Abstract
The Bcr-Abl and Lyn protein tyrosine kinases have been separately linked to the emergence of imatinib resistance in patients with chronic myelogenous leukemia. We have developed fluorescent sensors for these kinases that are enzymatically and photophysically distinct, allowing us to simultaneously, yet separately, visualize the tyrosine kinase activities of both Abl and Lyn. Multicolor monitoring revealed that an imatinib resistant cell line (MYL-R) displays a remarkable 13-fold enhancement in Lyn kinase activity relative to its imatinib sensitive counterpart (MYL). By contrast, both cell lines display nearly identical Abl activities. The upregulation of Lyn kinase phosphotransferase activity in MYL-R cells is linked to an overexpression of the Lyn B isoform. Furthermore, MYL-R cells possess a 4-fold higher level of activated Lyn and 5-fold lower level of autoinhibited Lyn than MYL cells. Furthermore, studies with an activating SH2 ligand revealed that Lyn from imatinib-resistant MYL-R cells is primed and active, whereas Lyn from imatinib-sensitive cells is dependent upon phosphorylated SH2 ligands for activity.
Simultaneous monitoring of two or more biochemical events offers the means to establish, in a temporally well-defined fashion, the influence of multiple effectors (e.g. second messengers, enzymes, pathways) on cellular behavior (1– 5). Several recent studies have focused on the use of various green fluorescent protein (GFP) derivatives to construct pairs of Förster resonance energy transfer (FRET) sensors (6– 9). A number of challenges have emerged in this regard, including the wide (and therefore overlapping) spectral width of the fluorescent proteins as well as the limited dynamic range (<40%) of the fluorescent response. Although GFP-based sensors are routinely employed in established cell lines, their application to primary cells is challenging and their use in non-cellular systems (e.g. lysates, pure enzymes, etc) would require purification and characterization of the protein construct. Many “small molecule” sensors offer a dynamic range that significantly exceeds what is possible via FRET, thereby providing an enhanced signal-to-noise ratio and thus reducing the amount of sensor required (i.e. limiting the likelihood of a sensor-induced “observer effect”) (10). Furthermore, the wide array of spectrally distinct synthetic fluorophores vastly exceeds that of the GFP family (11). These properties, along with their ease of large-scale preparation, have resulted in the cross-platform use of small molecule sensors in cell-, lysate-, and enzyme-based studies. However, their application to multicolor sensing of multiple protein kinases has not been, to the best of our knowledge, reported.
Multicolor sensing could be especially useful in the arena of medical diagnostics, where the relative activity of key enzymatic effectors and/or interacting pathways may furnish valuable information regarding the design of therapeutic protocols. A case in point is the emergence of imatinib (STI-571, Gleevec) resistance in chronic myelogenous leukemia (CML). Imatinib has transformed the management of patients with CML, which is characterized by expression of the deregulated protein tyrosine kinase Bcr-Abl (12). Although patients with CML and other leukemias benefit from imatinib treatment, relapse following therapy is common, particularly in individuals presenting advanced phase CML. Secondary mutations in the Abl active site and amplification of the fused Bcr-Abl gene were initially identified as the causative agents leading to resistance (13). However, abnormal activation of a Lyn tyrosine kinase (Lyn)-mediated signaling pathway is now recognized to facilitate imatinib resistance as well (13– 18). Although Lyn inhibitors have received significant attention as agents for the treatment of CML (19), enhanced Lyn kinase activity in imatinib-resistant CML remains to be verified. Personalized medicine, which is based on the principle of a tailor-made therapy targeting the individual patient’s needs, requires identification of the aberrant enzymatic activity responsible for the disorder and subsequent application of drugs that display the appropriate mechanism of action, potency, and selectivity. The latter requires a robust diagnostic system, involving the simultaneous comparative analysis of potential enzyme targets and the ability to screen therapeutic agents using the patient’s own cells or lysates from these cells.
Peptide-based fluorescent sensors of protein kinase activity have been primarily employed with purified enzymes, although a few examples of studies with lysates and intact cells have been reported (20, 21). In general, the phosphorylation-induced fluorescent changes in these systems are driven by electrostatics associated with the negatively charged phosphate in the phosphorylated product. The latter perturbs the photophysical properties of an environmentally sensitive fluorophore, which is commonly excited at relatively short wavelengths (<400 nm) or displays relatively modest extinction coefficients. Unfortunately, fluorophores with photophysical properties such as these are generally inappropriate for cell-based systems, including lysates. In addition, the sole reliance upon fluorophores excited at less than 400 nm sharply compresses the spectral range and thus limits the ability to perform multicolor sensing. By contrast, we’ve recently found that phosphorylation of tyrosine mitigates the ability of this amino acid to serve as a fluorescent quencher, a phenomenon that can be potentially applied to a large array of fluorophores irrespective of their emission wavelengths (22). Based on this observation, we have now prepared a pair of orthogonal Lyn and Abl sensors and have used these to assess the relative activity of their respective enzymes from imatinib-resistant and-sensitive cell lines.
RESULTS AND DISCUSSION
Abl and Lyn Kinase Monitoring in CML
Two protein tyrosine kinases, namely Abl and Lyn, have been implicated in the mechanisms responsible for imatinib resistance (13–19). The construction of orthogonal sensors for these enzymes was fashioned using three specific attributes: (i) Lyn- and Abl-selective peptide substrates, (ii) fluorophores with non-overlapping emission properties, and (iii) fluorophore-peptide conjugates that respond to phosphorylation in a fluorescently sensitive fashion. Although orthogonal peptide substrates for these two tyrosine kinases have not been previously described, their well-defined substrate sequence preferences suggested that it should be possible to identify mutually exclusive substrates (21–23). Indeed, based upon these preferences, we designed the two sequences present in 9 and 11 and assessed their selectivity for their intended enzyme targets. Tyrosine is a known quencher of fluorescence and we recently showed that this property is sharply curtailed upon phosphorylation (22). The quenching mechanism proceeds via a dynamic collisional process (24), possibly involving a phosphorylation-induced change in tyrosine’s redox potential (25). Consequently, and in marked contrast to FRET-based fluorescence quenching, tyrosine’s presumed mechanism of action should be independent of the excitation and emission wavelengths of the fluorophore and thus applicable to an array of flurophores. Indeed, this strategy had been successfully employed with a short wavelength fluorophore (pyrene) (22) and, in this study, now extended to include mid (cascade yellow) and long (oxazine) wavelength fluorophores. The dynamic range associated with these sensors (from 2 – 7-fold) (22) is approximately an order of magnitude greater than what has been described for the GFP FRET-based reporters (6, 8, 9). Furthermore, since previously described GFP-based sensors are composed of two GFP proteins, each of which have broad excitation and emission bands, much of the spectral bandwidth is occupied. Single fluorophore constructs, with relatively narrow bandwidths, preserve spectral space for additional sensors.
Our sensors reveal that the imatinib resistant cell line MYL-R and its imatinib sensitive counterpart MYL exhibit essentially identical Abl kinase activity, but markedly differ in their Lyn kinase activity. In short, a significantly higher fraction of Lyn is primed and active in the MYL-R versus MYL cell line. Furthermore, our studies reveal that although Lyn is present as two isoforms, A and B, it is the latter that is dramatically up-regulated in MYL-R cells.
Design of the Lyn/Abl sensor pair
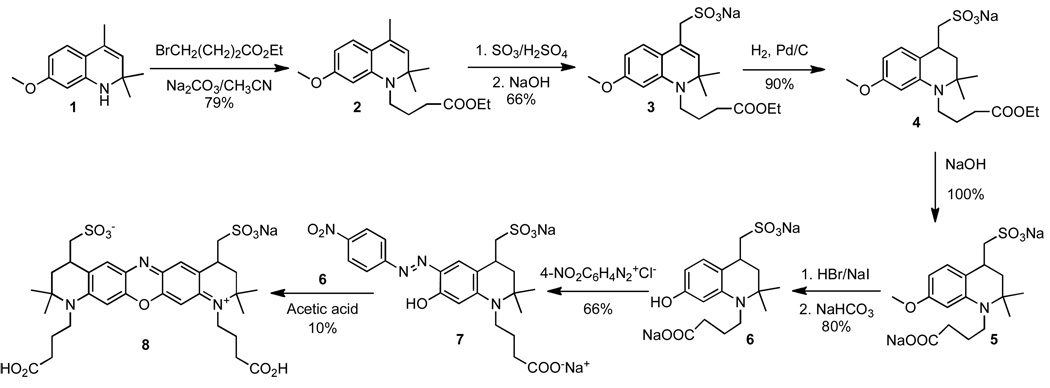
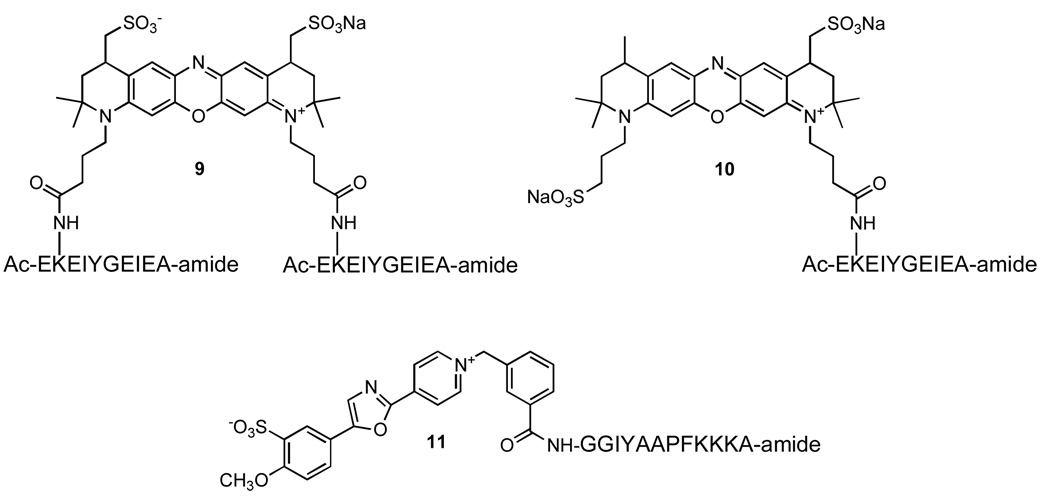
Ac-Glu-Lys-Glu-Ile-Tyr-Gly-Glu-Ile-Glu-Ala-amide, where the Lys side chain serves as the fluorophore attachment site and Tyr serves as the phosphoryl acceptor, is a sequence phosphorylated by Lyn (22). As noted above, Tyr also acts as a fluorescent quencher. We’ve previously noted that effective fluorescence quenching occurs when the fluorophore is positioned on the third residue removed from the Tyr moiety (22). However, it occurred to us that deeper quenching might be observed if the fluorophore were exposed to two phosphorylatable tyrosine moieties. Consequently, we designed and synthesized the oxazine fluorophore 8 [λex = 666 nm, λem = 682 nm, ε = 120,000 M−1 cm−1, Φ = 0.29; Fig. 1], which contains two carboxylate moieties and therefore two attachment sites for the peptide substrate. Oxazine 8 was prepared from the starting 1,2-dihydroquinoline 1 in seven steps and subsequently doubly condensed with the Lys side chain of the active site-directed peptide substrate to furnish the 1:2 fluorophore-peptide construct 9 (Fig. 2). The latter displays a Lyn-mediated 3.3-fold increase in fluorescence (Fig. 3a). A corresponding mono-carboxylate oxazine (λex = 667 nm, λem= 684 nm, ε = 118,000 M−1 cm−1, Φ = 0.27) was also prepared (data not shown) and attached to the Lys side chain of the active site-directed peptide to furnish the 1:1 fluorophore-peptide adduct 10. The latter also serves as a Lyn substrate and displays a modestly reduced (relative to 9) 2.5-fold enhancement in fluorescence in response to kinase-catalyzed phosphorylation.
Fig. 1.
Synthesis of the oxazine diacid 8. The latter was coupled to the Lys side chain of the Lyn peptide substrate Ac-Glu-Lys-Glu-Ile-Tyr-Gly-Glu-Ile-Glu-Ala-amide, to furnish the 2:1 peptide:oxazine conjugate 9.
Fig. 2.
Structures of the Lyn (9 and 10) and Abl (11) substrates.
Fig. 3.
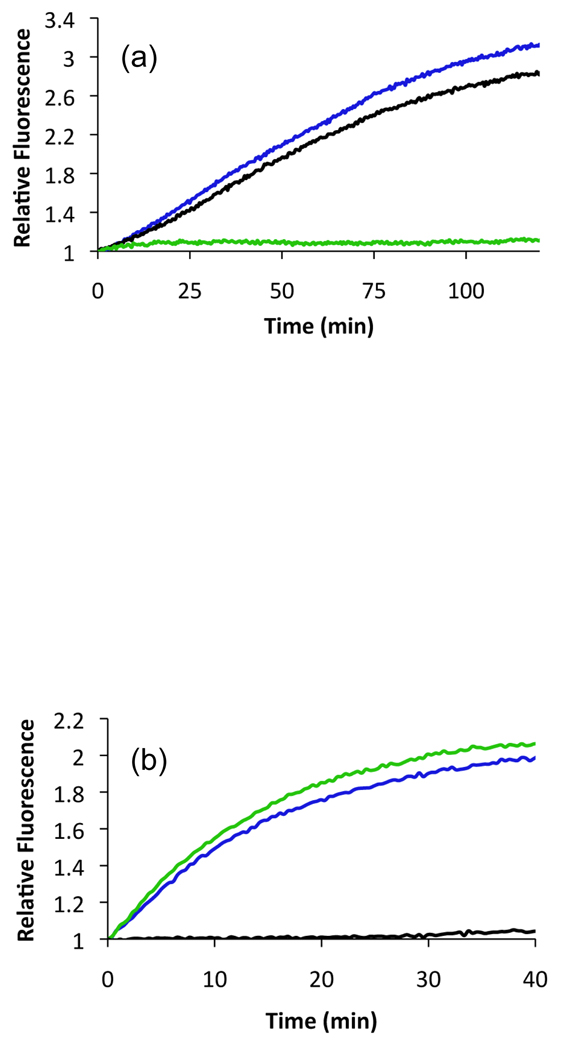
(a) Phosphorylation of 9 (λex = 665 nm, λem = 705 nm) by purified Lyn B (black), Lyn B/Abl (blue), and Abl (green). (b) Phosphorylation of 11 (λex = 400 nm, λem = 525 nm) by purified Lyn B (black), Lyn B/Abl (blue), and Abl (green).
The Abl sensor was derived from an Abl recognition motif, Gly-Gly-Ile-Tyr-Ala-Ala-Pro-Phe-Lys-Lys-Lys-Ala (23). The Cascade Yellow fluorophore (ε = 25,000 cm−1 M−1, Φ = 0.56) possesses photophysical properties (λex = 400 nm and λem = 525 nm) distinct from those of the oxazines. Abl-catalyzed phosphorylation of (Cascade Yellow)-Gly-Gly-Ile-Tyr-Ala-Ala-Pro-Phe-Lys-Lys-Lys-Ala 11 (Fig. 2) induces a 2.2-fold enhancement in fluorescence (Fig. 3b). Lyn-catalyzed phosphorylation is only detected (λem= 705 nm) with the oxazine-modified peptide and Abl activity is only observed (λem= 525 nm) with the Cascade Yellow-modified peptide (Fig. 2a–b). The Abl kinase does not interfere with the ability of Lyn to phosphorylate its sensor nor does the Lyn kinase impede Abl’s activity with its designated sensor.
The inhibitory efficacy of imatinib and dasatinib
The observed IC50 of imatinib for Abl (0.48 ± 0.04 µM) is essentially the same as previously reported values using a radioactive assay (Supplementary Figure 1a) (26). These initial studies were performed at a non-physiological ATP concentration of 50 µM. Since the mechanism of action of imatinib is competitive with respect to ATP, we anticipated a sharp rise in IC50 (i.e. decrease in inhibitory efficacy) at a more physiologically relevant ATP level (5 mM). Indeed, under these conditions, the IC50 of imatinib is approximately 40-fold higher (19 ± 1 µM) (Supplementary Figure 1b). Furthermore, in contrast to previous studies (26, 27), we find that imatinib does not significantly discriminate between Abl and Lyn at either low (0.48 ± 0.04 µM versus 0.25 ± 0.02 µM, respectively) or normal (19 ± 1 µM versus 55.0 ± 2.5 µM, respectively) ATP levels (Supplementary Figures 1c and 1d).
Dasatinib has recently received significant attention for the treatment of imatinib-resistant CML (19). Indeed, dasatinib is a more potent inhibitor of both Lyn B and Abl than imatinib (Supplementary Figures 1e–1h). At physiological ATP levels, dasatinib inhibits both Abl (0.088 ± 0.003 µM) and Lyn (0.17 ± 0.03 µM) two orders of magnitude more potently than imatinib (cf. 19 ± 1 µM and 55.0 ± 2.5 µM, respectively). As expected, at sub-physiological ATP levels, dasatinib acts as an even more powerful inhibitor of Abl (0.047 ± 0.003 µM) and Lyn (0.021 ± 0.004 µM). However, we do note that the effect of high:low ATP concentration (2-fold for Abl and 8-fold for Lyn) is not quite as dramatic as that observed with imatinib (40-fold for Abl and 20-fold for Lyn). We believe that this may be a consequence of an exceptionally high affinity of desatinib for both Abl and Lyn, one that cannot be accurately quantified, especially under low ATP conditions. In particular, others have reported sub-nM IC50 values for dasatinib. However, in these instances, the concentration of the kinase significantly exceeded that of dasatinib itself (26). Since the IC50 we obtained for Lyn at 50 µM ATP is nearly identical to the enzyme concentration employed, this raises the possibility that only a fraction of the total enzyme present is in an active, inhibitor-sensitive state. The latter was confirmed by western blot analysis for Tyr 416 (pY416), an autophosphorylation site required for Lyn kinase activity (Supplementary Figure 2). Only 25% of the commercial enzyme is present in the catalytically active pY416 state.
Lyn and Abl kinase activity in MYL and MYL-R lysates
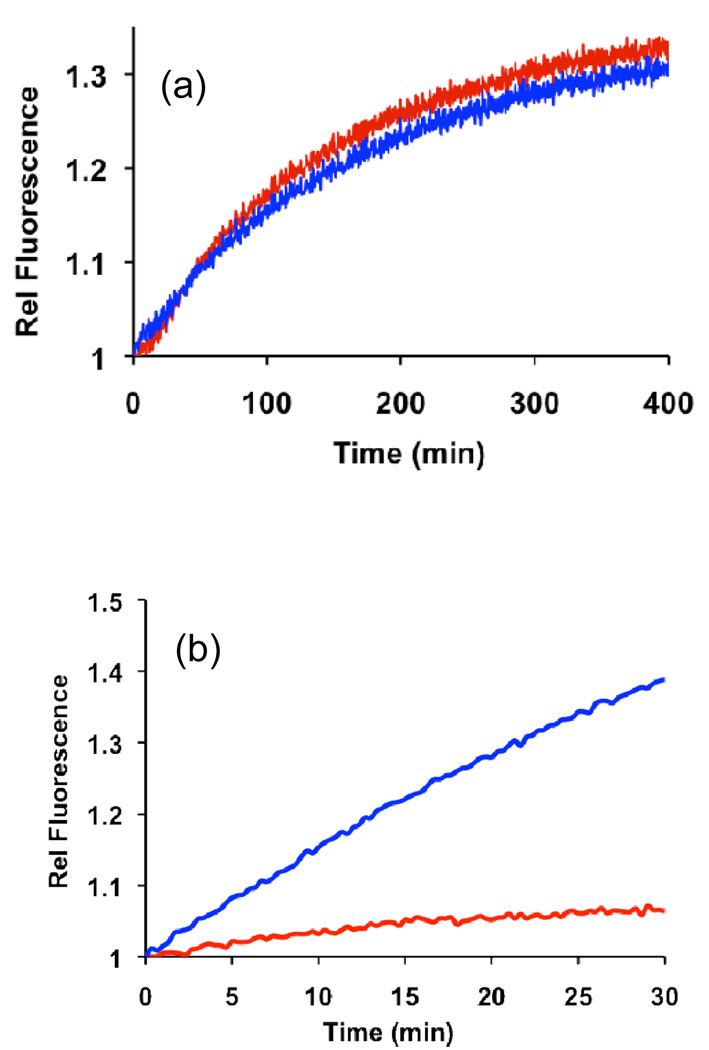
MYL is a previously described bone marrow mononuclear cell line derived from a 24-year-old female CML patient (17). Prior studies reported that Lyn mRNA, protein, and phosphoprotein, are overexpressed in MYL-R cells, an imatinib-resistant cell line sub-cloned from MYL cells (17). We probed for Abl and Lyn activity in lysates from both the sensitive and resistant cell lines. Both cell lines display similar Abl activity (Fig. 4a). We note that near-complete removal of Abl from lysates via immunodepletion significantly reduces Abl sensor activity, consistent with the notion that compound 11 serves as a selective indicator of Abl activity in cell lysates (Supplementary Figure 4). In contrast to the similar Abl kinase activity in both MYL and MYL-R cell lines, we found that MYL-R cells display dramatically more Lyn kinase (~13-fold) activity than their MYL counterparts (Fig. 4b). Reassuringly, sensor 9 phosphorylation is lost in Lyn-immunodepleted MYL-R lysates, directly demonstrating that sensor 9 is selective for Lyn kinase activity (Supplementary Figure 3).
Fig. 4.
(a) Phosphorylation of the Abl sensor 11 (λex = 400 nm, λem = 525 nm) by MYL (red) and MYL-R (blue) lysates. (b) Phosphorylation of the Lyn sensor 9 (λex = 665 nm, λem = 705 nm) by MYL lysates (red) MYL-R lysates (blue). Lysates contain all endogenous protein kinases and were normalized for total protein content as described in the Methods Section (Protein Kinase Assays). Abl and Lyn kinase activities were monitored simultaneously during the course of the experiments.
Assessment of Lyn sensor efficacy in cell lysates
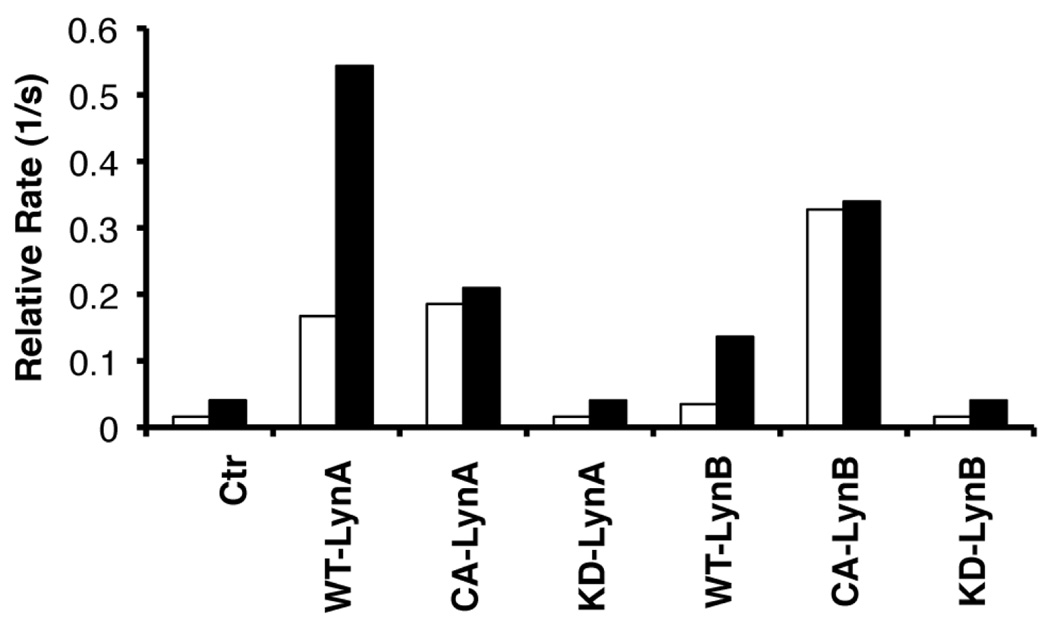
Since the activity of Lyn, but not Abl, differs in MYL and MYL-R cells, we focused our attention on a more detailed assessment of the Lyn kinase sensor. HEK293 cells were employed as a negative background expression system, since lysates from these cells (empty pcDNA3.1 control vector) exhibit minimal activity toward sensor 9 (Fig. 5). By contrast, lysates from cells transiently transfected with a plasmid encoding the wild type Lyn A or Lyn B isoform (a splice variant missing residues 22–42) (28, 29) phosphorylate sensor 9 in a time-dependent fashion. The wild type enzyme can occupy an inactive conformation, in which pY527 (Src numbering; Y508, LynA; Y487, LynB) is intramolecularly bound to the SH2 domain of the kinase. Peptides containing pTyr embedded within an appropriate amino acid sequence compete for the SH2 domain, thereby releasing the intramolecularly bound phosphorylated residue, ultimately leading to enzyme activation (30). The SH2 ligand Ac-pTyr-Glu-Glu-Ile-Glu dramatically increases the rate of peptide 9 phosphorylation in both Lyn A and Lyn B transfected cells (Fig. 5). A slight enhancement over background is also observed with lysates from the control-transfected cells, suggesting the presence of endogenous kinases at minimally detectable levels that respond to the SH2 ligand. Constitutively active mutants of Lyn A and B (Y508F and Y487F, respectively) are likewise detectable. As expected, the phosphotransferase activity of these mutants is not enhanced by pTyr-bearing peptides. Finally, expression of the corresponding inactive “kinase dead” (KD) versions of Lyn A and B (K275E and K254E, respectively) fail to display sensor 9 phosphorylation activity above control-transfected cells (+/− SH2 ligand).
Fig. 5.
Initial rate of the phosphorylation of Lyn sensor 9 [without (unfilled) and with (filled) SH2 ligand (200 µM Ac-pTyr-Glu-Glu-Ile-Glu)] by HEK293 cells transfected with the indicated constructs (where Ctl = control-transfected HEK293 cells). Lysates contain were normalized for total protein content as described in the Methods Section (Protein Kinase Assays).
Lyn expression and activity in MYL and MYL-R
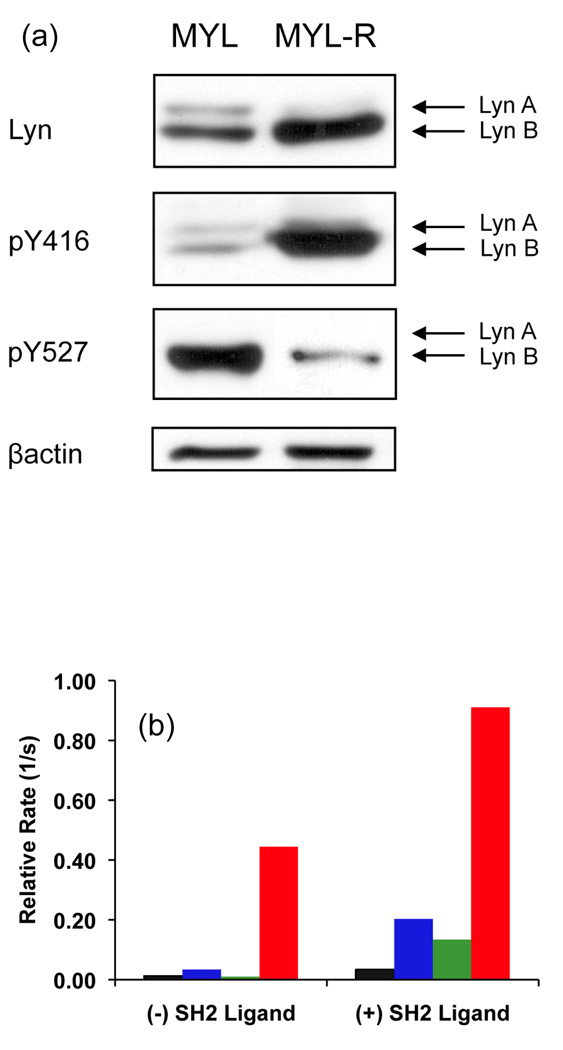
Western blot analysis revealed that MYL-R cells possess 50% more total Lyn (A + B) than MYL cells, which is primarily due to a dramatic increase in Lyn B content (Fig. 6a). MYL-R cells possess a 4-fold higher level of activated Lyn (pY416) and 5-fold lower level of autoinhibited Lyn (pY527) than MYL cells. The SH2 ligand, Ac-pTyr-Glu-Glu-Ile-Glu, enhances Lyn kinase activity 7-fold in MYL lysates, but only 2-fold in MYL-R lysates (Fig. 6b and Supplementary Figure 5). This reflects the relative amount of Lyn present in the autoinhibited state in these cell lines. These results reveal that Lyn from imatinib-resistant cells is primed and active, whereas Lyn from imatinib-sensitive cells is dependent upon phosphorylated SH2 ligands for activity.
Fig. 6.
(a) Total Lyn A and B, pY416, and pY527 content in MYL and MYL-R cell lines. (b) Initial phosphorylation rate of Lyn sensor 9 with Lyn-precleared MYL lysates (black), MYL lysates (blue), Lyn-precleared MYL-R lysates (green), and MYL-R lysates (red).
Conclusions and Outlook
We have constructed a pair of orthogonal Abl and Lyn sensors. Multiwavelength monitoring revealed that Lyn, not Abl, activity is dramatically upregulated in an imatinib resistant cell line. Furthermore, the extent of catalytic activation (13-fold) is even greater than what would have been predicted on the basis of enhanced protein content alone (1.5-fold). The simultaneous measurement of multiple protein kinases may prove useful in a diagnostic setting since, as in the case of imatinib resistance, several enzymes are often implicated as the potential biochemical trigger for aberrant cellular behavior. The latter should be feasible using leukocytes purified from patient peripheral blood or bone marrow aspirates. Furthermore, multicolor sensing of catalytic activity has therapeutic implications since the majority of drugs that target protein kinases primarily interfere with catalytic activity. Consequently, sensors that furnish an activity readout offer the means to assess the efficacy of drug cocktails and dosing regimens, prior to therapy, using the patient’s own cells.
METHODS
General Aspects
Analytical grade reagents were purchased from Sigma-Aldrich or Fisher, except for 1H-benzotriazolium 1-[bis(dimethylamino)methylene]-5-chlorohexafluorophosphate (1-),3-oxide (HCTU), benzotriazole-1-yloxytrispyrrolidinophosphonium hexafluorophosphate (PyBop), amino acids, and TGR resins, which were obtained from NovaBiochem. Amine-reactive fluorophores were obtained from Aldrich or Invitrogen. The SH2 ligand, Ac-pTyr-Glu-Glu-Ile-Glu, was purchased from Bachem. UV-vis spectra were measured using a Biochrome Libra S22 spectrophotometer. Emission and excitation spectra were obtained using a Horiba Jobin Yvon Fluorolog 3 spectrofluorometer at 23 °C. Quantum yields were measured using indodicarbocyanine iodide as an internal standard. Solutions were not deaerated because control experiments showed that oxygen did not quench the fluorescence of these dyes, due perhaps to their short lifetimes. Mass spectra were obtained on an Applied Biosystems API 2000 LC/MS/MS system. See Supporting Information for Mass Spectral data. 1H and 13C NMR spectra were recorded on Bruker DRX-300 spectrometer. See Supporting Information for NMR data. All operations with dyes were performed under dim light. Flasks containing dyes were wrapped with aluminum foil. Chemical names for compounds were obtaining using ChemDraw Ultra 7. Human CML cells (imatinib-sensitive Myl and imatinib-resistant MylR) were a generous gift from Dr. Hideo Tanaka (Dept. of Haematology and Oncology, Hiroshima University, Hiroshima, Japan). Cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and 1% penicillin/streptomycin (P/S; Invitrogen). HEK 293 cells were acquired from ATCC. Cells were cultured in DMEM medium (Invitrogen) supplemented with 10% FBS and 1% P/S. Cells were grown in a humidified incubator at 37°C with 5% CO2. The Lyn A and Lyn B expression constructs were a generous gift from Dr. Juan Rivera (Mol. Immunol. and Inflammation Branch, NIH, Bethesda, MD). Constructs were cloned from the pLenti6/V5 vector (Invitrogen) into the pCDNA3.1 vector (Invitrogen). Point mutations were made in the Lyn A (K275E, Y508F) and Lyn B (K254E, Y487F) constructs using the Quickchange Site-directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Expression vectors were introduced into HEK 293 cells by Lipofectamine transfection (Invitrogen) according to the manufacturer’s instructions. 36 h post-transfection cells were washed with PBS and pelleted; cells were stored at −80 °C until experimentation.
Chemical synthesis. 4-(7-methoxy-2,2,4-trimethyl-2H-quinolin-1-yl)-butyric acid ethyl ester (2)
A 500 mL round-bottom flask was set up with a magnetic stir bar, a reflux condenser, and a nitrogen line. The flask was charged with 7-methoxy-2,2,4-trimethyl-1,2-dihydro-quinoline 1 (31) (43 g, 0.21 mol), ethyl 4-bromobutyrate (88 g, 0.44 mol), sodium iodide (11 g, 0.073 mol), acetonitrile (150 mL, 2.9 mol), and sodium carbonate (52 g, 0.48 mol). The flask was heated at reflux with stirring for 72 h. The reaction mixture was filtered and the acetonitrile was removed with a rotary evaporator. The unreacted starting compound was removed from the product by vacuum distillation using an oil pump. The recovery of 7-methoxy-2,2,4-trimethyl-1,2-dihydro-quinoline was 7 g (16%, b.p. 120–125°C, 0.7 Torr). The viscous yellow residue in the distillation flask was the pure product. The yield was 53.0 g as an oil (79% yield).
Sodium [1-(3-ethoxycarbonyl-propyl)-7-methoxy-2,2-dimethyl-1,2-dihydro-quinolin-4-yl]-methanesulfonate (3)
A 500 mL round-bottom, three-necked flask was set up with a nitrogen line, a mechanical stirrer, and a thermometer. The flask was immersed in a salt-ice cooling bath and 4-(7-methoxy-2,2,4-trimethyl-2H-quinolin-1-yl)-butyric acid ethyl ester (2, 30 g, 0.09 mol) was placed in the flask and the stirrer was started. Sulfuric acid (30 mL, 0.6 mol) was added slowly to the flask over the course of about 5 min. The reaction mixture was then stirred for additional 30 min with cooling. When the temperature inside the flask was about −10°C, fuming sulfuric acid (17 mL, 0.18 mol, 30% of free SO3) was added quickly to the flask. The stirring was continued and the flask was allowed to slowly warm up to room temperature while in cooling bath. The reaction mixture was stirred for another 48 h at room temperature. The reaction mixture was poured into a 2 L beaker containing 500 g of ice and the beaker was immersed in a salt-ice cooling bath. The acid was neutralized with 10% solution of sodium hydroxide in water until the pH of the reaction mixture was about 7. The temperature of the reaction mixture was kept below 10 °C during the neutralization process, at the end of which the product crystallized. The crystals were filtered using a Büchner funnel, washed with 50 mL of ice-cold water, and air-dried. The crystals were placed in 2 L flask equipped with reflux condenser and 1 L of 100% ethanol was added. The mixture was stirred with heating until the ethanol started to boil and some of the solid material dissolved. The mixture was filtered while still hot using a Buchner funnel. The crystals (Na2SO4 hydrate) were washed with three portions of 100 mL of hot ethanol. A combined filtrate solution was evaporated using a rotary evaporator and the resultant white solid was dried under high vacuum overnight. The yield was 26 g (66% yield).
Sodium [1-(3-ethoxycarbonyl-propyl)-7-methoxy-2,2-dimethyl-1,2-dihydro-quinolin-4-yl]-methanesulfonate (4)
Sodium [1-(3-ethoxycarbonyl-propyl)-7-methoxy-2,2-dimethyl-1,2-dihydro-quinolin-4-yl]-methanesulfonate (3, 22 g, 0.052 mol), methanol (250 mL, 6.2), and Pd on carbon (10%, 1.0 g) were added to a 1 L high pressure, heavy wall Parr hydrogenation bottle. The bottle was attached to a high-pressure Parr hydrogenation apparatus and the system was evacuated using a water aspirator and then filled with hydrogen gas. The evacuation/filling procedure was repeated three more times. Then the system was filled with hydrogen at 50 psi pressure and the shaker was activated. The bottle was shaken at room temperature for 4 h until hydrogen consumption stopped. The remaining pressure was carefully released and the catalyst was removed by vacuum filtration through a Celite pad. The solvent was removed under vacuum to give the final product as a white solid. The yield was 20 g (90% yield).
Disodium 4-(7-methoxy-2,2-dimethyl-4-(sulfonatomethyl)-3,4-dihydroquinolin-1(2H)-yl)butanoate) (5)
A 500 mL round-bottomed, one-necked flask was set up with a magnetic stir bar and a reflux condenser. The flask was charged with sodium [1-(3-ethoxycarbonyl-propyl)-7-methoxy-2,2-dimethyl-1,2-dihydro-quinolin-4-yl]-methane-sulfonate (4, 20 g, 0.05 mol), sodium hydroxide (2.09 g, 0.0522 mol), and 100 mL water. The mixture was stirred with heating (50°C) for 1 h and then cooled to room temperature. The solvent was removed under vacuum and the solid was stirred with 100 mL of MeOH at reflux for 30 min. The hot suspension was filtered, the solid was washed with hot methanol, and dried. The yield of the final product (white solid) was 20 g (100% yield).
Disodium 4-[7-hydroxy-2,2-dimethyl-4-(sulfonatomethyl)-3,4-dihydroquinolin-1(2H)-yl]butanoate (6)
A 250 mL round-bottomed, one-necked flask was set up with a magnetic stir bar, a reflux condenser, and a nitrogen line. The flask was charged with disodium 4-(7-methoxy-2,2-dimethyl-4-(sulfonatomethyl)-3,4-dihydroquinolin-1(2H)-yl)butanoate) (5, 12 g, 0.029 mol), sodium iodide (10 g, 0.07 mol), and a solution of 9.0 M hydrogen bromide in water (70 mL). The flask was flushed with nitrogen and then was heated at 105 °C (oil bath) with stirring for 18 h. The reaction mixture was cooled to room temperature and the excess hydrobromic acid was removed using rotavap. To ensure a complete removal of the hydrobromic acid, 100 mL of water was added to the residue and the solvent was removed. The yellow solid was dissolved in 50 mL of water and the solution was neutralized by addition of solid sodium bicarbonate. The solvent was removed by vacuum and the solid was refluxed with stirring with 500 mL of acetone overnight under nitrogen to remove sodium iodide and sodium bromide that are soluble in acetone. The mixture was cooled to room temperature and filtered. The solid was dried was washed with acetone and dried. The yield was 9.3 g (80% yield) as a tan solid.
The 4-nitrophenyldiazenylphenol 7
Step 1. p-Nitroaniline (0.546 g, 0.00395 mol) was added to a 100 mL round-bottomed, one-necked flask equipped with a magnetic stir bar and an addition funnel. 10 mL of 10% HCl was added to the flask. The mixture was stirred at room temperature until a clear solution formed, and then the flask was immersed in an ice-water bath. A solution of sodium nitrite (0.273 g, 0.00395 mol) in 5 mL of water was added drop-wise to the flask with stirring. When the addition was complete, the mixture was stirred for another 30 min at 0°C. Step 2. Disodium 4-[7-hydroxy-2,2-dimethyl-4-(sulfonatomethyl)-3,4-dihydroquinolin-1(2H)-yl]butanoate, 6) (1.59 g, 0.00395 mol) was added to a 250 mL conical flask, followed by 10 mL of 10% HCl. The flask was immersed in an ice-water cooling bath and stirring was started. The solution of p-nitrophenyldiazonium salt (from step 1) was added to the flask in small portions (1–2 mL) over the course of 10 min. The reaction mixture was stirred for 1 h at 0°C. The precipitated red solid was filtered, washed with water (3×15 mL). The red solid was placed in a 500 mL round-bottomed, one-necked flask equipped with a magnetic stirrer. Water (150 mL) was added to the flask and the stirring was started. Solid sodium bicarbonate (1.125 g) was added to the flask in five portions to avoid excessive foaming. The solvent was removed using a rotary evaporator. The solid residue was re-dissolved in methanol. The solution was filtered to remove inorganic salts and the methanol was removed using a rotary evaporator to give a red solid. The residue was re-dissolved in 25 mL of water, and the volume was reduced to 10 mL using the rotary evaporator (this operation was necessary to remove traces of methanol). The solution was frozen and lyophilized overnight. The yield of red solid product was 1.38 g (66% yield).
Sodium (1,11-bis(3-carboxypropyl)-2,2,10,10-tetramethyl-2,3,4,8,9,10-hexahydro-1H-dipyrido[3,2-b:2',3'-i]phenoxazine-11-ium-4,8-diyl)dimethanesulfonate (8)
(E)-(7-hydroxy-1-(3-carboxypropyl)-2,2-dimethyl-6-((4-nitrophenyl)diazenyl)-1,2,3,4-tetra-hydroquinolinium-4-yl)methanesulfonate) (7, 0.671 g, 0.00132 mol), disodium 4-[7-hydroxy-2,2-dimethyl-4-(sulfonatomethyl)-3,4-dihydroquinolin-1(2H)-yl]butanoate) (6, 0.532 g, 0.00132 mol), and acetic acid (25 mL, 0.44 mol) were added to a 100 mL round-bottomed, one-necked flask set up with a magnetic stir bar and a reflux condenser. The reaction mixture was heated at reflux for 24 h. TLC showed complete consumption of starting material and formation of deeply blue-colored product (SiO2, eluent: acetone-water (1:1), Rf product = 0.1, Rf starting material = 0, and 0.4). The reaction mixture was evaporated and the residue was dissolved in 50 mL of a mixture of acetone-water (4:1). The solution was mixed with 10 g silica and the solvent was removed under vacuum. The dye/silica mixture was loaded onto a silica column prepared in acetone. The column was eluted with an acetone-water gradient (0 – 10%). The dye containing fractions were combined and the solvent was removed using a rotary evaporator. NMR analysis showed the presence of impurities, and the dye was further purified on a preparative RP HPLC column, which was eluted with water and acetonitrile. (Program: 0 – 60 min 100% water, 60 – 120 min 10% acetonitrile/90% water, 120 – 150 min 50% acetonitrile/50% water, 150 – 180 min 80% acetonitrile/20% water, 180 – 200 min 100% water.) The dye eluted at 105–115 min. The dye-containing fractions were combined and reduced in volume from about 120 mL to 10 mL using a rotary evaporator. The dye solution was frozen and lyophilized. The yield of pure dye (blue solid) was 100 mg (10% yield).
Peptide synthesis. Lyn Sensor Peptide (9)
The peptide-resin Ac-Glu(tBu)-Lys(Mtt)-Glu(tBu)-Ile-Tyr(tBu)-Gly-Glu(tBu)-Ile-Glu(tBu)-Ala-amide-resin was synthesized using standard Fmoc peptide synthesis protocol [amino acids 5 eq, HCTU 5 eq, DIPEA (N,N-diisopropylethylamine) 10 eq] as previously described.(21) The side chains of Glu and Tyr were protected with t-Bu. The side chain of Lys was protected with the acid sensitive 4-methyltrityl (Mtt) group. Tetramethylfluoroform-amidinium (TFFH) (5 eq) and DIPEA (10 eq) were used to ensure successful coupling of Fmoc-Lys(Mtt)-OH (5 eq) at the Y-3 position. After synthesis of the peptide on the resin and subsequent acetylation of the N-terminus, the Mtt on Lys was selectively removed with 2% TFA. The side chain deprotected Lys amine was reacted with the doubly activated oxazine diacid 8 (oxazine 1 eq, PyBop 10 eq, DIPEA 20 eq) in DMF. The peptide was cleaved with TFA:H2O:TIS (triisopropylsilane) in a ratio of 95:2.5:2.5, and purified via HPLC.
Abl Sensor Peptide (11)
NH2-Gly-Gly-Ile-Tyr(tBu)-Ala-Ala-Pro-Phe-Lys(Boc)-Lys(Boc)-Lys(Boc)-Ala-amide-resin was synthesized via a standard Fmoc peptide synthesis protocol using a Prelude automatic peptide synthesizer from Protein Technology (amino acids 5 eq, HCTU 5 eq, DIPEA 10 eq). The free N-terminus was reacted with Cascade Yellow succinimidyl ester (2 eq) and DIPEA (4 eq) in DMF. The peptide was cleaved with TFA:H2O:TIS (triisopropylsilane) in a ratio of 95:2.5:2.5, and purified via HPLC.
Protein kinase assays
Assays were generally performed following a previously reported protocol (22) but with a total volume of 25 µl from the following stock solutions: 10 µl H2O, 2.5 µL 0.1 mM peptide stock solution, 2.5 µL 1M Tris (pH 7.5), 2.5 µL 200 mM MgCl2, 2.5 µL 0.1 mg mL−1 BSA, 0.5 µL 50 mM dithiothreitol (DTT), 2 µL 0.3 µM kinase and 2.5 µL 50 mM ATP. Final concentration of the assay solution was: 100 mM Tris (pH 7.5), 0.01 mg mL−1 BSA, 1 mM DTT, 24 nM kinase, 20 mM MgCl2, 5 mM ATP and 10 µM peptide sensor. Assays were initiated either by adding ATP or peptide and the fluorescence was monitored using a Gemini EM fluorescence microplate reader from Molecular Devices with 384-well microplates at 30°C. Peptide 9: (λex = 665 nm; λem = 705 nm); Peptide 11: (λex = 400 nm; λem = 525 nm). Dual monitoring of Abl and Lyn kinase activity was performed in an analogous fashion, but in the presence of both 9 and 11, and either Abl or Lyn alone or in combination. IC50 determinations were performed as described above with varying concentrations of imatinib or dasatinib. Cell lysate assays were likewise performed as described above using the following conditions: 100 mM Tris (pH 7.5), 5 mM ATP, 20 mM MgCl2, 1/100 (volume) protease inhibitor cocktail (P8340, Sigma-Aldrich), 1/20 phosphatase inhibitor cocktail 2 (P5726, Sigma-Aldrich), 10 µM peptide sensor. Protein concentration was determined using Bradford reagent (Thermo Scientific), and the concentration of each sample was equilibrated to 2.5 µg µL−1 by addition of lysis buffer.
Supplementary Material
Acknowledgements
We thank the National Institutes of Health (RO1-CA79954 and RO1-GM069976) for financial support.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
REFERENCES
- 1.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 2.Grant DM, Zhang W, McGhee EJ, Bunney TD, Talbot CB, Kumar S, Munro I, Dunsby C, Neil MA, Katan M, French PM. Multiplexed FRET to image multiple signaling events in live cells. Biophys J. 2008;95:L69–L71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niino Y, Hotta K, Oka K. Simultaneous live cell imaging using dual FRET sensors with a single excitation light. PLoS One. 2009;4:e6036. doi: 10.1371/journal.pone.0006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 6.Berrera M, Dodoni G, Monterisi S, Pertegato V, Zamparo I, Zaccolo M. A toolkit for real-time detection of cAMP: insights into compartmentalized signaling. Handb Exp Pharmacol. 2008:285–298. doi: 10.1007/978-3-540-72843-6_12. [DOI] [PubMed] [Google Scholar]
- 7.Ni Q, Titov DV, Zhang J. Analyzing protein kinase dynamics in living cells with FRET reporters. Methods. 2006;40:279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 9.Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 10.Sharma V, Lawrence DS. Uber-responsive peptide-based sensors of signaling proteins. Angew Chem Int Ed Engl. 2009;48:7290–7292. doi: 10.1002/anie.200903717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gora-Tybor J, Robak T. Targeted drugs in chronic myeloid leukemia. Curr Med Chem. 2008;15:3036–3051. doi: 10.2174/092986708786848578. [DOI] [PubMed] [Google Scholar]
- 13.Breccia M, Alimena G. Resistance to imatinib in chronic myeloid leukemia and therapeutic approaches to circumvent the problem. Cardiovasc Hematol Disord Drug Targets. 2009;9:21–28. doi: 10.2174/187152909787581363. [DOI] [PubMed] [Google Scholar]
- 14.Dai Y, Rahmani M, Corey SJ, Dent P, Grant S. A Bcr/Abl-independent, Lyn-dependent form of imatinib mesylate (STI-571) resistance is associated with altered expression of Bcl-2. J Biol Chem. 2004;279:34227–34239. doi: 10.1074/jbc.M402290200. [DOI] [PubMed] [Google Scholar]
- 15.Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, Talpaz M. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101:690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 16.Donato NJ, Wu JY, Stapley J, Lin H, Arlinghaus R, Aggarwal BB, Shishodia S, Albitar M, Hayes K, Kantarjian H, Talpaz M. Imatinib mesylate resistance through BCR-ABL independence in chronic myelogenous leukemia. Cancer Res. 2004;64:672–677. doi: 10.1158/0008-5472.can-03-1484. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Tanaka H, Kimura A. Establishment and characterization of a novel imatinib-sensitive chronic myeloid leukemia cell line MYL, and an imatinib-resistant subline MYL-R showing overexpression of Lyn. Eur J Haematol. 2007;78:417–431. doi: 10.1111/j.1600-0609.2007.00835.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG, Darnay BG, Lamothe B, Sun H, Talpaz M, Donato NJ. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst. 2008;100:926–939. doi: 10.1093/jnci/djn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee F, Fandi A, Voi M. Overcoming kinase resistance in chronic myeloid leukemia. Int J Biochem Cell Biol. 2008;40:334–343. doi: 10.1016/j.biocel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Dai Z, Dulyaninova NG, Kumar S, Bresnick AR, Lawrence DS. Visual snapshots of intracellular kinase activity at the onset of mitosis. Chem Biol. 2007;14:1254–1260. doi: 10.1016/j.chembiol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Dai Z, Cahill SM, Blumenstein M, Lawrence DS. Light-regulated sampling of protein tyrosine kinase activity. J Am Chem Soc. 2006;128:14016–14017. doi: 10.1021/ja065852z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Cahill SM, Blumenstein M, Lawrence DS. Self-reporting fluorescent substrates of protein tyrosine kinases. J Am Chem Soc. 2006;128:1808–1809. doi: 10.1021/ja0577692. [DOI] [PubMed] [Google Scholar]
- 23.Songyang Z, Carraway KL, 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 24.Wakata A, Cahill SM, Blumenstein M, Gunby RH, Jockusch S, Marti AA, Cimbro B, Gambacorti-Passerini C, Donella-Deana A, Pinna LA, Turro NJ, Lawrence DS. A mechanistic design principle for protein tyrosine kinase sensors: application to a validated cancer target. Org Lett. 2008;10:301–304. doi: 10.1021/ol702675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doose S, Neuweiler H, Sauer M. Fluorescence quenching by photoinduced electron transfer: a reporter for conformational dynamics of macromolecules. ChemPhysChem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 26.O'Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, Cowan-Jacob SW, Lee FY, Heinrich MC, Deininger MW, Druker BJ. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 27.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 28.Stanley E, Ralph S, McEwen S, Boulet I, Holtzman DA, Lock P, Dunn AR. Alternatively spliced murine lyn mRNAs encode distinct proteins. Mol Cell Biol. 1991;11:3399–3406. doi: 10.1128/mcb.11.7.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi TL, Bolen JB, Ihle JN. Hematopoietic cells express two forms of lyn kinase differing by 21 amino acids in the amino terminus. Mol Cell Biol. 1991;11:2391–2398. doi: 10.1128/mcb.11.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Brodeur SR, Gish G, Songyang Z, Cantley LC, Laudano AP, Pawson T. Regulation of c-Src tyrosine kinase activity by the Src SH2 domain. Oncogene. 1993;8:1119–1126. [PubMed] [Google Scholar]
- 31.Rosowsky A, Modest EJ. 2,2,4-Trimethyl-1,2-dihydroquinolines. Preparation and Nuclear Magnetic Resonance Studies. The Journal of Organic Chemistry. 1965;30:1832–1837. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.