Abstract
MicroRNAs (miRNAs) are small, non-coding RNA molecules that post-transcriptionally regulate gene expression. Evidence has shown that miRNAs play important roles in various cellular processes, including proliferation, differentiation and survival. The intestinal epithelium is regenerated throughout life, and enterocytes undergo differentiation during migration along the crypt/villus axis. Our study aimed at establishing the expression profiles of miRNAs during intestinal epithelial cell (IEC) differentiation and determining a miRNA “signature” that distinguishes between small and large IECs. MiRNA arrays were employed to profile miRNA expression in two IEC models: the enterocyte-like Caco2-BBE and the colonocyte-like HT29-Cl.19A cell lines. Microarray data showed that in both cell lineages, the differentiated stage exhibited a different miRNA expression profile from undifferentiated stage. Interestingly, Caco2-BBE cells were distinguished from HT29-Cl.19A cells by their unique miRNA expression profile. Notably, HT29-Cl.19A cells exhibited down-regulation of miR-1269 and up-regulation of miR-99b and miR-125a-5p compared with Caco2-BBE cells. Most importantly, transfection of Caco2-BBE cells with mature miR-99b, mature miR-125a-5p and antisense of mature miR-1269 decreased growth rate and trans-epithelial resistance of the cells, indicating their shift toward HT29-Cl.19A cell phenotype. In conclusion, our study shows that miRNAs might play a role in determining the unique physiological characteristics of IECs.
Keywords: microRNA, Caco2-BBE, HT29-Cl.19A, differentiation, human intestinal epithelial cells
Introduction
MicroRNAs (MiRNAs) are small (18–25 nucleotides), non-coding RNAs that negatively regulate target mRNAs by binding to their 3’-untranslated region (UTR) (Ambros, 2001). MiRNAs act by triggering degradation of mRNAs or repression of translation (Filipowicz et al., 2008). The interactions between miRNAs and their mRNA targets are imperfect, allowing a single miRNA to potentially regulate multiple genes (Lim et al., 2005). It is estimated that miRNAs could regulate up to 30% of the protein-coding genes in the human genome (Lewis et al., 2003). Although the specific functions of most miRNAs are currently unknown, these molecules have been implicated in various biological processes, such as immunity, proliferation and development (Bueno et al., 2008; Taganov et al., 2007; Stefani and Slack, 2008). It has been also reported that expression of miRNAs is deregulated during several diseases, such as in various types of cancer, suggesting their critical roles in modulating cell behavior (Bartels and Tsongalis, 2009).
Gastrointestinal tract is a dynamic organ, in which cell proliferation, differentiation, migration and apoptosis are tightly regulated. MiRNAs are implicated in differentiation of various cell types, such as embryonic stem cells (Hinton et al., 2009), dendritic cells (Hashimi et al., 2009), and mesenchymal stem cells (Schoolmeesters et al., 2009). Therefore, it would be of interest to determine if these molecules are involved in intestinal epithelial cell differentiation. For this study, we used two well-established intestinal epithelial cell lines: the enterocyte-like Caco2-BBE and the colonocyte-like HT29-Cl.19A. Caco2-BBE cells derived from the parental Caco2 strain that displays the ability to develop a morphologically homogenous brush border expressing microvillar proteins comparable to human enterocytes (Peterson and Mooseker, 1992). Extensive studies have disclosed that Caco2-BBE cells exhibit characteristic features of enterocytes, such as they spontaneously differentiate as absorptive epithelial cells (Peterson and Mooseker, 1992; Merlin et al., 1998). Caco2-BBE cells therefore represent a good in vitro model to study human enterocytes (Peterson and Mooseker, 1992; Merlin et al., 1998). The human colonic cancer HT29 cells are undifferentiated in standard conditions. Augeron and co-workers generated two new HT29 clones in 1984 by treating the cells with sodium butyrate. One of them, named as HT29-Cl.19A, exhibits a permanent colonocyte phenotype and has been used as a model of human colonic cell line (Augeron and Laboisse, 1984).
It has been shown that a number of miRNAs are expressed in a highly tissue-specific manner, and that their expression pattern drives the specificity of protein profiles characteristic for each organ (Lim et al., 2005; Lagos-Quintana et al., 2002). Here, we identified unique miRNA expression profiles to distinguish between undifferentiated and differentiated stages of intestinal epithelial cells, as well as between small and large intestinal epithelial cells. Our study showed that miRNAs could play an important role in determining the distinct physiological characteristics of intestinal epithelial cells.
Materials and Methods
Cell culture
Caco2-BBE and HT-29Cl.19A cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 1.5 μg/ml plasmocin (Invitrogen). Cells were kept at 37ºC in 5% CO2 atmosphere and 90% humidity. Cells were split at a density of 2 × 104 cells/ml and were grown for 2 days (undifferentiated stage) or 14 days (differentiated stage) on filters (pore size: 0.4 μm; Costar).
MiRNA expression analysis by miRNA array
Total RNAs were extracted from cells using the RNeasy mini kit (Quiagen) according to the manufacturer’s instruction. Yield and purity of the RNA were verified. MiRNA array was performed in triplicate using the humanMI_V2 chip (Illumina), which contains up to 1145 miRNAs.
MiRNA target prediction
To determine the potential target genes of detected miRNAs, three different miRNA target prediction algorithms were used: PicTar (http://pictar.mdc-berlin.de) (Krek et al., 2005), miRanda (http://microrna.sanger.ac.uk/sequences/) (John et al., 2004) and TargetScan (http://www.targetscan.org/) (Grimson et al., 2007). The Matchminer program (http://discover.nci.nih.gov/matchminer/index.jsp) (Bussey et al., 2003) was then used to determine genes that were identified by at least two algorithms.
Real-time RT-PCR
Total RNAs isolated using the RNeasy mini kit as described above were reversely transcribed using the NCodeTM miRNA first-strand cDNA synthesis kit (Invitrogen) to quantify mature miRNA expression, or using the first-strand cDNA synthesis kit (Fermentas) to quantify gene expression according to the manufacturer’s instruction. Levels of mature miRNAs expression were quantified by real-time RT-PCR using the universal primer provided in the NCodeTM miRNA first-strand cDNA synthesis kit and the specific forward primers. Expression levels of potential target genes were quantified using specific forward and reverse primers. 18S was used as housekeeping gene. Fold-induction was calculated using the Ct method as follows: , and the final data were derived from 2 ΔΔCT. All primers used for real-time RT-PCR were described in Table S1.
MiRNAs and transfection
hsa-miR-99b (Pre-miRNA precursor AM17100, Product ID: PM11021), hsa-miR-125a-5p (Pre-miRNA precursor AM17100, Product ID: PM12561) and anti-hsa-miR-1269 (Pre-miRNA precursor AM17000, Product ID: AM13388) were purchased from Ambion. Caco2-BBE cells were transfected with miRNAs using the siPORTTM NeoFXTM transfection agent (Ambion) according to the manufacturer’s instruction.
Measurement of cell proliferation and cell resistance
Caco2-BBE cell barrier function was assessed by measuring the trans-epithelial resistance (TER). Briefly, Caco2-BBE cells were trypsinized, transfected in suspension with vehicle or with a mixture of miR-99b, miR-125a-5b, and anti-miR-1269 using the SiPORT transfecting agent (Ambion), and seeded on polycarbonate membrane supports (pore size 0.4 μm, Costar, Corning). TER of the cells was measured daily with an equilibrated Epithelial Voltohmmeter (World Precision Instruments, Berlin, Germany). Values were subtracted from TER value of blank filters (without cells) and were expressed as Ω.cm2.
Cell proliferation rate was also examined by measuring cell resistance in real-time using the electric cell-substrate impedance sensing (ECIS) 1600R device (Applied BioPhysics). The ideal frequency used to measure cell resistance was first determined as previously described (Charrier et al., 2005). Briefly, Caco2-BBE or HT29-Cl.19A cells were seeded on ECIS 8W1E electrodes at a density of 2 × 104 cells/400 μl/electrode. When the cells reached confluency, a scan of the resistance using different frequencies was applied. Cells were then trypsinized, and the frequency scan was repeated on naked electrodes. The ratio of the log of resistance with cells over the log of resistance without cells was calculated, and the frequency at which this ratio reached the maximum value was determined as the ideal frequency. Resistance of cells was measured in real-time at their ideal frequency and a voltage of 1 V.
Statistical analysis
Values were expressed as means ± S.E.M. Statistical analysis was performed using unpaired two-tailed Student's t-test by InStat v3.06 (GraphPad) software. P < 0.05 was considered statistically significant.
Results
MiRNA expression pattern during differentiation of enterocyte-like Caco2-BBE cells
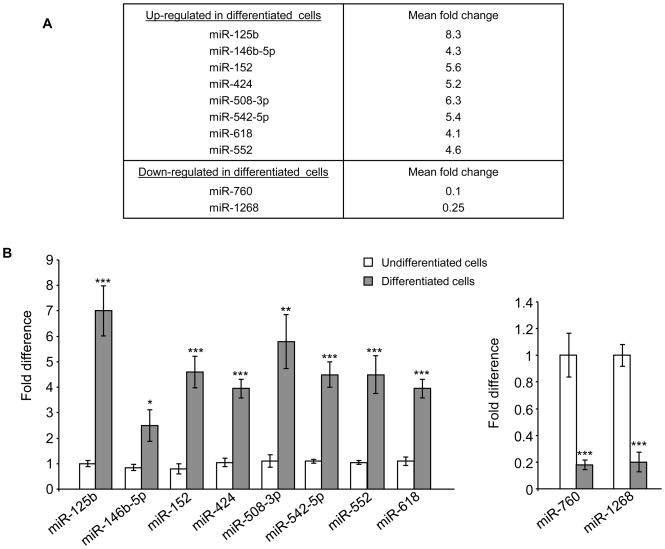
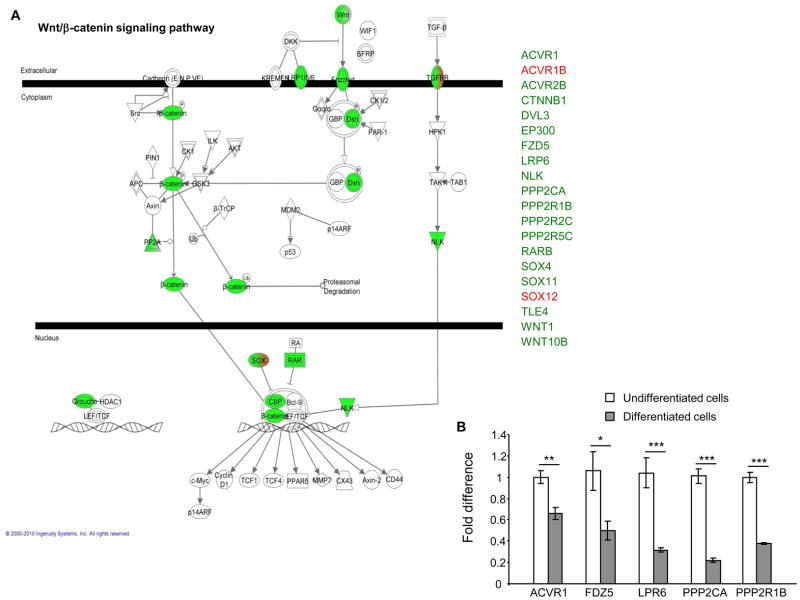
Since miRNAs have been implicated in differentiation process of various cell types, we aimed at identifying a miRNA profile for intestinal epithelial cells during their differentiation. For that, Caco2-BBE cells were grown on filters for 2 or 14 days to mimic the undifferentiated or differentiated stage, respectively, of enterocytes. Total RNAs of the cells were extracted and applied to a miRNA array analysis using the Illumina humanMI_V2 chip, which contains almost the entire of miRNAs identified in humans. We found that in differentiated Caco2-BBE cells, 84 miRNAs were significantly up-regulated and 36 miRNAs were significantly down-regulated compared to undifferentiated cells (Table S2). The miRNAs markedly deregulated with a mean fold change ≥ 4 were selected for further analysis. Using this threshold, 8 up-regulated miRNAs (miR-125b, miR-146b-5p, miR-152, miR-424, miR-508-3p, miR-542-5p, miR-618 and miR-552) and 2 down-regulated miRNAs (miR-760 and miR-1268) were identified when Caco2-BBE cells became differentiated (Figure 1A). Quantification of expression levels of these miRNAs by real-time RT-PCR confirmed the miRNA array data (Figure 1B). Since a single miRNA can target hundreds of mRNAs (Lim et al., 2005), we identify the potential mRNA targets of these miRNAs using three miRNA target prediction algorithms: miRanda, PicTar and TargetScan. The Matchminer program was then used to determine genes that were predicted by at least two algorithms. This bioinformatic approach revealed 1262 genes potentially regulated by the 10 miRNAs that were mostly deregulated in differentiated Caco2-BBE cells compared to undifferentiated cells (Table S3). Using the Ingenuity Pathways software, we determined the most enriched functional groups of genes among the miRNA targets, and the canonical pathways in which they may be involved. Interestingly, the Wnt/β-catenin pathway was revealed among those regulated by the selected miRNAs. Among the 20 proteins involved in this pathway that were potentially targeted by the 10 miRNAs identified here, 18 were down-regulated (shown in green) when cells were differentiated (Figure 2A). Crossing these genes with previously established deregulated genes during IEC differentiation (Fleet et al., 2003; Gregorieff et al., 2005) revealed ACVR1, PPP2CA, PPP2R1B, FDZ5, and LRP6 as highly potential targets which are down-regulated during IEC differentiation. Quantitative RT-PCR analysis further showed significant decreases in expression levels of these target genes in differentiated Caco2-BBE cells compared to undifferentiated cells (Figure 2B). In addition, all of the members of the eukaryotic translation initiation factor family potentially targeted by these miRNAs were found to be down-regulated (Table S4). Among them, EIF2B5, EIF2C2, EIF4EBP1 and EIF4G2 were previously reported to be down-regulated during Caco2-BBE cell differentiation (Fleet et al., 2003), and EIF2B3 was down-regulated during enterocyte maturation (Mariadason et al., 2005). Expression levels of these putative target genes were verified by quantitative RT-PCR. As shown in Figure S1, their expression levels were significantly decreased in differentiated Caco2-BBE cells compared to undifferentiated cells. Our results collectively indicate that miRNAs might be involved in the differentiation of enterocyte-like Caco2-BBE cells.
Figure 1. MiRNA expression pattern during differentiation of Caco2-BBE cells.
A) MiRNAs differentially expressed with ≥ 4 fold changes in differentiated Caco2-BBE cells compared to undifferentiated cells determined by miRNA array analysis. B) Real-time RT-PCR analysis of miRNA expression levels. Values represent means ± S.E.M. of n = 6/group. *P < 0.05; **P < 0.005; ***P < 0.001.
Figure 2. Functional relationships of miRNA target genes involved in the Wnt/β-catenin pathway predicted by using the Ingenuity Pathways software.
A) The genes potentially targeted by the miRNAs that were deregulated during Caco2-BBE cell differentiation (miR-125b, miR-146b-5p, miR-152, miR-424, miR-508-3p, miR-542-5p, miR-618, miR-552, miR-760 and miR-1268) were predicted to be involved in Wnt/β-catenin pathway. Potentially down- and up-regulated genes are presented in green and red, respectively. B) Real-time RT-PCR quantification of mRNA expression levels of miRNAs putative target genes. Values represent means ± S.E.M. of n = 6/condition. *P < 0.05; **P < 0.005; ***P < 0.001.
MiRNA expression pattern during differentiation of colonocyte-like HT29-Cl.19A cells
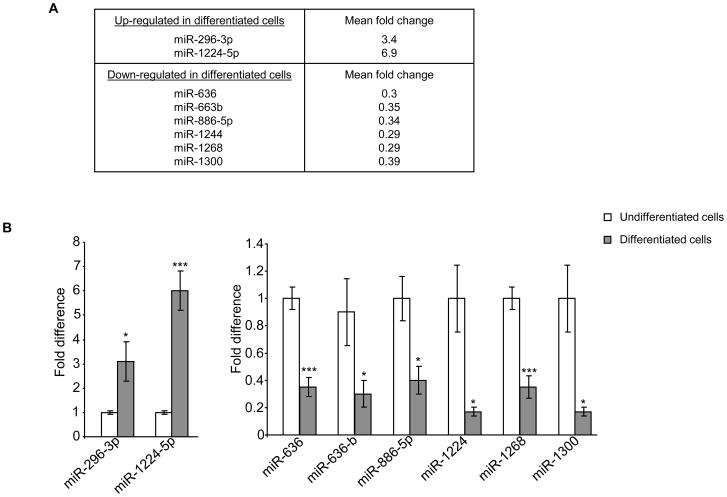
We next investigated miRNA expression profiles for HT29-Cl.19A cells during their differentiation using the same approach applied for Caco2-BBE cells. MiRNA array analysis revealed that 56 miRNAs were up-regulated and 67 miRNAs were down-regulated in well-differentiated HT29-Cl.19A cells compared to undifferentiated cells (Table S5). Among them, two up-regulated miRNAs (miR-296-3p and miR-1224-5p) and 6 down-regulated miRNAs (miR-636, miR-663b, miR-886-5p, miR-1244, miR-1268 and miR-1300) were the most markedly deregulated miRNAs with fold changes of ≥ 2.9 (Figure 3A). Real-time RT-PCR data were in agreement with miRNA array results, confirming changes in expression levels of these miRNAs during differentiation of HT29-Cl.19A cells (Figure 3B). MiR-1300 was not considered since it was recently reported that this miRNA is, in fact, a fragment of the EEF1A mRNA (see http://www.mirbase.org/). MiRNA target prediction analysis using miRanda, PicTar and TargetScan algorithms identified 397 genes potentially regulated by these seven most deregulated miRNAs (Table S6). The most enriched functional groups of genes among the miRNA targets and the canonical pathways in which they were potentially involved were determined using the Ingenuity Pathways software. Interestingly, among the predicted pathways with highest scores, several members of the integrin and integrin-linked kinase (ILK) signaling pathways were up-regulated. Ten proteins of the ILK signaling pathway were potentially regulated by those miRNAs and all of them were up-regulated (Table S7). Similar results were obtained for the integrin signaling pathway, of which 8 over 9 proteins were predicted to be up-regulated (Table S8). Quantification of mRNA expression levels of these potentially up-regulated genes by real-time RT-PCR revealed that FOS, TMSL3 and ARF1 expression levels were significantly increased in differentiated HT29-Cl.19A cells compared to undifferentiated cells (Figure S2). Several members of these pathways have been previously shown by in vivo studies to be highly expressed at the bottom of the crypts (Gagne et al.; Desloges et al., 1998). It is also known that HT29-Cl.19A cells display the features of deep-crypt secretory cells once differentiated, such as the electrophysiological characteristics of chloride-secreting cells that respond to neuropeptidergic stimulation (Rouyer-Fessard et al., 1989). Our findings are therefore consistent with the characteristics of colonocytes, suggesting that miRNAs play a role in the differentiation of this intestinal epithelial cell type.
Figure 3. MiRNAs expression pattern during differentiation of HT29-Cl.19A cells.
A) MiRNAs differentially expressed with ≥ 2.9 fold changes in differentiated HT29-Cl.19A cells compared to undifferentiated cells determined by miRNA array analysis. B) Real-time RT-PCR analysis of miRNA expression levels. Values represent means ± S.E.M. of n = 6/group. *P < 0.05; ***P < 0.001.
MiR-99b, miR-125a-5p and miR-1269 have a role in determining intestinal epithelial cell fate
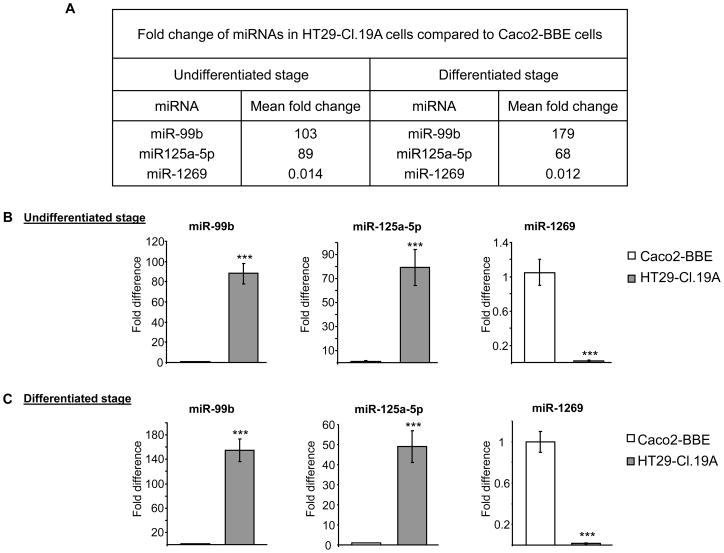
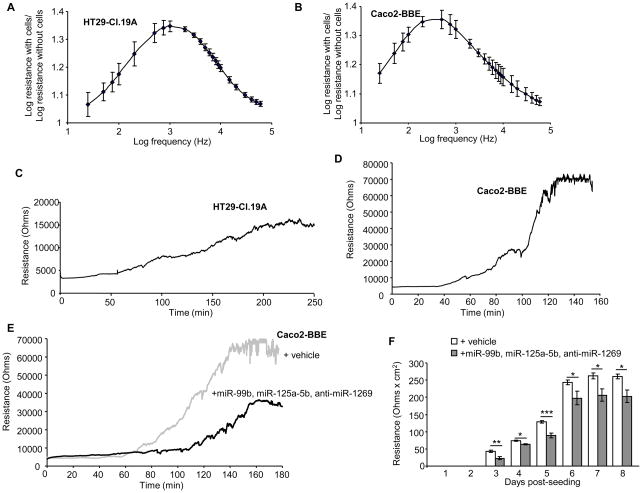
Comparing miRNA expression profiles of Caco2-BBE versus HT29-Cl.19A cells revealed miRNAs with significantly different expression at both undifferentiated and differentiated stages. At the undifferentiated stage, 138 miRNAs were up-regulated and 103 miRNAs were down-regulated in HT29-Cl.19A cells compared to Caco2-BBE cells (Table S9). At the differentiated stage, HT29-Cl.19A cells exhibited 171 up-regulated and 120 down-regulated miRNAs compared to Caco2-BBE cells (Table S10). Interestingly, expression levels of miR-99b, miR-125a-5p and miR-1269 were found to be markedly different (> 50 folds) between HT29-Cl.19A and Caco2-BBE cells, independently of the differentiation status (Figure 4A). Real-time RT-PCR analysis confirmed up-regulation of miR-99b and miR-125a-5p and down-regulation of miR-1269 in HT29-Cl.19A cells compared to Caco2-BBE cells (Figure 4B, C). Given the difference in miRNA profiles between these two intestinal epithelial cell types, we tested if exogenous miRNAs could shift characteristics of Caco2-BBE cells toward HT29-Cl.19A cells. Caco2-BBE cells were therefore transfected with mature miR-99b, mature miR-125a-5p and antisense of mature miR-1269 (anti-miR-1269), and growth rate and resistance of the cells were monitored in real-time using the electric cell-substrate impedance sensing (ECIS) technique. The ideal frequency used to measure cell resistance was first determined by scanning cell resistance with different frequencies on electrodes with confluent cells and naked electrodes. The ratio of the log of resistance with cells over the log of resistance without cells was calculated, and the frequency at which this ratio reached a maximum value was determined as the ideal frequency. For HT29-Cl.19A cells, the ratio peak was found at a log [frequency (Hz)] of 3.3, corresponding to a frequency of 2000 Hz (Figure 5A). For Caco2-BBE cells, the ideal frequency was found to be 500 Hz (Figure 5B), which is consistent with our previous study (Charrier et al., 2005). Measurement of cell resistance at their ideal frequency showed that Caco2-BBE cells grew and developed trans-epithelial resistance (TER) more rapidly than HT29-Cl.19A cells (Figure 5C, D). Caco2-BBE cells reached confluency at ~130 h post-seeding (Figure 5D), whereas HT29-Cl.19A cells required up to ~220 h to develop their maximal TER (Figure 5C). Notably, the TER value of confluent Caco2-BBE monolayers was 4-fold higher than that of confluent HT29-Cl.19A monolayers (Figure 5C, D). Most importantly, transfection of Caco2-BBE cells with a combination of miR-99b, miR-125a-5p and anti-miR-1269 shifted them toward HT29-Cl.19A cells (Figure 5E) by decreasing the growth rate and TER of the cells. In particular, vehicle-transfected Caco2-BBE cells started to grow and develop the TER at ~70 h post-seeding, whereas it was delayed till 120 h post-seeding in miRNAs-transfected cells. Furthermore, these miRNAs decreased the maximum TER of confluent Caco2-BBE cells by ~2 folds (Figure 5E). In addition, the effect of these miRNAs on TER of the cells was verified on polarized monolayers cultured on filters. For this purpose, Caco2-BBE cells were transfected in suspension with vehicle or with the miRNA mixture and seeded on filters. TER of the cells was measured daily with an Epithelial Voltohmmeter. As shown in Figure 5F, Caco2-BBE cells transfected miR-99b, miR-125a-5b and anti-miR-1269 developed significantly decreased TER compared to vehicle-transfected cells. This result was consistent with TER data measured by ECIS technique, further supporting the effect of miRNAs on Caco2-BBE cell resistance. To better understand the mechanisms by which miR-99b, miR-125a-5p and miR-1269 could regulate intestinal epithelial cell fate, we searched their potential target genes using the computational approach (Table S11). We found interestingly that among them, up to 104 genes were involved in cellular growth and proliferation as predicted by the Ingenuity Pathways software (Table S12), and the majority (> 85%) was up-regulated in Caco2-BBE cells compared to HT29-Cl.19A cells. This may explain the decreases in growth rate and TER of Caco2-BBE cells by transfection with miR-99b, miR-125a-5p and anti-miR-1269. Altogether, our results raise this miRNA pattern as a potential regulator of intestinal epithelial cell fate.
Figure 4. MiRNAs expression profile distinguishing Caco2-BBE from HT29-Cl.19A cells.
A) MiRNAs differentially expressed with > 50 fold changes in HT29-Cl.19A cells compared to Caco2-BBE cells, at both differentiated and undifferentiated stages, determined by miRNA array analysis. B) Real-time RT-PCR analysis of miRNA expression levels. Values represent means ± S.E.M. of n = 6/group. ***P < 0.001.
Figure 5. MiR-99b, miR-125a-5p and miR-1269 determine intestinal epithelial cell fate.
A, B) Determination of the ideal frequency to measure resistance of HT29-Cl.19A (A) and Caco2-BBE cells (B) by scanning of the resistance using different frequencies on ECIS 8W1E electrodes with confluent cells or without cells (naked electrodes). The ratio of the log of resistance with cells over the log of resistance without cells was calculated, and the frequency at which this ratio reached a maximum value was determined as the ideal frequency. C, D) Cells were seeded on ECIS electrodes at a density of 2 × 104 cells/400 μl/electrode. Resistance of HT29-Cl.19A cells was measured at 2000 Hz (C), and resistance of Caco2-BBE cells was measured at 500 Hz (D). E) Caco2-BBE cells were transfected with vehicle (grey line) or a mixture of mature miR-99b, mature miR-125a-5p and anti-miR-1269 (black line), and cell resistance was measured in real-time. F) Caco2-BBE cells were trypsinised, transfected with vehicle or with a mixture of miR-99b, miR-125a-5p and anti-miR-1269, and seeded on filters. TER of the cells was measured daily with an equilibrated Epithelial Voltohmmeter. Values were subtracted from TER value of blank filters (without cells) and were expressed as Ω.cm2. Data represent means ± S.E.M. of n = 6/condition. *P < 0.05; **P < 0.005; ***P < 0.001.
Discussion
Since their discovery in Caenorhabditis elegans in 1993 (Lee et al., 1993; Wightman et al., 1993), miRNAs have been implicated in gene expression regulation at most stages of development, including cell differentiation and organ formation (Stefani and Slack, 2008). One approach to profile miRNA expression is the use of miRNA arrays. This method has been extensively used for cells or tissues, and various miRNAs have been found to be involved in many cellular processes. For example, using such approach, studies in mesenchymal stem cells or human embryonic stem cells have shown that miRNAs are involved in the differentiation process (Hinton et al., 2009; Schoolmeesters et al., 2009). MiRNA profiling studies also led to the identification of miRNAs that are involved in the differentiation of dendritic cells or liver cells (Hashimi et al., 2009; Tzur et al., 2009).
Gastrointestinal tract is a dynamic organ containing cells displaying specific protein profiles in relation to their localization along the crypt/villus axis. To date, limited studies have investigated the role of miRNAs in intestinal epithelial cell differentiation. A study conducted on zebrafish showed that miR-145 is crucial for the gut development as its downregulation repressed the gut epithelial maturation (Zeng et al., 2009). A recent study from our group showed that miR-7 is implicated in intestinal epithelial cell differentiation via its ability to regulate CD98 expression (Nguyen et al.). To our knowledge, only one study to date has focused on the role of miRNAs in human intestinal epithelial cell maturation. In this study, Tsuchiya et al. showed that miR-338-3p and miR-451 contribute to the formation of basolateral polarity of colonic epithelial T84 cells by facilitating translocalization of β1-integrin to the basolateral membrane (Tsuchiya et al., 2009).
In the present study, using well-established enterocyte-like (Caco2-BBE) and colonocyte-like (HT29-Cl.19A) cell models and the most powerful miRNA array available, we have shown that differentiated cells exhibit a distinct miRNA expression profile from undifferentiated cells. The computational approach helped us to reveal the Wnt/β-catenin or ILK and integrin signaling pathways as the most potential pathways regulated by miRNAs during the differentiation process of Caco2-BBE or HT29-Cl.19A cells, respectively. It is known that in the small intestine, the Wnt/β-catenin pathway proteins are expressed at the bottom of the crypt where cells are proliferating, but not at the top where cells are highly differentiated (Mariadason et al., 2005). A gene expression profiling of Caco2-BBE cells also showed that the Wnt/β-catenin signaling pathway is suppressed during the differentiation process (Fleet et al., 2003). Several members of the ILK and integrin signaling pathways have been also shown by to be highly expressed at the bottom of the crypts (Gagne et al.; Desloges et al., 1998). Given the well-known function of these pathways in the development of the gut epithelium, our results suggest that miRNAs might be involved in the regulation of intestinal epithelial cell differentiation.
Our findings also suggest that differences between enterocytes and colonocytes may be explained, at least in part, by their miRNA expression profiles. Notably, expression of miR-99b, miR-125a-5p and miR-1269 miRNAs were markedy different between Caco2-BBE and HT29-Cl.19A cells independently of cell differentiation stage. Interestingly, modifying expression levels of these miRNAs changed the growth rate and TER of cells, leading to a shift from Caco2-BBE toward HT29-Cl.19A cells. The roles of these miRNAs are mostly unknown. MiR-99b was found to be deregulated in endometrioid adenocarcinoma (Wu et al., 2009) as well as in ectopic versus eutopic endometrial tissue (Ohlsson Teague et al., 2009). MiR-125a-5p is deregulated in multiple myeloma (Lionetti et al., 2009). Interestingly, a recent study showed that miR-125a-5p can inhibit migration and invasion of lung cancer cells and therefore may function as a metastatic suppressor (Wang et al., 2009). MiR-1269 has not been investigated so far. Since these miRNAs might play a role in IEC fate, it would be interesting to compare their predicted target genes with a global gene expression profile given by a cDNA microarray to identify the most potential miRNA targets.
To our knowledge, our study is the first supported case suggesting a role for these miRNAs in determining intestinal epithelial cell fate. Our findings are in agreement with previous studies showing that expression patterns of some miRNAs could determine the tissue identity (Lagos-Quintana et al., 2002; Lim et al., 2005; Wienholds et al., 2005). For example, transfection with miR-124, which preferentially expressed in the brain, shifted the expression profile of HeLa cells (cervix epithelial cells) toward that of brain cells (Lim et al., 2005). In conclusion, our study raises miRNAs as a potential regulator of human intestinal epithelial cell differentiation. Furthermore, the distinct physiological characteristics of each intestinal epithelial cell type are determined, at least in part, by a unique miRNA profile.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grants R24-DK-064399 center grant, R01-DK-071594 (to D. M.), R01-DK-080684 (to S. S) and R01-DK-55850 (to S. V. S.) from the NIDDK. This work was also supported by the Crohn’s and Colitis Foundation of America by a research fellowship award (to G. D.). We thank Dr. Chandrasekharan B. P for her help in the use of Ingenuity Pathways software.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Augeron C, Laboisse CL. Emergence of permanently differentiated cell clones in a human colonic cancer cell line in culture after treatment with sodium butyrate. Cancer Res. 1984;44:3961–3969. [PubMed] [Google Scholar]
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, de Castro IP, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- Bussey KJ, Kane D, Sunshine M, Narasimhan S, Nishizuka S, Reinhold WC, Zeeberg B, Ajay W, Weinstein JN. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4:R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier L, Yan Y, Driss A, Laboisse CL, Sitaraman SV, Merlin D. ADAM-15 inhibits wound healing in human intestinal epithelial cell monolayers. Am J Physiol Gastrointest Liver Physiol. 2005;288:G346–353. doi: 10.1152/ajpgi.00262.2004. [DOI] [PubMed] [Google Scholar]
- Desloges N, Basora N, Perreault N, Bouatrouss Y, Sheppard D, Beaulieu JF. Regulated expression of the integrin alpha9beta1 in the epithelium of the developing human gut and in intestinal cell lines: relation with cell proliferation. J Cell Biochem. 1998;71:536–545. [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fleet JC, Wang L, Vitek O, Craig BA, Edenberg HJ. Gene expression profiling of Caco-2 BBe cells suggests a role for specific signaling pathways during intestinal differentiation. Physiol Genomics. 2003;13:57–68. doi: 10.1152/physiolgenomics.00152.2002. [DOI] [PubMed] [Google Scholar]
- Gagne D, Groulx JF, Benoit YD, Basora N, Herring E, Vachon PH, Beaulieu JF. Integrin-linked kinase regulates migration and proliferation of human intestinal cells under a fibronectin-dependent mechanism. J Cell Physiol. 222:387–400. doi: 10.1002/jcp.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton A, Afrikanova I, Wilson M, King C, Maurer B, Yeo G, Hayek A, Pasquinelli A. A Distinct MicroRNA Signature for Definitive Endoderm Derived From Human Embryonic Stem Cells. Stem Cells Dev. 2009 doi: 10.1089/scd.2009.0224. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S, Lombardi L, Bortoluzzi S, Neri A. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Nicholas C, L'Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, Arango D, Augenlicht LH. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Merlin D, Steel A, Gewirtz AT, Si-Tahar M, Hediger MA, Madara JL. hPepT1-mediated epithelial transport of bacteria-derived chemotactic peptides enhances neutrophil-epithelial interactions. J Clin Invest. 1998;102:2011–2018. doi: 10.1172/JCI4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. 285:1479–1489. doi: 10.1074/jbc.M109.057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, Mooseker MS. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J Cell Sci. 1992;102 ( Pt 3):581–600. doi: 10.1242/jcs.102.3.581. [DOI] [PubMed] [Google Scholar]
- Rouyer-Fessard C, Augeron C, Grasset E, Maoret JJ, Laboisse CL, Laburthe M. VIP receptors and control of short circuit current in the human intestinal clonal cell line Cl.19A. Experientia. 1989;45:1102–1105. doi: 10.1007/BF01950169. [DOI] [PubMed] [Google Scholar]
- Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force Aldred S, Fedorov Y. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, Sato F, Tsujimoto G, Shimizu K. MicroRNA-338–3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37:3821–3827. doi: 10.1093/nar/gkp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur G, Israel A, Levy A, Benjamin H, Meiri E, Shufaro Y, Meir K, Khvalevsky E, Spector Y, Rojansky N, Bentwich Z, Reubinoff BE, Galun E. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS One. 2009;4:e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p--a metastatic inhibitor of lung cancer. FEBS J. 2009;276:5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- Zeng L, Carter AD, Childs SJ. miR-145 directs intestinal maturation in zebrafish. Proc Natl Acad Sci U S A. 2009;106:17793–17798. doi: 10.1073/pnas.0903693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.