given).
Identification of Breast Cancer Candidate Genes within the Defined Mammary Tumor QTL Regions
Manual literature search was performed to identify genes related to the disease of cancer, and specifically breast cancer, in the defined toxicity- and mammary tumor QTL regions of (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101). As indicated in Table 1, the (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101) regions contain a total of 838, 241 and 85 genes, respectively. The gene density (e.g. genes per Kbp sequence) was two to three times higher in the (D13Rat123-D13Rat101) region (1.9%) as compared to the regions of (D6Rat149-D6Rat11) (0.7%) and (D6Rat119-D6RatArb3) (0.9%), correspondingly so were the percentage of cancer-related genes in the regions. Accordingly, 11.9%, 16.6% and 32.9% cancer-related genes were identified in the (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101) regions. Supplementary Table 1 lists all cancer- and breast cancer-related genes, and also highlights obvious candidates, such as Cyp1b1, Msh2, Twist1, Ahr, Foxa1, Esrβ, Akt1, Mta1 and Kiss1, relevant to the present mammary tumor model.
Table 1.
Gene, SNP and CNV information in the (D6Rat149-D6Rat11), (D6Rat-D6RatArb3) and (D13Rat123-D13Rat101) regions.
| QTL regions | |||
|---|---|---|---|
| (D6Rat149-D6Rat11) | (D6Rat119-D6RatArb3) | (D13Rat123-D13Rat101) | |
| Size (bp) | 115,241,142 | 27,023,952 | 4,468,645 |
| Total genes | 838 | 241 | 85 |
| Genes per Kbp (%) | 0.7% | 0.9% | 1.9% |
| Cancer-related genes per Kbp (%) | 100 (11.9%) | 40 (16.6%) | 28 (32.9%) |
| Breast cancer-related genes per Kbp (%) | 33 (3.9%) | 15 (6.2%) | 8 (9.4%) |
| Total SNPs | 864 | 248 | 49 |
| SNPs per Kbp (%) | 0.7% | 0.9% | 1.1% |
| Polymorphic SNPs between SS and BN | 474 (54.9%) | 129 (52.0%) | 31 (63.3%) |
| Candidate disease SNPs a | 47 (5.4%) | 5 (2.0%) | 9 (18.4%) |
| CNVs between SS and BN | 2 | 1 | 0 |
, genotypes shared between two susceptible to DMBA induced mammary tumor strains (SS, SPRD) and different from genotypes of three resistant strains (COP, WKY, BN).
We next used the SNPlotyper tool to compare the haplotype structure across the (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101) QTL regions in 5 inbred rat strains, 2 of which are susceptible to DMBA-induced mammary tumor development( SS (Adamovic et al., 2008) and SPRD (Quan et al., 2006)), and 3 (BN (Adamovic et al., 2008), COP (Shepel et al., 1998) and WKY (Quan et al., 2006)) that are resistant, in order to identify potentially important SNPs in mammary carcinogenesis. In total, 864, 248, and 49 SNPs were found, yielding 0.7%, 0.9% and 1.1% SNPs per Kbp sequence in the (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101) regions (see Table 1), reflecting a rather uniform number of SNPs present per region. Correspondingly so were the presence of polymorphic SNPs between the SS and BN strains in the (D6Rat149-D6Rat11) (54.9%), (D6Rat119-D6RatArb3) (52.0%) and (D13Rat123-D13Rat101) (63.3%) regions, similarly to what was found for the entire rat genome (57.3%), see Table 1. Interestingly, a total of 18.4% possible disease SNPs were identified in the (D13Rat123- D13Rat101) QTL region, compared to only 5.4% and 2.0% in the (D6Rat149-D6Rat11) and (D6Rat119-D6RatArb3) regions (see Table 1). Supplementary Table 2 lists potential candidate disease alleles, and genes that included these SNPs. Breast cancer genes such as Elm4, Vrk1 and Chirl1, appeared as obvious candidates. We further identified Fut8 and Actn1, known cancer-related genes, and Rtn1, Wdr22 and Ppfia, genes so far not related to cancer, as other positional candidates, because of the clustering of SNPs in the (D6Rat149-D6Rat11) and (D13Rat123-D13Rat101) QTL region (see Supplementary Table 2). Interestingly, clustering of candidate disease SNPs that may be of importance were also found in non-coding regions.
Furthermore, we used our recently mapped rat CNV data (Guryev et al., 2008), another form of sequence variation that has been implicated in disease susceptibility and resistance, to identify additional candidates that may contribute to the mammary cancer phenotype. A total of 2, 1 and 0 CNVs were found between the susceptible SS and the resistant BN rat strains in the (D6Rat149-D6Rat11), (D6Rat119-D6RatArb3) and (D13Rat123-D13Rat101) QTL regions (see Table 1). Supplementary Table 3 lists potential candidate genes that co-localize with the mapped CNVs. We included genes located in the relative vicinity of CNVs (0.5 Mbp sequence on each side of the CNV) as potential disease candidates, as our previous study (Guryev et al., 2008) suggested long-range dosage effects of regulatory elements that reside in the CNVs. A total of 73 and 35 genes were identified to reside within, and in the near vicinity of, the structurally rearranged DNA regions in the (D6Rat149-D6Rat11) and (D6Rat119-D6RatArb3) QTLs (see Supplementary Table 3). Nine are known cancer-related genes, 4 of which are breast cancer-related (Zfp36l1, Serpina1, Serpina5 and Dicer1), enhancing their role as interesting mammary tumor candidates in the present model.
DISCUSSION
We previously reported the use of a novel rat strain pair, SS (sensitive) and BN (resistant), to identify chromosomes with mammary cancer phenotypes in response to chemical carcinogenesis (DMBA) (Adamovic et al., 2008). We now report the complete chromosomal screening for DMBA-induced rat mammary cancer phenotypes in this consomic rat model system, using BN chromosome substitutions on to the SS genomic background. Seven chromosomes (RNO3, 5, 9, 13, 14, 16 and 19) were found to influence mammary cancer phenotype. In addition, a novel QTL controlling a toxic phenotype in response to DMBA was unexpectedly identified in RNO6, with a 95% mortality rate. This QTL seemed to co-exist with a mammary tumor phenotype, as all survivors developed aggressive tumors. Upon further investigation with congenic lines, we identified 1) a novel mammary tumor QTL to a region of 27.1 Mbp in the distal part of RNO6 (SS.BN-(D6Rat119-D6RatArb3) congenic), a region that is entirely separated from the toxicity phenotype, and 2) a novel and powerful mammary tumor susceptibility locus of 4.5 Mbp (SS.BN-(D13Rat123-D13Rat101) congenic) that mapped to the proximal q-arm of RNO13.
In this study, we show that the chromosome substitution technique, when applied to DMBA-induced rat mammary cancer, is a good complement to existing techniques for studying the genetics of breast cancer susceptibility and resistance, with the advantage of allowing the effect of QTLs to be investigated within, and between, individual chromosomes in a homogeneous population. Shao et al. (2008) recently reported that an average of 6-7 chromosomes are involved per multi-factorial trait in rat and mouse chromosome substitution strains (range 3 to 11 significant chromosomes per trait in the rat), which is in concordance with our finding of 7 significant chromosomes (3 for tumor multiplicity, 2 for tumor incidence and 4 for the tumor latency phenotype). It is likely that within each chromosome with a mammary cancer phenotype, multiple QTLs affecting the phenotype exist, as suggested by Samuelson et al. (2005). In our study, substitution of the BN chromosome 3 demonstrated a powerful effect on reducing tumor incidence at week 15 by 69% compared with the parental strain. Schaffer et al. (2006) previously identified a rat mammary QTL reducing tumor susceptibility to estrogen induced carcinomas on chromosome 3 from an ACI × BN cross. Further study with congenic animals will elucidate the relationship between this carcinogen-induced QTL and the estrogen-induced QTL. On RNO5, there have been reported at least 6 QTLs reducing susceptibility to estrogen-induced (2 QTLs (Gould et al., 2004; Schaffer et al., 2006)) and DMBA-induced (4 QTLs (Samuelson et al., 2005; Quan et al., 2006)) mammary carcinogenesis in other rat strain pairs. Our study showed a 42% reduction in tumor incidence at week 15, consistent with data from other groups. Nonetheless, our screening of the consomic panel confirms the presence of multiple important hereditary factors underlying the trait of mammary cancer that can be further elucidated and studied in this model system.
Upon further investigation of the significant chromosomes 6 and 13 using congenic rats, three unique and well-defined QTLs were identified. The toxic in response to DMBA QTL, was narrowed to the proximal-mid part of the BN chromosome 6 (the region of (D6Rat149-D6Rat11)), a region which harbors multiple estrogen and breast cancer-related genes, including key players in the activation of polycyclic aromatic hydrocarbons to active metabolites. Of note, the Ahr gene is known to be up-regulated upon oral administration of DMBA, expressing a receptor that induces gene transcription of Cyp1b1 (Nebert and Dalton, 2006), also located within the (D6Rat149-D6Rat11) region. The coding enzyme of Cyp1b1 is known to 1) metabolize DMBA into a mutagenic epoxide intermediate that attacks DNA, and causes malignant transformation in polycyclic aromatic hydrocarbon-mediated carcinogenesis (Nebert and Dalton, 2006; Shimada, 2006) and 2) to hydroxylate estrogen, altering the tumor-promoting effects of estrogen in breast cancer (Clemons and Goss, 2001). Moreover, the (D6Rat149-D6Rat11) region also contains the Esr-β gene, of which the coding receptor directly interacts with Ahr to generate estrogenic effects leading to abnormal growth and, ultimately tumor progression (Ohtake et al., 2003). Esr-β may also be involved in breast cancer susceptibility (Maguire et al., 2005). Foxa1 in this region codes for a co-factor required for the association of the estrogen receptor with chromatin in mediating an estrogen response in breast cancer cells (Carroll et al., 2005). It is intriguing to speculate that differences in these genes in the BN RNO6 could have caused differences in metabolism resulting in death of the animals. Remarkably, the toxicity phenotype was completely eliminated in the SS.BN-(D6Rat119-D6RatArb3) congenic animals, enabling us to map a mammary tumor QTL in the (D6Rat119-D6RatArb3) region.
On RNO13, not previously known to have a QTL for rat mammary cancer, the BN alleles of the (D13Rat123-D13Rat101) region were associated with decreased latency and increased multiplicity. This 4.5 Mbp region awaits additional analysis with finer congenic mapping.
It is notable that the minimal region of (D13Rat123-D13Rat101) (4.5 Mbp) contained twice as many cancer-related genes as the regions of (D6Rat149-D6Rat11) (115.2 Mbp) and (D6Rat119-D6RatArb3) (27.1 Mbp) (Table 1), respectively. In addition, it encompassed approximately 4 and 9 times as many potential cancer-disease SNPs as the (D6Rat149-D6Rat11) and (D6Rat119-D6RatArb3) regions, respectively. It is tempting to associate these findings with the fact that the (D13Rat123-D13Rat101) region accomplished an almost identical effect regarding the tumor multiplicity and latency phenotypes, respectively, as the 6 times larger (D6Rat119-D6RatArb3) region. Interestingly, we also found potential disease SNPs that did not reside within protein coding sequence, suggesting the presence of non-coding elements that may be important in the development of mammary tumors. Such elements were previously reported to alter breast cancer risk in both rats and humans (Samuelson et al., 2007).
Finally, the significant SS-9BN, SS-13BN, SS-14BN, SS-16BN and SS-19BN consomic-, and the SS.BN-(D6Rat119-D6RatArb3) and SS.BN-(D13Rat123-D13Rat101) congenic-strains, exhibited a higher mammary tumor susceptibility phenotype (for either the tumor latency or multiplicity phenotype, or in some cases for both phenotypes) when compared to the phenotype of the susceptible SS parental strain, suggesting the presence of susceptibility genes or lack of resistance genes on the BN chromosome/chromosomal segment introgressed in the SS genomic background, or could reflect the action of modifying genes in the SS background on BN. The high number of strains exhibiting this particular feature cannot easily be explained, however, this kind of effect has been described previously for rat mammary tumor QTLs mapped in other rat strain pairs (Shepel et al., 1998; Quan et al., 2006; Schaffer et al., 2006). Furthermore, no metastases were detected in the parental SS-, consomic- or congenic females, which is in concordance with other published data (Russo and Russo, 2000), and may partly be due to the fact that rats usually are not followed for more than 24 months.
In conclusion, using chromosomal substitution rat strains, we have identified 7 chromosomes affecting mammary carcinogenesis in the rat. We extended our findings using congenic animals, further identifying novel mammary cancer and DMBA toxicity QTLs. In addition, comparison of known genetic strain differences using existing rat genomic databases enabled us to construct priority lists containing single breast cancer candidate genes within the defined QTLs, serving as potential functional variants for future testing. We have demonstrated the potential of this method for investigating breast cancer susceptibility and resistance and aim to identify additional mammary tumor susceptibility loci and genes, which ultimately may prove important in breast cancer prevention.
Supplementary Material
Acknowledgments
Supported by: Grants from the Healthier Wisconsin Research Initiative (SLS, HJJ); Lady Harley Riders Breast Cancer Research Fund (SLS); Medical College of Wisconsin Breast Cancer Research Fund (TA); Scott and Peggy Sampson Memorial Breast Cancer Research Fellowship (TA). Consomic rats were developed by a Program in Genomic Applications (Grant U01HL066579). Congenic animals were developed by two NHLBI grants (SCOR PPG P01HL082798 and MI R01HL089930).
REFERENCES
- Adamovic T, McAllister D, Rowe J, Wang T, Jacob HJ, Sugg SLS. Genetic mapping of mammary tumor traits to rat chromosome 10 using a novel panel of consomic rats. Cancer Genet Cytogenet. 2008;186:41–48. doi: 10.1016/j.cancergencyto.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Adamovic T, McAllister D, Guryev V, Wang X, Andrae JW, Cuppen E, Jacob HJ, Sugg SL. Microalterations of inherently unstable genomic regions in rat mammary carcinomas as revealed by long oligonucleotide array-based comparative genomic hybridization. Cancer Res. 2009;69:5159–5167. doi: 10.1158/0008-5472.CAN-08-4038. [DOI] [PubMed] [Google Scholar]
- Aitman T, Critser J, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, Holmdahl R, Hubner N, Izsvák Z, Jacob HJ, Kuramoto T, Kwitek AE, Marrone A, Mashimo T, Moreno C, Mullins J, Mullins L, Olsson T, Pravenec M, Riley L, Saar K, Serikawa T, Shull JD, Szpirer C, Twigger SN, Voigt B, Worley K. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- Cowley AW, Jr, Roman RJ, Jacob HJ. Application of chromosomal substitution techniques in gene-function discovery. J Physiol. 2004;554:46–55. doi: 10.1113/jphysiol.2003.052613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar AlbáM, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gould MN, Wang B, Moore CJ. Modulation of Mammary Carcinogenesis by Enhancer and Suppressor Genes. In: Burn GL, editor. Genes and signal transduction in multistep carcinogenesis. M. Decker; New York: 1989. pp. 19–38. [Google Scholar]
- Gould KA, Tochacek M, Schaffer BS, Reindl TM, Murrin CR, Lachel CM, VanderWoude EA, Pennington KL, Flood LA, Bynote KK, Meza JL, Newton MA, Shull JD. Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics. 2004;168:2113–2125. doi: 10.1534/genetics.104.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SA, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Selection bias and functional impact of DNA copy number variation in the rat. Nat Genet. 2008;40:538–545. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- Huggins C, Grand L, Brillantes F. Mammary cancer induced by a single feeding of polynuclear hydrocarbons, and its suppression. Nature. 1961;189:204–207. doi: 10.1038/189204a0. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang W, Sahin AA, Hittelman WN. DNA adducts in normal tissue adjacent to breast cancer: a review. Cancer Detect Prev. 1999;23:454–462. doi: 10.1046/j.1525-1500.1999.99059.x. [DOI] [PubMed] [Google Scholar]
- Maguire P, Margolin S, Skoglund J, Sun XF, Gustafsson JA, Børresen-Dale AL, Lindblom A. Estrogen receptor beta (ESR2) polymorphisms in familial and sporadic breast cancer. Breast Cancer Res Treat. 2005;94:145–152. doi: 10.1007/s10549-005-7697-7. [DOI] [PubMed] [Google Scholar]
- Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW., Jr Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics. 2007;31:228–235. doi: 10.1152/physiolgenomics.00280.2006. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Singer JB, Matin A, Lander ES. Analyzing complex genetic traits with chromosome substitution strains. Nat Genet. 2000;24:221–225. doi: 10.1038/73427. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, Yanagisawa J, Fujii-Kuriyama Y, Kato S. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423:545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- Quan X, Laes JF, Stieber D, Riviére M, Russo J, Wedekind D, Coppieters W, Farnir F, Georges M, Szpirer J, Szpirer C. Genetic identification of distinct loci controlling mammary tumor multiplicity, latency, and aggressiveness in the rat. Mamm Genome. 2006;17:310–321. doi: 10.1007/s00335-005-0125-9. [DOI] [PubMed] [Google Scholar]
- Russo J, Russo I. Biology of disease: biological and molecular bases of mammary carcinogenesis. Lab Invest. 1987;57:112–137. [PubMed] [Google Scholar]
- Russo J, Gusterson B, Rogers A, Russo I, Wellings S, Van Zwieten M. Biology of disease: comparative study of human and rat mammary tumorigenesis. Lab Invest. 1990;62:224–278. [PubMed] [Google Scholar]
- Russo J, Russo I. Atlas and histologic classification of tumors of the rat mammary gland. J Mammary Gland Biol Neoplasia. 2000;5:187–200. doi: 10.1023/a:1026443305758. [DOI] [PubMed] [Google Scholar]
- Samuelson DJ, Aperavich BA, Haag JD, Gould MN. Fine mapping reveals multiple loci and a possible epistatic interaction within the mammary carcinoma susceptibility quantitative trait locus, Mcs5. Cancer Res. 2005;65:9637–9642. doi: 10.1158/0008-5472.CAN-05-1498. [DOI] [PubMed] [Google Scholar]
- Samuelson DJ, Hesselson SE, Aperavich BA, Zan Y, Haag JD, Trentham-Dietz A, Hampton JM, Mau B, Chen KS, Baynes C, Khaw KT, Luben R, Perkins B, Shah M, Pharoah PD, Dunning AM, Easton DF, Ponder BA, Gould MN. Rat Mcs5a is a compound quantitative trait locus with orthologous human loci that associate with breast cancer risk. Proc Natl Acad Sci U S A. 2007;69:6299–6304. doi: 10.1073/pnas.0701687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BS, Lachel CM, Pennington KL, Murrin CR, Strecker TE, Tochacek M, Gould KA, Meza JL, McComb RD, Shull JD. Genetic bases of estrogen-induced tumorigenesis in the rat: mapping of loci controlling susceptibility to mammary cancer in a Brown Norway × ACI intercross. Cancer Res. 2006;66:7793–7800. doi: 10.1158/0008-5472.CAN-06-0143. [DOI] [PubMed] [Google Scholar]
- Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O’Brien W, Courtland HW, Jepsen KJ, Kirby A, Kulbokas EJ, Daly MJ, Broman KW, Lander ES, Nadeau JH. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc Natl Acad Sci U S A. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepel LA, Lan H, Haag JD, Brasic GM, Gheen ME, Simon JS, Hoff P, Newton MA, Gould MN. Genetic identification of multiple loci that control breast cancer susceptibility in the rat. Genetics. 1998;149:289–299. doi: 10.1093/genetics/149.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- Steck SE, Gaudet MM, Eng SM, Britton JA, Teitelbaum SL, Neugut AI, Santella RM, Gammon MD. Cooked meat and risk of breast cancer: lifetime versus recent dietary intake. Epidemiology. 2007;18:373–382. doi: 10.1097/01.ede.0000259968.11151.06. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.