Abstract
Genetic instability is one of the principal hallmarks and causative factors in cancer. Human transposable elements (TE) have been reported to cause human diseases, including several types of cancer through insertional mutagenesis of genes critical for preventing or driving malignant transformation. In addition to retrotransposition-associated mutagenesis, TEs have been found to contribute even more genomic rearrangements through non-allelic homologous recombination. TEs also have the potential to generate a wide range of mutations derivation of which is difficult to directly trace to mobile elements, including double-strand breaks that may trigger mutagenic genomic rearrangements. Genome-wide hypomethylation of TE promoters and significantly elevated TE expression in almost all human cancers often accompanied by the loss of critical DNA sensing and repair pathways suggests that the negative impact of mobile elements on genome stability should increase as human tumors evolve. The biological consequences of elevated retroelement expression, such as the rate of their amplification, in human cancers remain obscure, particularly, how this increase translates into disease-relevant mutations. This review is focused on the cellular mechanisms that control human TE-associated mutagenesis in cancer and summarizes the current understanding of TE contribution to genetic instability in human malignancies.
Keywords: retroelements, transposable elements, cancer, genetic instability, recombination
Introduction
Genetic instability is one of the key features associated with cancer causation and progression [1-5]. A variety of factors can contribute to this instability, including exposure to DNA-damaging mutagens, as well as heritable or somatic genetic defects in the DNA repair response. Mobile elements can contribute to the assortment of mutagenic events commonly identified in human cancers. Despite their massive influence on the evolution of mammalian genome (reviewed in [6]), and numerous sporadic examples of mobile elements contributing to cancer, the full impact of TEs on the transformation process has been difficult to determine.
Human transposable elements include members of both DNA and RNA families of transposons, although there is no evidence of current activity of the DNA elements [7]. The RNA transposons, also referred to as retroelements, are further subdivided into long-terminal repeat (LTR)-containing or non-LTR groups (for the non-LTR group members see Figure 1; reviewed in [6,8]). Human endogenous retroviruses (HERVs) are the most well-known representatives of the LTR family in the human genome, but there is no documentation of their current activity. Non-LTR retrotransposons are the predominant source of TE-related mutagenesis in the human genome [reviewed in [8-10]). This is in contrast to our rodent relatives who retain high levels of LTR-element activity [reviewed in [11]]. Endogenous retroviruses have been reported to influence tumor progression in mice [12,13] and Xiphophorus hybrids [14], however, no such claims have been made for human cancers.
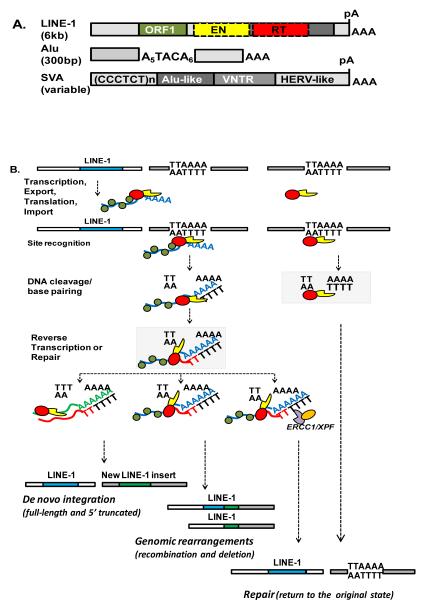
Figure 1.
A. Schematic of the genome organization of active human retroelements. LINE-1 consists of a 5′ UTR, an inter ORF region, and 3′UTR (light grey boxes) that ends in a polyadenylation signal (pA) and a run of adenosines (AAA). L1 encodes two open reading frames , ORF1 (green box) and ORF2. ORF2 encodes a protein with endonuclease (EN; yellow box) and reverse transcriptase (RT; red box) activities.
The ~300 bp human SINE, Alu, is composed of two non-identical 7SL-derived monomers (light grey boxes) that are connected by an adenosine (A)-rich linker. The Alu element ends in a variable length stretch of adenosines (AAA). SVA elements consist of four distinct regions: CCCTCT repeats, an inverted Alu-like sequence, VNTR variable number of tandem repeats, and human endogenous retrovirus (HERV) -like sequence. The approximate length of each element is shown in parentheses.
B. L1 integration steps and outcomes. TTAAAA is the consensus L1 endonuclease recognition site. A blue box represents a transcriptionally active and retrotranspositionally competent L1 locus in the human genome. The L1 RNP (ribonucleoprotein particle) is composed of the L1 mRNA (blue), L1 ORF1 protein (green) and L1 ORF2 protein that contains endonuclease (EN, yellow) and reverse transcriptase (RT, red) domains. The L1 RNP or L1 ORF2 recognizes an L1 target site in the human genome and generates a first DNA nick that at some point during retrotransposition becomes a DNA double strand break (DSB). The polyA tail of the L1 mRNA base pairs with the DNA at the nick that serves as a primer for TPRT (target primed reverse transcription) by L1 RT that generates a cDNA ‘flap’ (red). This intermediate can be resolved to generate de novo L1 integration event (green box), integration-associated genomic rearrangements, or it can be recognized and aborted by ERCC1/XPF complex, a component of the nucleotide excision repair machinery, leading to the restoration of the integration site.
The active, human non-LTR group includes Long Interspersed element-1 (LINE-1 or L1), short interspersed elements (SINE) represented by Alu, and the more recently characterized SVA elements [15-18] (Figure 1A). L1 elements have two open reading frames, ORF1 and ORF2, both of which are essential for L1 retrotransposition [19]. L1 ORF2 protein encodes endonuclease and reverse transcriptase enzymatic activities [20-22] that are responsible for host DNA nicking and L1 cDNA synthesis (see Figure 1B), respectively. While LINE-1 retrotransposons encode proteins necessary for their mobilization [19], Alu and SVA elements increase their copy number by parasitizing the L1 machinery [23,24]. The overall mobilization of retroelements declined significantly over the past tens of millions of years [7]. However, their extreme success in populating host genomes has allowed them to occupy about a third of the human genome resulting in a total of 500,000 copies of L1, 1,000,000 copies of Alu, and about 3,000 copies of SVA elements [7,15,17].
A. Transposable elements expression in normal and cancer cells
Early studies of L1 expression in mice put too strong an emphasis on primarily germ line expression of these elements [25,26]. These findings suggested that retroelement expression in normal human cells is completely suppressed by a variety of mechanisms, the number of which is still growing (see below), because of the potential harmful impact of these elements on genome stability. Most recent studies, however, report co-expression of L1 ORF1 and ORF2 proteins in human somatic cells, such as Leydig, Sertoli, and vascular endothelial cells [27], the presence of hypomethylated (i.e. likely transcriptionally active) L1 loci in normal human brain and skin [28], and various levels of L1mRNA expression in a variety of human somatic tissues [29]. A low level of L1 RNA was detected in adult tissues of transgenic mice expressing an L1 element driven by its endogenous promoter and L1 retrotransposition during embryogenesis created somatic and germ line mosaicism in these animals [30]. Recovery and analysis of expressed sequence tags from immortalized human lymphoblastoid cell lines identified low expression of multiple L1 loci with noticeable variation among different individuals [31]. These observations support a concept that human somatic cells experience ongoing expression of endogenous L1 elements. Little is known about somatic expression of human SINE and SVA elements. However, similar to L1, Alu expression is strongly controlled by promoter methylation [32,33] that exhibits tissue specificity with significant hypomethylation reported in the male germ line [34]. It is, therefore, likely that human SINEs may also be expressed at low-levels in normal tissues.
In contrast to normal cells, the majority of human cancers, and cancer-derived cell lines, support variable, but typically much higher endogenous full-length L1 mRNA expression [35-38]. One of the major controlling mechanisms of this upregulation is the loss of promoter methylation [36,39,40]. Aberrant DNA methylation, which would be expected to increase L1 expression, is reported to occur very early in the step-wise transformation process [41-43]. Silencing of retrotransposons by methylation is at least in part controlled by the argonaute family of proteins [44,45]. Changes in the L1 and Alu promoter methylation status between normal and cancer cells with the tendency toward hypomethylation even in the early stages of malignant transformation [41] have been extensively reported. Among the most informative are studies that assess methylation status and activity of retroelements in the same cellular environment [28] and studies that detail retroelement promoter methylation condition at different stages during cancer progression, such as progressive hypomethylation of the Alu promoter [46] or human-specific L1 loci [47]. Once the promoter suppression is released, production of retrotranspositionally competent L1 mRNA is further controlled by mRNA processing through extensive splicing and polyadenylation [48-50]. L1 expression may be additionally regulated by siRNA activity [51]. Elevated expression of Alu elements in human cancers has also been reported [33,52] and it can be further upregulated by stress and translational inhibitors [53,54]. These observations, combined with the reported augmentation of mutation rates in the majority of human cancers, suggest that retroelement expression and presumably activity could be one of the forces driving genetic instability that may accelerate tumor evolution. Indeed, an involvement of L1-encoded ORF2 endonuclease in nuclear receptor-dependent tumor translocations that are associated with aggressive prostate cancer phenotype has been reported [55]. This observation establishes a direct connection between L1 endonuclease function and the progression of prostate cancer. Even though the association of retroelement expression with genetic instability in cancer remains largely correlative [56-58], it is serendipitous that human cancers vary drastically both in the levels of endogenous L1 expression and the amount and type of genomic instability [59,60].
B. Polymorphism of retroelement activity
In addition to the regulation of transposable element expression and insertion, variation in the combined damage resulting from TE activity in any given genome is influenced by the presence of polymorphic elements [61], as well as by variation in the retrotransposition activity of the same TE locus in different individuals [62,63]. For example, the potential for L1 activity is estimated to vary as much as 300-fold between individuals within a population [63]. No similar studies evaluating combined Alu activity have been reported. However, polymorphism of Alu elements in the human genome and the profound effect of cis-acting sequences within and adjacent to Alu on its ability to retrotranspose [64-67] strongly suggest that total Alu activity within any given genome is likely to fluctuate extensively.
An additional level of TE activity regulation arises from the recently discovered involvement of various cellular processes in the modulation of L1 and Alu retrotransposition. Examples of proteins involved in such regulation include members of the APOBEC family of proteins and Nucleotide Excision and Non-Homologous End Joining DNA repair pathways (NER and NHEJ) (for more detail see DNA repair pathways affecting TE activity section) [68-73].
Even though the number of the L1- and Alu-associated diseases including cancer, continues to grow, there is no clear understanding of the extent of the contribution of these elements to the origin and progression of the specific human cancers. Nevertheless, retroelements polymorphism, substantial variation in their activity in any given genome, and the diverse types of damage that they can cause (see below) raises a possibility for significant variation in the potential TE-associated contribution to cancer in the population.
C. Genetic instability associated with transposable elements in human cancers
Insertional mutagenesis
Human retroelements (L1, Alu, and SVA) amplify in the host genome by generating new integration events using an RNA intermediate, which also leaves the parental copy intact (Figure 1B). This genome-wide, relatively random spread of transposable elements often inserts them in genomic regions with crucial biological functions resulting in insertional mutagenesis (a summary of TE insertion-mediated diseases is in [8] and relative retrotransposition is shown in Figure 2). Among the examples of human cancers caused by Alu-and L1-mediated insertional mutagenesis are disruptions of BRCA or APC genes leading to breast and colon cancers, respectively (Table 1). Occasionally, resolution of L1 integration intermediates causes deletion of genomic sequences [74-76](Figure 1B) causing mutations that may not be easily attributed to L1 activity in the genome-wide sequencing analysis. Overall, TE-associated insertional mutagenesis can contribute to cancer via germ-line and somatic mutations, either of which can directly lead, or predispose the cells, to the onset of malignant transformation.
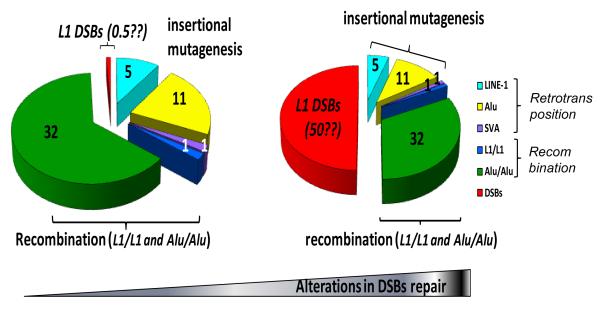
Figure 2. Estimated contribution of various types of retroelement-induced mutagenesis to genomic instability.
The three main types of L1-, Alu-, and SVA-related mutagenesis such as insertional , recombination, and error-prone DSB repair are represented. The relative contribution of L1, Alu, and SVA (with SVA set at 1) to insertional mutagenesis (shown in light blue, yellow, and purple, respectively) is estimated based on the reported disease incidence (reviewed in Belancio et.al., 2008). The relative contribution of L1 and Alu to recombination (shown in dark blue and green, respectively) is estimated based on disease and recombination-associated deletions (Deininger and Batzer, 1999, Han K. et.al., 2007, Han K. et.al., 2008). The potential relative mutagenic contribution of the L1-induced DSBs (shown in red) is essentially unknown and example estimates of 0.5 and 50 relative to insertion events are presented for comparison.
Table 1.
CANCER INSERTIONS
| RETROELEMENT | GENE | DISEASE | REFERENCE |
|---|---|---|---|
| Alu | CRB1 | retinoblastoma | Den Hollander et al, 1999 |
| 3x BRCA2 | breast cancer | Miki et al, 1996; Teugels et al, 2005; Machado et al, 2007 |
|
| BRCA1 | breast cancer | Teugels et al, 2005 | |
| NF1 | Neurofibromatosis | Wallace et al, 1991 | |
| L1 | APC | Colon cancer | Miki et al, 1992 |
| MYC | Breast cancer | Morse et al, 1988 |
Despite the relative decline in mobilization activity over recent evolutionary times [7], human retroelement insertions are still responsible for approximately 0.3% of new human germ-line diseases, which translates into about one germ line retrotransposition event of Alu, L1, and SVA in 20, 100, and 900 births, respectively [77-79]. Currently there is no reliable estimation of the extent of TE-associated insertional mutagenesis in somatic cells or any specific cancer. Data obtained in tissue culture demonstrate that a number of cell lines originated from different types of cancer support retrotransposition of exogenous L1 and Alu elements with varying efficiency [70,80,81]. In general, it has been found that cells that repress apoptosis [80], or cells that have defects in certain DNA repair pathways [69,82], both of which are common changes in human cancers, support higher retrotransposition. Thus, although the endogenous activity of mobile elements in cancers has not been determined, combined with L1 and Alu overexpression detected in many tumor cells, there are many reasons to believe that the insertion rate of these elements could be much higher in tumors than in corresponding normal cells.
Recombination
Even those Alu and L1 elements that appear to have inserted ‘harmlessly’ have continuing opportunity to wreak havoc on the stability of the human genome. They do so predominately through their contributions to recombination, which is frequently detected in cancer[2]. The most obvious impact on genome stability is through their tendency to serves as sources of homology for non-allelic homologous recombination (NAHR) that preferentially involve Alu elements (Table 2). NAHR events have been well recognized as a major source of DNA damage (reviewed in [83]) that leads to either duplication or deletion of the sequences between the two participating Alu elements. Bioinformatics and reporter gene studies provide experimental evidence that the presence of repetitive sequences, such as Alu elements, within intronic regions of genes promotes 40 to 300 times more recombination-induced deletions in the mutant p53 background relative to the wild type genomic environment [84]. Moreover, it has been found that Alu elements in close proximity with inverted orientations are particularly prone to recombination [84,85]. In a reporter gene assay these inverted Alus are 10,000x more unstable in the absence of functional p53 than in the wild type genetic background [84] providing strong support for significant contribution of repetitive elements to genetic instability during tumorigenesis (Figure 2). In yeast, introduction of a single DSB promotes a boost in genomic instability via recombination events between inverted repetitive elements located as far as 21 kb from one another [86] and other studies show that inverted repeats are unstable even without DSBs [87]. Recombination between inverted Alu repeats frequently does not lead to a clean, homologous recombination event. Instead, it produces recombination events with break points occurring in the vicinity of the two Alu elements, apparently through resolution in a non-homologous manner. Thus, for any individual recombination outcome, it is impossible to determine whether the Alus located in the vicinity of the break point contributed to that particular genomic rearrangement event or not. A growing body of evidence suggests that both direct and inverted repeats are considerable contributors to recombination-associated mutagenic events, such as loss-of-heterozygosity and other rearrangements identified during cancer progression, particularly in the presence of p53 mutations and probably many other mutations that contribute to genetic instability in cancer.
Table 2.
Alu/Alu RECOMBINATION
| GENE | DISEASE | MUTATION ORIGIN | REFERENCE |
|---|---|---|---|
| 10x MLH1 | HNPCC | Germline | Reviewed in Viel et al, 2002; Aissi-Ben Moussa et al, 2009; Li et al, 2006; van der Klift et al, 2005; Wang et al, 2003 |
| 32x MSH2 | HNPCC | Germline | Li et al, 2006; Akrami et al, 2005; van der Klift et al, 2005; Apessos et al, 2005; Wang et al, 2003; Charbonnier et al, 2005 |
| 30x VHL | VHL | Germline | Franke et al, 2009; Casarin et al 2006 |
| 15x hCAD | hepatoma | Somatic | Hsieh et al, 2005 |
| 5x MYB | T-All | Somatic and normal | O’Neil et al, 2007 |
| 7x MLL1 | AML | Somatic and normal | Strout et al, 1998 |
| 23x BRCA1 | Breast cancer | Germline | Reviewed in Mazoyer S., 2005 |
Numerous examples of Alu/Alu NAHR contributing to cancer have been reported. Table 2 lists a few specific cases of cancers where Alu/Alu NAHR recombination events have led to the same disease numerous times and illustrate a number of interesting points regarding Alu-promoted recombination in cancer. First, several genes that have a large number of Alu elements within their sequence seem to have a high proportion of genetic rearrangements involving Alu/Alu NAHR [88-90]. This phenomenon is specific to Alu repeats because the same formula does not apply to L1 elements. The discrepancy is presumably because of the depletion of L1 sequences in human genes relative to Alu elements that are enriched in human introns and because of the longer distance between adjacent L1 inserts within genes compared to the average proximity of neighboring Alus [7]. For example, 23 out of 29 reported recombination events in the BRCA1 gene, which has an unusually high density of Alu elements (41.5% of the gene) [89] involve Alu elements (reviewed in [88]). Additionally, although a number of instances of hereditary non-polyposis colorectal cancer (HNPCC) caused by Alu/Alu-induced rearrangements in the mutL homolog 1 (mlh1) gene that is relatively Alu-poor have been reported [90,91](Table 2), the majority of rearrangements in this gene are caused by non-homologous recombination that does not involve Alu elements. On the other hand, the majority of rearrangements of the more Alu-rich Mut-S-homolog-2 (msh2) gene, causing HNPCC, are due to Alu/Alu recombination events (Table 2). Two other cancer-promoting genes also have the majority of their rearrangements caused by Alu/Alu NAHR. These are the von Hippel-Lindau tumor suppressor (VHL) gene in the syndrome of the same name [92,93], and the human caspase-activated DNase gene, the key enzyme for nucleosome fragmentation during apoptosis, frequently harboring deletions in hepatoma [94]. Just for the 7 genes shown in Table 2 there have been over 120 different rearrangements reported that involve Alu/Alu recombination. This number far surpasses the number of total reported diseases caused by Alu insertions (reviewed in [8] and Figure 2), making it clear that in the human genome recombination events involving Alu have a much more profound impact on genomic instability than insertion of these elements.
A second important point is that Alu/Alu cancer-related recombination events occur not only in the germ line but also somatically, with some somatic rearrangements occurring on a regular basis. Among the above examples, the HNPCC and VHL mutations represent germ line defects in tumor suppressor genes that greatly increase the risk of those cancers in carriers. On the other hand, the hepatoma data [94] represent somatic recombination cases. The contribution of somatic Alu/Alu recombination events is also demonstrated by a recurring duplication of the MYB (an acronym derived from “myeloblastosis ”) locus that can contribute to T-cell acute lymphoblastic leukemia, T-ALL [95], and partial tandem duplications of exons in the myeloid/lymphoid or mixed-lineage leukemia (MLL), a histone methyltransferase gene that contributes to acute myeloid leukemia AML [96]. In the case of MLL, the majority of AML cases that do not show any chromosomal aberrations have this duplication caused by Alu/Alu NAHR [96]. In all of these examples, there are generally multiple Alu elements contributing to these mutagenic events. Even though genes containing large numbers of Alus are more prone to genomic rearrangements via recombination, it is also emerging that some of the Alu elements have a higher propensity than others to participate in recombination events, possibly due to higher levels of sequence identity with nearby Alus.
Finally, one of the more remarkable findings regarding both the MYB and MLL duplication events is that very similar duplication events appear to be occurring at low levels in the blood of the majority, if not all, unaffected humans. This suggests that somatic cells are frequently undergoing recombination events leading to copy number variations. It seems likely that the lack of disease in these apparently healthy individuals with the MYB or MLL1 duplications normally associated with cancer is either because the mutations are occurring at a late enough stage in the differentiation of the blood cells that prevent them from sufficient proliferation to manifest the disease, or possibly because there are more additional mutations that must occur in these cells before they can become transformed. This observation of somatic copy number variation associated with TE-induced recombination supports the hypothesis that transposable elements can be one of the factors contributing to the step-wise mutagenic process of malignant transformation.
DNA double-strand breaks (DSBs)
The finding that excess DNA double strand breaks (DSBs) are generated by expression of L1 ORF2 [68], opens the possibility that L1 contributes to genomic instability in human cancers in another major mechanism. This supposition is based on the observation that the estimated number of the L1-induced DSBs generated in HeLa cells is at least 10-100-fold higher than the number of the L1 retrotransposition events [68]. At this point, it is not known whether the large surplus of the observed DSBs is the result of abortive insertion events, or simply excess endonuclease expressed from the elements that gets free to attack the genome (Figure 1B).
Very little is known about the fate of the L1 endonuclease-induced DSBs. However, there is strong evidence of a correlation between L1 ORF2 expression in prostate cancer cells and formation of chromosomal translocations that lead to transcripts coding for fusion proteins [55]. Thus, it seems very likely that these elements represent a significant source of DSBs leading to genetic rearrangements in cancer. Because retroelements duplicate the ORF2-endonuclease cleavage site in their flanking regions [68,97], one hypothesized result of these L1-generated DSBs would be the prediction of increased Alu/Alu recombination. Additionally, the loss of check-point mechanisms preventing genome replication in the presence of unrepaired DNA damage that exist in normal cells allows cancer cells to be more successful in moving through the cell cycle even in the presence of DNA lesions some of which could be generated by L1 [98]. Thus, it is possible that the load of mutations associated with the L1-induced DSBs is significantly increased in cancer cells relative to their normal counterparts (Figure 2).
D. DNA repair pathways affecting TE activity
A critical step in the majority of malignant transformations involves defects in various DNA repair pathways that increase genetic instability, leading to more rapid tumor evolution. Thus, it is important to understand how these various genetic components interact with mobile elements expressed in tumors. An indication that cellular environment can fundamentally change L1′s ability to retrotranspose originated from the report of efficient retrotransposition of L1 lacking endonuclease activity in chinese hamster ovary cells deficient in non-homologous end joining (NHEJ) repair [82]. NHEJ involvement in L1 retrotransposition is further supported by the recent report of diminished L1 retrotransposition in chicken cells defective in NHEJ repair [73]. Furthermore, cells deficient in ATM did not support efficient L1 integration suggesting that the cellular ‘sensing’ of the L1 insertion intermediates is important in the L1 life cycle [68].
While some NHEJ signaling appears to be required for successful L1 retrotransposition, another cellular DNA repair protein usually involved in nucleotide excision repair (NER) exhibits the opposite effect on L1 mobilization [69]. Cells deficient in functional ERCC1 support significantly higher L1 retrotransposition than cells that express wild type ERCC1 [69] strongly suggesting that this protein may provide protection against L1 retrotransposition-induced damage to host DNA. Based on the known ERCC1 function [99], together with XPF it most likely acts on L1 intermediates by cleaving the single strand DNA branch structure or “flap” generated during L1 cDNA synthesis, thus, suppressing L1 retrotransposition (Figure 1B). Interestingly, Alu retrotransposition does not increase in the absence of ERCC1 (Roy-Engel AM., unpublished), even though Alu elements rely on L1 ORF2 protein for their mobilization [23]. This observation parallels previous reports of differential effect of APOBEC proteins on L1 and Alu retrotransposition [70-72] and the discrepancy in the timing required for the completion of Alu and L1 retrotransposition [100]. Because the involvement of DNA repair machinery in the retrotransposition process is only beginning to emerge, many specific details about the role they play in aiding or down-regulating L1, Alu, and SVA mobilization remain unidentified. Selective loss of specific DNA repair pathways in various human cancers may provide an additional level of variability in the TE contribution to genetic instability in a cancer type-specific manner. For example, newly discovered ERCC1 involvement in suppression of L1 retrotransposition becomes particularly relevant for the possibility of increased L1 contribution to genetic instability in human lung cancers that often lack functional ERCC1 [101]. Involvement of DNA repair in TE retrotransposition also suggests that polymorphisms within DNA repair genes may potentially contribute to variation in cancer evolution associated with TE activity.
E. TE sequences and gene expression: changes in the cancer transcriptome
The presence of mobile elements near genes or within the introns of human genes may cause a number of more subtle changes in gene expression, some of which appear to be specific to the transformed state (some major mechanisms are summarized in Figure 3). TE interference with gene expression is orientation dependent and varies relative to the size of the insert [102-104]. As expected, full-length L1 inserts are subject to negative selection because they appear to interfere with normal gene expression the most, particularly in the forward orientation [7,102,103]. They also introduce sense and anti-sense promoter encoded by the L1 5′ untranslated region (UTR; Figure 3A) that can drive expression of adjacent genes both in normal and tumor cells [105,106]. Cancer-specific L1-driven hybrid transcripts were detected in two breast cancer and three colon cancer cell lines [107]. Much like L1, the internal pol-III promoter present in Alu elements can also transcribe adjacent sequences (Figure 3B) and is proposed to drive the expression of miRNAs [108]. Expression of some of the normally silenced by methylation miRNAs occurs in cancer cells with hypomethylated genomes [109,110]. Given that cellular transformation has been shown to relax suppression of transcription of SINE RNA polymerase III promoters [33,52], it is plausible that Alu-driven miRNAs could be up-regulated in transformed cells. Although, the biological relevance of the composite L1- and Alu-driven transcripts in cancer causation or progression is unclear, these RNAs could represent transcriptional interference with cellular genes important for cancer progression.
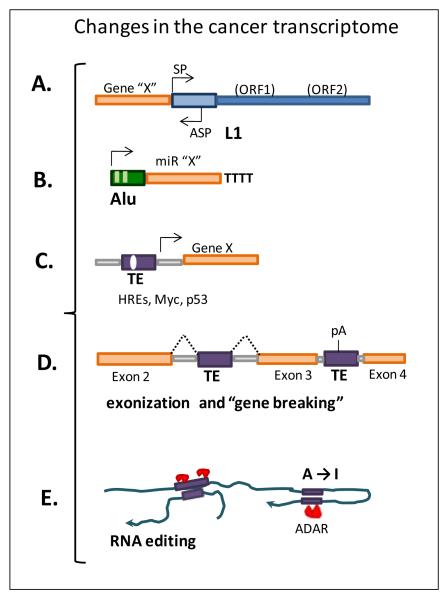
Figure 3. TE-derived changes in the cancer transcriptome.
Gene expression can be affected by the presence of mobile element sequences near, or within, genes. The unmethylated status observed in cancer cells usually leads to the loss of transcriptional regulation of TE promoters affecting nearby genes as shown in A) where the antisense promoter (ASP) activity of a full-length L1 element (blue) can drive expression of genes immediately adjacent to these sequences (Nigumann et.al. 2002;Speek 2001) and B) by the expression of miRs from upstream pol III-containing Alu elements (Borchert et.al. 2006). C) TE-derived sequences upstream of genes can supply regulatory sequences such as p53 sites affecting gene expression (Zemojtel et al., 2009); or D) introduce splicing and polyadenylation signals changing transcriptional products leading to events such as exonization s that can alter protein function (Yi et al., 2003). E) The presence of repeated sequences in opposite orientation within transcripts can form double stranded RNA (dsRNA) which is modified by RNA editing enzymes with a potential impact on expression of the gene.
It is well-established that TEs have played a significant part in reshaping the mammalian genome in the course of evolution (reviewed in [6]). A genome-wide survey of human genes determined that about one fourth of the analyzed human promoter regions contains TE-derived sequences within the 500 bp region upstream of its annotated initiation site [111]. The drift in Alu promoter sequence due to methylation and deamination of CpGs is also reported to generated p53-binding sites [112] suggesting a potential alteration in the transcriptional regulation of the genes harboring these events (Figure 3C). Perhaps this is the reason that Alu sequences are reported to be underrepresented in the first 1,000 bp of human promoters relative to introns, on the other hand, upstream regions beyond 1 kb appear to be enriched in Alus [113] and over 1000 of the human genes may be regulated by Alu-associated CpGs [114].
TE sequences, particularly L1 elements, contain internal polyadenylation (pA) signals, and splice donor or acceptor sites that can be utilized during transcription and as a result contribute to changes in gene expression and produce hybrid L1 transcripts [48-50,106,115,116]. Alu elements are known to acquire these cis-regulatory signal post insertionally through accumulation of random mutations that introduce functional polyadenylation sites [117-119] or lead to Alu-exonization events [120-122] (Figure 3D). A comprehensive survey of disease-associated cryptic exons demonstrated that about two thirds of them contained TE sequences [122]. Evaluation of the human genome by an automated approach (SERpredict) that detects specific exonized retroelements predicted expression of several RNAs from genes with TE-derived tumor specific isoforms [123]. Recently SVA elements have been reported to provide functional splice sites to generated hybrid transcripts with human genes [124,125]. Although little is known about tumor specific TE-generated RNA or protein isoforms and even less about their potential role in tumorigenesis, there is a potential that some of them may affect cellular response to specific anti-cancer therapeutic agents. For example, a 98 bp Alu-like sequence in the Bcl-rambo beta gene acts as an exon creating a new protein variant that promotes etoposide- and taxol- induced death, which upon removal of the Alu-like sequence loses this function [126].
Because TEs are dispersed throughout many unprocessed transcripts, adjacent repetitive sequences present in the inverted orientation can form double strand RNA (dsRNA) in the nucleus. Double stranded pre-mRNA can be postranscriptionally modified by RNA editing enzymes. The diversity of RNA editing mechanisms includes nucleoside modifications such as cytosine to uridine (C to U) introduced by the APOBEC family of proteins and adenine to inosine (A to I) deaminations by the adenosine deaminase enzymes or ADARs. Inverted Alu sequences within transcripts are subjected to extensive editing by ADARs with a potential impact on the expression of the gene [127,128]. Loss of RNA editing appears to be common in tumors with reported global cancer-related hypo-editing of Alus in tissues from brain, prostate, lung, kidney and testis and with correlation between defects in ADAR activity and cancer progression [129]. Although there are links between RNA editing and cancer, the mechanism and biological consequences of the relationship between these processes remain unclear (reviewed in [130]).
F. Cancer treatment and transposable elements
TE-associated toxicity
Early activation of L1 and Alu elements during tumorigenesis and the reported ability of L1 expression to induce cell cycle arrest, apoptosis, and a senescence-like phenotype in cancer cells [68,80,131,132] could raise the possibility of a “double-edged sword” hypothesis associated with TE expression during cancer evolution (Figure 4). Increased DNA damage from up-regulated L1 expression in normal cells or during early stages of transformation may initiate the cascade of events leading to apoptosis or senescence, therefore, potentially preventing cellular progression to malignancy. On the other hand, if the cells have lost the ability to arrest cell cycle progression or apoptosis, L1-induced DNA damage is likely to be one of the factors contributing to genetic instability and potentially cancer evolution toward a more aggressive phenotype. This prospective dual role of L1-associated damage during transformation is supported by the findings that ectopic L1 expression in normal human cells leads to a senescence-like phenotype [29] and that L1-induced toxicity in breast cancer cells can be overcome by suppression of p53 function, Bcl2 expression or treatment with caspase inhibitor, which suppress apoptotic response in these cells [80,131,133]. These studies offer insights into the types of extreme cellular responses to L1 over-expression and provide the means to investigate L1 interaction with cellular environment. The caveat of these findings is that the reported outcomes occurred as a response to acute, transient expression of L1, leaving understanding of the cellular reaction(s) to chronic exposure to L1-induced damage largely unexplored.
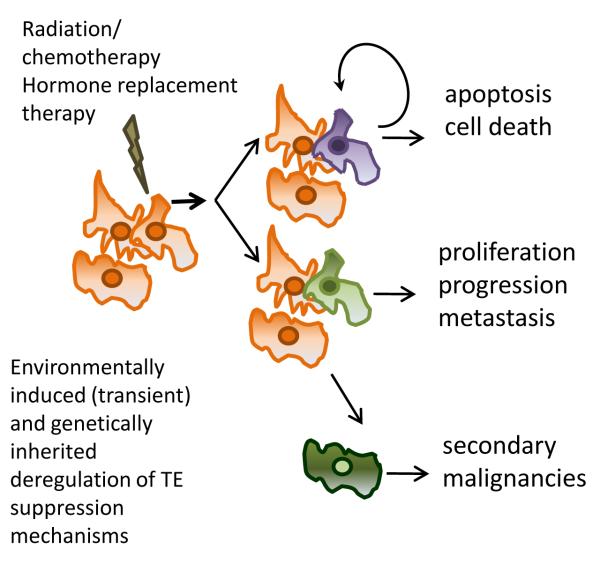
Figure 4. the “double-edge sword” hypothesis.
Environmentally induced changes either due to transient exposures (e.g. demethylating agents) alone or in combination with genetic defects, such as DNA repair deficiencies in cancerous cells, can lead to a diminished regulation of mobile elements. The increase in activity and/or deregulated transcription of TE promoters can have an agonistic effect such as promoting apoptosis or alternatively have an antagonistic effect by inducing changes leading to chemotherapeutic resistance, proliferation or even the generation of secondary malignancies.
TE regulation is not limited to genomic context and programmed responses. Their expression and activity can also be stimulated by cellular changes induced by exposures to external stimuli (reviewed in [134]. For example, Alu expression increases after heat shock, treatments with puromycin, cycloheximide [54], etoposide and cisplatin [135], exposure to UV radiation [135], and in cells infected with adenovirus type 2 [136]. However, an increase in Alu retrotransposition rates has only been suggested for the treatment with etoposide [137]. Similarly, L1 activity has been reported to increase after treatment with either benzo(a)pyrene (BaP) [138] or exposure to ionizing radiation [132]. In addition to these reports, exposures to a number of heavy metals such as nickel, cadmium and mercury increase L1 activity without changing steady-state mRNA levels, acting most likely through the inhibition of cellular factors that limit L1 insertion [139,140]. The TE response to external stimuli brings a question of the potential effects currently used cancer therapeutics may have on TE activity and TE-driven instability. As an example, external stimuli causing genetic damage (such as double strand breaks generated by chemotherapeutic agents) induce a p53 response [98]. Recent data indicate that p53 can activate L1 transcription through p53 binding sites that emerged within L1 promoter of young L1 subfamilies [141]. This p53-driven increase in L1 expression is likely to introduce more DSBs and amplify the p53-mediated DNA damage response (Figure 4). Our current knowledge predicts that many of these treatments may increase TE activity suggesting the possibility of the TE-induced instability contributing to the evolution of the therapy-resistant cancer phenotypes.
It is well-established that transcription of transposable elements is heavily suppressed by methylation and likely other epigenetic modifications [39,142]. One of the most dramatic examples of the loss of epigenetic control of retroelements was reported in the Dnmt3L knockout mouse that presented increased TE transcription due to the loss of de novo methylation of these elements [143]. Many cancer therapies include chromatin-modifying and methylation reducing agents such as 5-aza-2′-deoxicytidine (5-Aza-CdR) that inhibit DNA methylation and histone deacetylases (HDAC) [144-146]. The use of “epigenetic therapies” in cancer treatment is reported to have an impact on mobile element regulation [56,147] with potential synergistic and antagonistic results. For example, demethylation of Alu elements in response to 5-Aza-CdR and 4-phenylbutyric acid (PBA) treatment of gastric and bladder cancer cells has been associated with the induction of several miRNAs (Figure 3B) [109,110], one of which, miR-512-5, when expressed, suppresses the anti-apoptotic protein Mcl-1 resulting in the death of the treated gastric cancer cells [110]. Other conventional agents routinely used in cancer therapies such as tamoxifen can also lead to progressive hypomethylation of L1 elements and their increased expression [148]. As epigenetic treatments are common in cancer management, current data on their effect on the regulation of TE expression strongly advocate that a more detailed insight into the potential reactivation of silent retrotransposons is needed in order to understand their role in cancer therapy outcomes.
E. Domesticated Transposable Elements and Cancer
Telomerase reverse transcriptase (TERT) has been proposed to be the product of a domesticated transposable element [149-151]. The reverse transcriptase domain of telomerase enzyme is a relative of the reverse transcriptase of the non-LTR elements, like L1 [152]. In addition, telomere formation uses an RNA encoded at one locus (TER) that combines with a reverse transcriptase (telomerase) made at a different genomic location. This is similar to other non-autonomous retroelements, like Alu, that encode their RNA at one location and steal the reverse transcriptase produced from another location. Much like, L1 and Alu, whose activity is upregulated in cancer cells, the majority of human cancers reactivate expression of telomerase in order to sustain uncontrolled proliferation. Additionally, in species like Drosophila, retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres (reviewed in [153]).
Humans have also domesticated transposable elements by design and have used them as tools in gene therapy and studies of carcinogenesis [154,155]. Of particular relevance have been a broad range of studies with a DNA transposon named sleeping beauty, that has been engineered to create insertional mutagenesis in mouse model systems to study cancer [156,157]. This type of tool is being used to create animal models of disease, including cancer, and also to utilize the transposon as a genetic tag to isolate the mutated genetic loci that contributed to the specific cancers [158-160].
The Promise of Next Generation Sequencing
The full level of the impact of mobile elements on human genome instability has been extremely difficult to measure, particularly in terms of somatic instability. One of the major difficulties is the presence of massive numbers of L1 and Alu elements in the genome that obscure most approaches to detect small differences in copy number or arrangement between genomes. Next generation sequencing holds tremendous promise for obtaining truly quantitative measures of the amount of insertional mutagenesis and NAHR caused by mobile elements in cancer. Even then, it may be difficult to assess rearrangements caused by recombination events triggered by inverted Alu elements or caused by L1 endonuclease-induced DSBs, as neither of these types of events can be clearly identified on an individual basis as having been induced by the mobile elements. Furthermore, even with next generation sequencing, many of the approaches, like paired-end sequencing, may be biased in detection of certain classes of mobile element-related damage. In addition, until the appropriate bioinformatic pipelines are developed for in depth analysis of sequencing data involving mobile elements, there are likely to be continued difficulties in examinations involving the mobile elements.
Summary
The biological changes associated with malignant transformation are likely to amplify the impact of transposable elements on the stability of the human genome and cancer progression. The rate and scope of TE contribution to the origin and progression of human cancers is poorly quantified to date, although we know a great deal about the types of insertion and recombination events that contribute to this process. However, the field of transposable elements is evolving with accelerated rate toward uncovering a multitude of novel effects of TEs on the stability of the host genome and interactions with various cellular pathways. It is also likely that a considerably better understanding of TE-associated contribution to mutagenesis in cancer will emerge in the near future with accumulation of data generated by the human cancer genome sequencing project. Though the difficulties associated with the analysis of repetitive sequences significantly hinder the progress in identifying specific instances of TE involvement in human cancer, a fast- growing body of studies, heavily focused on the cancer cases with unknown etiology, and genome-wide association studies strongly suggest that transposable elements could be a contributory factor in a number of human cancer with unidentified causation.
TABLE 3.
Alterations in gene expression by TE sequences.
| Insertion | Examples of the type of impact |
|---|---|
|
5′ UTR,
promoter region |
Sources of new methylation sites (CpGs) |
| Alternate promoter sequences (Speek, 2001; Matlik et al., 2006) | |
| Introduction of potential regulatory sequences (Thomson et al., 2009) and potential response elements (Norris et al., 1995) |
|
| Altered expression of nearby genes (Nigumann et al., 2002; Laperriere et al., 2007) |
|
| Introns | Alternative splicing/ exonization (Sorek et al., 2002; Lev-Maor et al., 2008; Lin et al., 2009) |
| Introduction of polyadenylation signals and splice sites(Perepelitsa-Belancio and Deininger, 2003; Han et al., 2004; Wheelan et al., 2005; Belancio et al., 2006; Belancio et al., 2008b) | |
| RNA editing (Kim et al., 2004) | |
| Introduction of promoters: expression of mIRs (Bochert et al., 2006; Saito et al., 2009) | |
| 3′ UTR | Regulation of nuclear export of mRNAs (Chen et al., 2008; Chen and Carmichael, 2009) |
| Exons | Disruption of genes (see Table 1 and reviewed in Belancio et.al.,2008a ) |
Acknowledgements
This publication was made possible by Grants Number P20RR020152 (PLD , VPB and AMR-E), R01GM45668 and EPSCOR/BORSF grant (PLD) and R01GM079709A (AMR-E) from the National Institutes of Health (NIH). VPB is supported by NIH/NIA 5K01AG030074 and The Ellison Medical Foundation New Scholar in Aging award, AG-NS-0447-08. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Competitive Advantage Funds (2006) from the Louisiana Cancer Research Consortium (LCRC) were also awarded to AMR-E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kinzler KW, Vogelstein B. Landscaping the cancer terrain. Science. 1998;280:1036–1037. doi: 10.1126/science.280.5366.1036. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Lin J, et al. A multidimensional analysis of genes mutated in breast and colorectal cancers. Genome Res. 2007;17:1304–1318. doi: 10.1101/gr.6431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris PL, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum.Mol.Genet. 2004;13:1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 5.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 6.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat.Rev.Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 9.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum.Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 10.Deininger PL, Batzer MA. Alu repeats and human disease. Mol.Genet.Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 11.Maksakova IA, et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS.Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangeney M, Pothlichet J, Renard M, Ducos B, Heidmann T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. 2005;65:2588–2591. doi: 10.1158/0008-5472.CAN-04-4231. [DOI] [PubMed] [Google Scholar]
- 13.Pothlichet J, Mangeney M, Heidmann T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int.J.Cancer. 2006;119:1869–1877. doi: 10.1002/ijc.22066. [DOI] [PubMed] [Google Scholar]
- 14.Schartl M, Hornung U, Gutbrod H, Volff JN, Wittbrodt J. Melanoma loss-of-function mutants in Xiphophorus caused by Xmrk-oncogene deletion and gene disruption by a transposable element. Genetics. 1999;153:1385–1394. doi: 10.1093/genetics/153.3.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am.J.Hum.Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid CW, Deininger PL. Sequence organization of the human genome. Cell. 1975;6:345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. SVA elements: a hominid-specific retroposon family. J.Mol.Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 18.Woods-Samuels P, et al. Characterization of a nondeleterious L1 insertion in an intron of the human factor VIII gene and further evidence of open reading frames in functional L1 elements. Genomics. 1989;4:290–296. doi: 10.1016/0888-7543(89)90332-7. [DOI] [PubMed] [Google Scholar]
- 19.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 20.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 21.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 23.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat.Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 24.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol.Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nature Genetics. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 27.Ergun S, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J.Biol.Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 28.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangwala SH, Zhang L, Kazazian HH., Jr Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10:R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol.Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bratthauer GL, Fanning TG. Active Line-1 Retrotransposons in Human Testicular Cancer. Oncogene. 1992;7:507–510. [PubMed] [Google Scholar]
- 36.Chalitchagorn K, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 37.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol.Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skowronski J, Singer MF. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc.Natl.Acad.Sci.U.S.A. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 40.Tsutsumi Y. Hypomethylation of the retrotransposon LINE-1 in malignancy. Jpn.J Clin.Oncol. 2000;30:289–290. doi: 10.1093/jjco/30.7.289. [DOI] [PubMed] [Google Scholar]
- 41.Jackson K, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol.Ther. 2004;3:1225–1231. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 42.Kanai Y, Ushijima S, Tsuda H, Sakamoto M, Hirohashi S. Aberrant DNA methylation precedes loss of heterozygosity on chromosome 16 in chronic hepatitis and liver cirrhosis. Cancer Lett. 2000;148:73–80. doi: 10.1016/s0304-3835(99)00316-x. [DOI] [PubMed] [Google Scholar]
- 43.Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2009 doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 45.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watts GS, et al. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC.Med.Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szpakowski S, et al. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat.Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 51.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat.Struct.Mol.Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 52.Tang RB, et al. Increased level of polymerase III transcribed Alu RNA in hepatocellular carcinoma tissue. Mol.Carcinog. 2005;42:93–96. doi: 10.1002/mc.20057. [DOI] [PubMed] [Google Scholar]
- 53.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 54.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daskalos A, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int.J.Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 57.Richards KL, et al. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS.ONE. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roman-Gomez J, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 59.Boland CR, Shin SK, Goel A. Promoter methylation in the genesis of gastrointestinal cancer. Yonsei Med.J. 2009;50:309–321. doi: 10.3349/ymj.2009.50.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boland CR, Komarova NL, Goel A. Chromosomal instability and cancer: not just one CINgle mechanism. Gut. 2009;58:163–164. doi: 10.1136/gut.2008.160143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc.Natl.Acad.Sci.U.S.A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lutz SM, Vincent BJ, Kazazian HH, Jr., Batzer MA, Moran JV. Allelic heterogeneity in LINE-1 retrotransposition activity. Am.J.Hum.Genet. 2003;73:1431–1437. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seleme MC, et al. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc.Natl.Acad.Sci.U.S.A. 2006;103:6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bennett EA, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy-Engel AM, et al. Active Alu element “A-tails”: size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roy AM, et al. Upstream flanking sequences and transcription of SINEs. J.Mol.Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- 68.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J.Mol.Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muckenfuss H, et al. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J.Biol.Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 72.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J.Biol.Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki J, et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS.Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 75.Van de WN, Williams R, Ockelford P, Browett P. A 20.7 kb deletion within the factor VIII gene associated with LINE-1 element insertion. Thromb.Haemost. 1998;79:938–942. [PubMed] [Google Scholar]
- 76.Vincent BJ, et al. Following the LINEs: an analysis of primate genomic variation at human-specific LINE-1 insertion sites. Mol.Biol.Evol. 2003;20:1338–1348. doi: 10.1093/molbev/msg146. [DOI] [PubMed] [Google Scholar]
- 77.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 78.Kazazian HH. An estimated frequency of endogenous insertional mutations in humans. Nature Genetics. 1999;22:130–130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]
- 79.Xing J, et al. Mobile elements create structural variation: Analysis of a complete human genome. Genome Res. 2009 doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang N, Zhang L, Zhang Y, Kazazian HH. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Research. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 83.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat.Res. 2006 doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gebow D, Miselis N, Liber HL. Homologous and nonhomologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol.Cell Biol. 2000;20:4028–4035. doi: 10.1128/mcb.20.11.4028-4035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lobachev KS, et al. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.VanHulle K, et al. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol.Cell Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Branzei D, Foiani M. Leaping forks at inverted repeats. Genes Dev. 2010;24:5–9. doi: 10.1101/gad.1884810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazoyer S. Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum.Mutat. 2005;25:415–422. doi: 10.1002/humu.20169. [DOI] [PubMed] [Google Scholar]
- 89.Smith TM, et al. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 90.van der KH, et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes.Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
- 91.Viel A, et al. Different molecular mechanisms underlie genomic deletions in the MLH1 Gene. Hum.Mutat. 2002;20:368–374. doi: 10.1002/humu.10138. [DOI] [PubMed] [Google Scholar]
- 92.Casarin A, et al. Molecular characterization of large deletions in the von Hippel-Lindau (VHL) gene by quantitative real-time PCR: the hypothesis of an alu-mediated mechanism underlying VHL gene rearrangements. Mol.Diagn.Ther. 2006;10:243–249. doi: 10.1007/BF03256463. [DOI] [PubMed] [Google Scholar]
- 93.Franke G, et al. Alu-Alu recombination underlies the vast majority of large VHL germline deletions: Molecular characterization and genotype-phenotype correlations in VHL patients. Hum.Mutat. 2009;30:776–786. doi: 10.1002/humu.20948. [DOI] [PubMed] [Google Scholar]
- 94.Hsieh SY, Chen WY, Yeh TS, Sheen IS, Huang SF. High-frequency Alu-mediated genomic recombination/deletion within the caspase-activated DNase gene in human hepatoma. Oncogene. 2005;24:6584–6589. doi: 10.1038/sj.onc.1208803. [DOI] [PubMed] [Google Scholar]
- 95.O’Neil J, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J.Exp.Med. 2007;204:3059–3066. doi: 10.1084/jem.20071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc.Natl.Acad.Sci.U.S.A. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gentles AJ, Kohany O, Jurka J. Evolutionary diversity and potential recombinogenic role of integration targets of Non-LTR retrotransposons. Mol.Biol.Evol. 2005;22:1983–1991. doi: 10.1093/molbev/msi188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 99.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J.Biol.Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 100.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS.Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simon GR, Sharma S, Cantor A, Smith P, Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127:978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 102.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc.Natl.Acad.Sci.U.S.A. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum.Mol.Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 104.Ustyugova SV, Lebedev YB, Sverdlov ED. Long L1 insertions in human gene introns specifically reduce the content of corresponding primary transcripts. Genetica. 2006;128:261–272. doi: 10.1007/s10709-005-5967-2. [DOI] [PubMed] [Google Scholar]
- 105.Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 106.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Molecular and Cellular Biology. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics. 2009;94:397–406. doi: 10.1016/j.ygeno.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 108.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Structural & Molecular Biology. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 109.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 110.Saito Y, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 111.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 112.Zemojtel T, Kielbasa SM, Arndt PF, Chung HR, Vingron M. Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet. 2009;25:63–66. doi: 10.1016/j.tig.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 113.Tsirigos A, Rigoutsos I. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS.Comput.Biol. 2009;5:e1000610. doi: 10.1371/journal.pcbi.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oei SL, et al. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics. 2004;83:873–882. doi: 10.1016/j.ygeno.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 115.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 116.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen C, Ara T, Gautheret D. Using Alu elements as polyadenylation sites: A case of retroposon exaptation. Mol.Biol.Evol. 2009;26:327–334. doi: 10.1093/molbev/msn249. [DOI] [PubMed] [Google Scholar]
- 118.Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–5590. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roy-Engel AM, et al. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet.Genome Res. 2005;110:365–371. doi: 10.1159/000084968. [DOI] [PubMed] [Google Scholar]
- 120.Lin L, et al. Large-scale analysis of exonized mammalian-wide interspersed repeats in primate genomes. Hum.Mol.Genet. 2009;18:2204–2214. doi: 10.1093/hmg/ddp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum.Genet. 2009 doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 123.Mersch B, Sela N, Ast G, Suhai S, Hotz-Wagenblatt A. SERpredict: detection of tissue-or tumor-specific isoforms generated through exonization of transposable elements. BMC.Genet. 2007;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Damert A, et al. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH., Jr Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–1991. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yi P, et al. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 2003;534:61–68. doi: 10.1016/s0014-5793(02)03778-x. [DOI] [PubMed] [Google Scholar]
- 127.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 129.Paz N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Skarda J, Amariglio N, Rechavi G. RNA editing in human cancer: review. APMIS. 2009;117:551–557. doi: 10.1111/j.1600-0463.2009.02505.x. [DOI] [PubMed] [Google Scholar]
- 131.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53. J.Biomed.Biotechnol. 2004;2004:185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Belancio VP, Roy-Engel AM. Modulation of Human Mobile Elements and Genetic Instability by Environmental Factors. Encyclopedia of Environmental health. 2011 [Google Scholar]
- 135.Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes.Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- 136.Russanova VR, Driscoll CT, Howard BH. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol.Cell Biol. 1995;15:4282–4290. doi: 10.1128/mcb.15.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat.Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 138.Stribinskis V, Ramos KS. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 2006;66:2616–2620. doi: 10.1158/0008-5472.CAN-05-3478. [DOI] [PubMed] [Google Scholar]
- 139.El Sawy M, et al. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J.Mol.Biol. 2005;354:246–257. doi: 10.1016/j.jmb.2005.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kale SP, Moore L, Deininger PL, Roy-Engel AM. Heavy metals stimulate human LINE-1 retrotransposition. Int.J.Environ.Res.Public Health. 2005;2:14–23. doi: 10.3390/ijerph2005010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harris CR, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Montoya-Durango DE, et al. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat.Res. 2009;665:20–28. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 144.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu.Rev.Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 145.Yang AS, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 146.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat.Rev.Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 147.Liang G, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol.Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tryndyak VP, et al. Epigenetic reprogramming of liver cells in tamoxifen-induced rat hepatocarcinogenesis. Mol.Carcinog. 2007;46:187–197. doi: 10.1002/mc.20263. [DOI] [PubMed] [Google Scholar]
- 149.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 150.Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 151.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 152.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 153.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu.Rev.Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 154.Dupuy AJ, et al. Mammalian germ-line transgenesis by transposition. Proc.Natl.Acad.Sci.U.S.A. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ivics Z, Izsvak Z. Transposons for gene therapy! Curr. Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- 156.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 157.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J.Mol.Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 158.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Largaespada DA. Transposon-mediated mutagenesis of somatic cells in the mouse for cancer gene identification. Methods. 2009;49:282–286. doi: 10.1016/j.ymeth.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]