Abstract
Several nucleolar proteins, such as ARF, ribosomal protein (RP) L5, L11, L23 and S7, have been shown to induce p53 activation by inhibiting MDM2 E3 ligase activity and consequently to trigger cell cycle arrest and/or apoptosis. Our recent study revealed another nucleolar protein called nucleostemin (NS), a nucleolar GTP binding protein, as a novel regulator of the p53-MDM2 feedback loop. However, unlike other known nucleolar regulators of this loop, NS surprisingly plays a dual role, as both up and downregulations of its levels could turn on p53 activity. Here, we try to offer some prospective views for this unusual phenomenon by reconciling previously and recently published studies in the field in hoping to better depict the role of NS in linking the p53 pathway with ribosomal biogenesis during cell growth and proliferation as well as to propose NS as another potential molecular target for anti-cancer drug development.
Key words: ribosomal biogenesis, nucleolar stress, nucleostemin, p53, MDM2, cell cycle, cell growth
Introduction
Ribosomal biogenesis is an essential and tightly regulated process for making the cellular machinery required for protein synthesis during cell growth and proliferation. It involves a series of complicated steps including the synthesis and processing of rRNA, the assembly of rRNA with ribosomal proteins to form ribosomal subunits, and a series of further modifications before finally forming the mature ribosome. Perturbation of any key step in the ribosomal biogenesis process can trigger ribosomal stress and interfere with normal growth and development in organisms.1 Inhibiting rRNA synthesis with Actinomycin D2–4 or 5-Fluoruracil,5,6 malfunction or inhibiting proteins involved in ribosomal biogenesis, such as dominant negative protein Bop1,7 and nucleophosmin,8 nutritional or metabolite deficiency including serum9 or GTP depletion,10 have been shown to inhibit ribosome assembly and trigger ribosomal stress. During ribosomal stress, free ribosomal proteins (RPs) that are normally assembled into ribosomal subunits and eventually into mature 40S and 60S ribosomes are released into the nucleoplasm. The release of these ribosome-free forms of RPs including RPL5,11 RPL11,12,32 RPL23,2,3 and S7,13,14 can directly bind to MDM2, the negative regulator and E3 ubiquitin ligase of p53, and interfere with the p53-MDM2 regulation loop. As a result, p53, a normally unstable protein due to MDM2-mediated ubiquitylation and degradation, can be stabilized and activated to stimulate transcription of various genes that encode proteins responsible for cell cycle arrest or apoptosis in response to ribosomal stress.15–17 Interestingly, the function of RPL26 appears to be different from other RPs in interfering in the p53-MDM2 interaction. Binding of RPL26 to the 5′ untranslated region of p53 mRNA was shown to promote the translation of p53 protein rather than inhibiting p53 proteasomal degradation.18 MDM2 itself can promote the polyubiquitination and proteasomal degradation of RPL26, thus serving as an auto-regulatory mechanism to turn off p53 translation.19 Furthermore, global inhibition of transcription by Flavopiridol and Actinomycin D leads to reduced levels of MDM2 and consequently activates p53.20–22 Clearly, the relationship between p53 and MDM2 holds a crucial role in regulating the cellular response to ribosomal stress. The crosstalk between the ribosomal biogenesis and the p53 signaling pathway reveals an important surveillance mechanism to halt cell growth or induce cell death when cells are growing under poor or abnormal ribosome synthesis conditions.1 Now, joining this intrinsic surveillance is a new player called nucleostemin (NS) from the nucleolar compartment of the cell.23
Essentiality of Balanced NS Levels for Cell Proliferation
NS is a nucleolar GTPase initially identified in the nucleolus of rat neural stem cells,24 and subsequently found upregulated in various types of proliferating stem cells and cancer cells.24–27 Knockout studies demonstrate that NS is essential for cell proliferation and embryogenesis.28,29 During stem cell differentiation both in vivo and in vitro, NS levels were rapidly reduced prior to exiting the cell cycle, suggesting the expression of NS is associated with the proliferation potential of cells. Consistent with this conjecture, siRNA knockdown of NS also reduced cellular proliferation and triggered cell cycle arrest. However, surprisingly the cell cycle was also arrested when NS was overexpressed in cultured cells. In both of the cases, p53 was identified to play a key role as detailed below.23,24,30 Hence, maintaining a fine balance of NS levels is essential for cellular homeostasis, and when NS levels become imbalanced, p53 can be activated to halt cell growth and proliferation.23
The Role of NS in p53 Response to Ribosomal Stress
It is surprising to learn that either high or low levels of NS can induce cell cycle arrest via p53 activation.23,24 How would this happen? Although it was previously reported that NS could directly interact with p53,24 the functional meaning of this interaction had been obscure until the binding of NS to the p53 repressor, MDM2, was revealed later on.23 Since several stress signaling pathways lead to p53 activation by interfering with the p53-MDM2 regulation loop,31 identification of the MDM2-binding activity of NS provides the possibility for this nucleolar protein to modulate p53 activity in a similar manner. Indeed, ectopic and endogenous NS through its coiled-coiled domain directly bound to the acidic domain of MDM2,23 similar to the cases of ARF and RPs.2–4,12–14,32–34 Overexpression of NS inhibited MDM2-mediated ubiquitylation of p53 and led to its activation, consequently inducing p53-dependent cell cycle arrest.23 These results demonstrate that NS could serve as a suppressor of MDM2 when overexpressed.
Then, how could the nucleolar NS interact with nucleoplasmic MDM2? Previous studies suggested that NS localization to the nucleolus is determined by the its GTP binding activity. Mutation of the conserved residues in the G1 GTP binding domain of NS caused this protein redistributed to the nucleoplasm.35 Interestingly, this mutant NS bound to MDM2 better than did the wild type NS, suggesting that NS might interact with MDM2 within the nucleoplasm. Consistent with this result is the evidence showing that MDM2 does not co-localize with NS in the nucleolus.23 Unlike endogenous NS, which is primarily localized to the nucleolus under a physiological condition, excess NS proteins after overexpressed were often dispersed into the nucleoplasm and thus able to find and bind to MDM2 in the nucleoplasm.23 In such a way, overexpressed NS can activate p53 by binding to MDM2 and attenuating its activity toward p53 in the nucleoplasm.
However, knocking down endogenous NS using siRNA also induced cell cycle arrest through p53.23,24 Although this observation seems controversial to the aforementioned p53 activation by excess NS molecules, it is not unreasonable that reduction of endogenous NS levels might cause some stress. As detailed in the following sections, NS was recently found to be a component of the nucleolar protein complex critical for rRNA processing.36 Depletion of NS levels would retard the rRNA processing and subsequently reduce the yield of mature rRNAs for ribosome assembly, causing ribosomal stress. As a result, extra free, unassembled or ribosome-free RPs would be left in the nucleolus and then released to the nucleoplasm. Similar to the ribosomal stress caused by the treatment of cells with Actinomycin D2–4 or mycophenolic acid,2,10 nucleoplasmic RPs in response to NS knockdown might also find MDM2 to interact with and inhibit its activity, leading to p53 activation. Indeed, this was the case, as depletion of NS by siRNA induced the interaction of RPL5 and RPL11 with MDM2.23 Consistent with this, knockdown of either RPL5 or RPL11 reversed, at least partially, p53-dependent cell growth arrest caused by depletion of NS.23 Of note, TP53 knockout could not completely rescue the embryonic lethality caused by deleting the ns gene, suggesting that NS possesses additional p53-independent functions,28 which might be essential for cell proliferation and embryogenesis as described below.
The role of NS in ribosomal biogenesis.
Part or major part of the p53-independent functions of NS must be associated with its nucleolar residency. While little is known about the exact physiological role of NS, examining the molecular function of cGTPases bearing structural similarities to NS could provide some insight into the possible function of this nucleolar protein in ribosomal biogenesis. NS displays some protein features in its amino acid sequence similar to the YawG sub-family of cGTPases, which are characterized by its permutated order of GTP binding motifs G4-G5-G1-G2-G3, instead of the order of G1-G2-G3-G4-G5 as observed in conventional GTPases, such as the well-known Ras-like family. Many of these circularly permutated GTPases consist of an RNA binding domain and several bacterial and yeast cGTPase-like motifs and possess the ability to interact with ribosomal components in a nucleotide dependent manner.37 Of the several studied members in the YawG GTPase family, including yeast and bacterial cGTPases Nug1, Nog2p and Lsg1, all have been linked to the processes in ribosomal assembly.38–40 Nucleolar GTPase Nog2 in yeast localized to the nucleolus and nucleoplasm has been shown to be essential for the late step of 27S RNA processing and export of the pre-60S ribosomal complex out from the nucleus.39 Nug1, a yeast orthologue of human NS, can bind to RNA through its N-terminal basic domain and associate with pre-60S ribosomal particles to facilitate the assembly of the 60S ribosomal subunit. Knockdown of Nug1 caused a significant reduction of the large ribosome unit as well as reduced proliferation.38 These studies suggest that GTPases with their dynamic shuttling ability may function as a molecular chaperone or scaffold to shuttle ribosome subunits and factors, thus facilitating the process of ribosome assembly. However despite the structural and sequence similarity between Nug1 and NS, there are still distinct differences between the yeast and human nucleolar GTPases. The N-terminal basic domain of Nug1 primarily determines the nucleolar localization of Nug1. Deletion of the middle domain containing the 2 GTP binding motifs Nug1 had little influence on the nucleolar localization in yeast, whereas the disruption of GTP binding motifs in human NS triggers relocation of this protein from the nucleolus to the nucleoplasm. Furthermore, expression of NS in yeast was unable to rescue the defects of the nug1 deletion strain in yeast proliferation,38 suggesting while the two proteins share high sequence homology, their functions may have diverged. The GTPase binding activity of NS may play a more complicated role in mammalians than would that of Nug1 in yeast. Hence, it is necessary to further explore the function(s) of NS in ribosomal biogenesis.
A recent study began to offer some clues as to the role of NS in ribosomal biogenesis by employing biochemical approaches. In this study,36 NS was co-purified with a large complex of ∼700 kda consisting of several ribosomal proteins, including RPS6, RPS8, RPS24, RPL13 and RPL14, pre-RNA processing proteins, including Pes1, DDX21 and EBP2, and a translation initiation factor EIF2B1α. Pes1, DDX21 and EBP2 have been implicated in the late steps of pre-RNA processing. DDX21 functions as RNA helicase and is involved in the processing of 20S pre-RNA into 18S rRNA in Xenopus41 and in the production of 28S and 18S rRNA in human.42 Pes1 is involved in the processing of 36S, 32S and 12S pre-RNA,43 while yeast EBP2 is involved the processing of 27SA to 27SB pre-RNA,44 suggesting a possible role of NS in pre-RNA processing as well. Indeed, when NS was knockdown with siRNA, global protein and rRNA synthesis was reduced as measured by metabolic labeling of rRNA and protein levels. Also, NS might play a role in retention of some rRNA processing factors in the nucleolus or serve as a scaffold or chaperone for ribosomal subunits and pre-RNA processing proteins during ribosomal biogenesis, as depletion of NS by siRNA caused relocation of DDX21 and EBP2 from the nucleolus to the nucleoplasm and cytoplasm.36 However, it still remains unclear how exactly NS facilitates this process. One possibility is that NS functions as a chaperone utilizing its GTP binding activity to shuttle RPs from the nucleoplasm into the nucleolus and to facilitate the proper assembly or arrangement of RPs with rRNA to form ribosome subunits. Alternatively, the NS GTP hydrolysis activity may provide energy for the conformation or structural modifications required for the formation of stable protein-protein or protein-RNA interactions in the 700 kda large complex. In the future, it would be interesting to determine whether and how the ablation of the GTP binding activity of NS would influence the rate of ribosome and protein synthesis as well as its ability to retain or interact with pre-RNA processing and RPs in the nucleolus. While the newly discovered p53-independent function of NS helps clarify the role of NS in regulating cellular proliferation and cell cycle progression through association with ribosome biogenesis, further studies will be needed to address the remaining questions concerning the detailed mechanism(s) NS utilizes in facilitating pre-RNA processing and ribosome assembly and how its GTP hydrolysis activity may fit in the big picture. In addition, unraveling the regulation of NS would also shed light on the mechanism(s) underlying its nucleolar role in ribosomal biogenesis.
Regulation of NS.
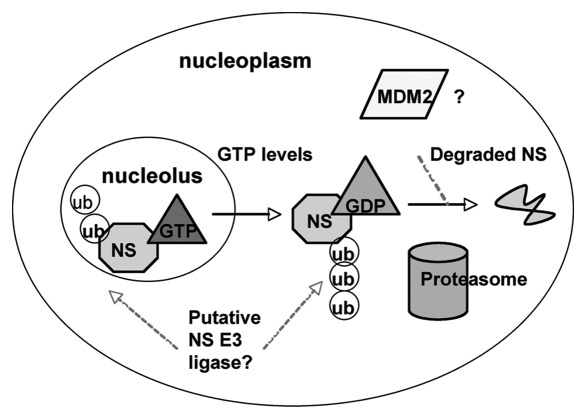
Although it has been demonstrated that the aberrant levels of NS due to depletion or overexpression result in cell cycle arrest through p53,23,24,30 physiological signals and mechanisms underlying the fine balance of NS to maintain cellular homeostasis or to regulate stem cell differentiation are completely unknown. Recent studies by determining which nuclear compartment NS resides in suggest that NS can be regulated by fluctuation in the intracellular levels of GTP.45 It was demonstrated that while the N-terminal basic domain of NS is sufficient to direct NS to its pre-dominant localization to the nucleolus, the intrinsic GTPase activity of NS is also necessary for NS retention in the nucleolus. Point mutations in or deletions of the GTP binding motifs disrupt nucleolar localization and re-distribute NS to the nucleoplasm.35 Furthermore, depleting intracellular GTP pools by inhibiting the rate-limiting enzyme for de nova guanine nucleotide synthesis, inosine monophosphate dehydrogenase, results in a reduction of NS protein levels independently of transcription of mRNA45 (our unpublished observations). Thus, the re-localization of NS in response to fluctuation in GTP levels may serve as a critical factor for regulating NS activity through post-translational modifications or NS stability by dictating NS protein interaction in the nucleolar or nucleoplasmic compartment (Fig. 1).
Figure 1.
Model to illustrate how GTP levels may regulate NS.
Indeed, our recent study showed that the nucleoplasmic form of NS with mutations in its GTP binding domain can bind to the acidic domain of MDM2 and inhibit its activity toward p53 degradation more efficiently than that of endogenous or wild type NS.23 The binding of NS to the E3 ubiquitin ligase MDM2 raises the question of whether NS itself can be a potential substrate for this E3 ligase. Despite the well-characterized role of MDM2 in mediating ubiquitin dependent proteasomal degradation of p53,15–17 other substrates for the MDM2 E3 ligase have been identified as well, such as Tip60,46 glucorticoid receptor,47 beta 2-adrenergic recptor,48 and MDMX.49 These findings indicate the versatile role of MDM2 as an E3 ligase. Intriguingly, a recent study suggested a potential role of MDM2 in regulating the levels of NS.45 Overexpression of MDM2 in cell culture reduced both exogenous and endogenous levels of NS. Moreover, knockdown of MDM2 with siRNA or inhibition of MDM2 with the drug Nutlin-3 was able to partially rescue GTP induced depletion of NS. Also, treatment of cell with a proteasome inhibitor MG132 was able to rescue GTP depletion-induced reduction of NS protein levels, suggesting that NS is indeed degraded by the proteasome.45 However, there is no direct evidence demonstrating the MDM2 can polyubiquitylate NS. Furthermore, the partial rescue of NS reduction due to depletion of GTP by knocking down MDM2 suggests that there might exist another crucial regulator of NS stability in cells. It is also possible that the influence of MDM2 on NS stability may occur through a mechanism independent of its E3 ligase activity. While the majority of protein degradation occurs through an ubiquitin-dependent mechanism, a subset of proteins has been shown to be degraded by the proteasome in an ubiquitin-independent fashion. For example, p21, a highly unstable protein that regulates the cell cycle progression, can be degraded by MDM2 independently of ubiquitylation and the status of E1.50,51 Thus, how NS stability and activity are regulated in response to GTP depletion or other pathological or physiological signals remains to be an outstanding question for future studies. Elucidation of this mechanism would certainly offer some hints for our better understanding the physiological role of NS in the nucleolus and in cell proliferation.
NS as a possible sensor of nucleolar stress.
The process of ribosomal biogenesis is highly coordinated with cell growth and proliferation. Cellular and environmental stress leading to compromised conditions for ribosome assembly often leads to nucleolar stress and the consequent release of nucleolar and ribosomal proteins into the nucleoplasm. While it has been established that the release of free forms of RPs into the nucleoplasm can lead to the inhibition of MDM2 and activation of p53,1 the mechanism for the release of these RPs remains unknown. Given that GTPases from the YawG family are highly involved in the export and import of ribosome subunits and in the process of ribosome assembly,37,39,40 it is likely that NS may play a role in mediating ribosomal stress leading to activation of p53. As discussed above, GTP depletion can reduce NS levels as well as an overall decrease in protein and ribosome synthesis, releasing unassembled RPs to the nucleoplasm to inhibit MDM2 activity and thus induce p53 activation.10,23,45 Is it possible that reduction of NS levels might serve as a trigger for the release of RPs from the nucleolus to the nucleoplasm if NS plays a role in retaining RPs in the nucleolus for ribosome assembly. Addressing this question would help us better understand why depletion of GTP by MPA induces the interaction of RPL11 and RPL5 with MDM2.10 Also, it is unclear whether other stress signals may influence the stability of NS and thus trigger ribosomal stress. For instance, treatment of cells with either an RNA pol I inhibitor Actinomycin D or Doxorubicin induces nucleolar stress and causes the release of RPs and other nucleolar proteins, such as NS, into the nucleoplasm, activating p53.45 However, it remains to be investigated whether the release of NS into the nucleoplasm itself serves as a signal or sensor of the nucleolar stress or is just a destination for its degradation. A large scale of screening stress signals or reagents that might cause fluctuation of NS would be conducive to clarification of the role of NS as a possible sensor or indicator of the nucleolar or ribosomal stress that leads to p53 activation and consequently attenuates the cell cycle.
Potential dual roles of NS in tumorigenesis.
Timely and proper production of ribosome subunits is critical to cell growth and proliferation, whereas aberrant ribosomal biogenesis could compromise both quality and quantity of protein production, leading to ribosomal stress and retarding cell growth and embryo development. Clearly, NS plays an essential role in warranting normal ribosomal biogenesis as discussed above. Further supporting this notion is a recent study showing that depletion of NS1, a Drosophila orthologue of human NS, markedly reduces pre-RNA processing, ribosome biogenesis and protein synthesis, leading to lethality in the majority of Drosophila progeny.52 However, among the few flies that survived to adulthood, the majority of them exhibited variable abnormal morphological growth patterns, such as malpositioned antennal growth, missing bristles, missing ocelli and even malignant tumor growth.52 These phenotypes not only emphasize the importance of NS in cell growth and animal development, but also suggest that NS might play a tumor suppression role. The latter is in line with the ability of NS to activate p53 via suppression of MDM2 function as discussed above.23 Even though Drosophila does not have MDM2, NS might turn on fly p53 by counteracting a p53 suppressor in response to the nucleolar stress that is possibly caused by knockdown of GTPBP4, a newly indentified nucleolar protein that interacts with p53 in the fly.53 Because of the essential function of NS in rRNA processing and ribosome assembly, which are essential for cell proliferation,36 this nucleolar protein must be critical for tumor cell growth and proliferation. From this aspect, NS may play an oncogenic role in favoring cell transformation and neoplasia. This is probably why some human cancers harbor high levels of NS25,54,55 (our own unpublished data). Hence, NS could have a dual role in tumorigenesis probably depending upon microenvironments within a cell or specific genetic and epigenetic circumstances. Although there has not yet been any mutation identified in the mammalian ns gene, it would not be surprising if there might exist cancer-associated mutations or single nucleotide polymorphisms (SNPs) that could change the amino acids within the domain of NS critical for MDM2 binding, consequently surrendering its ability to activate the p53 pathway. Thus, it would be interesting to screen genetic mutations or SNPs of NS, which are associated with cancer pathogenesis.
Ending marks.
In summary, it is clear that the balance of NS levels is critical for the maintenance of cellular homeostasis. NS plays a dual role in ribosomal biogenesis and the p53 response when this essential process in the nucleolus is disturbed. This unique characteristic of NS makes this nucleolar protein as an attractive molecular target for developing anti-cancer therapy. Reducing NS level or inhibiting its activity would cause ribosomal stress that can initiate the activation of p53,23 leading to cell growth arrest and apoptosis. Hence, targeting NS would offer a way for eradicating growing cancer cells that harbor wild type p53 by not only activating the tumor suppression function of p53, but also cutting off the ribosome supplies for protein production necessary for active growing cancer cells. Also, NS is often found in malignant cancer cells25,54,56 and overexpression of NS in cancer cells is associated with increased cellular proliferation and possibly tumorigenic potential through its role in pre-RNA processing and ribosomal biogenesis.36 Therefore, even though for many of those cancer cells at their late stage often with mutated or lost p53, targeting NS or repressing its activity can attenuate ribosomal biogenesis and consequently prevent the growth of cancer cells.23 Regardless of the status of p53, inhibition of NS function could offer another good opportunity for developing therapeutic strategies against all types of human cancers.
In addition to the potential of using NS as an anti-cancer target, the identification of NS as a crucial player in pre-RNA processing and ribosomal biogenesis also provides much insight into the physiological role of NS.36 However, this also raises more questions regarding the upstream signaling pathways regulating NS and how NS modulates rRNA processing and ribosome assembly exactly. For instance, how does NS GTPase activity regulate NS protein levels or activity in pre-RNA processing and ribosomal biogenesis? What are other proteins that might be involved in mediating this regulation? Addressing these questions by more systematically studying the role and upstream regulators of NS in ribosomal biogenesis at molecular and biochemical levels would not only shed light on our fully understanding this mysterious biogenesis process in the tiny nucleolar structure, but also uncover more potential protein molecules for developing anti-cancer drugs in the future.
Acknowledgements
This work was supported in part by NIH-NCI grants CA127724, CA095441 and CA129828 to H.L.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12605
References
- 1.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:15–17. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 5.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 6.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 9.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun XX, MS D, Lu H. Mycophenolic Acid Activation of p53 Requires Ribosomal Proteins L5 and L11. J Biol Chem. 2008;283:12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daigle DM, Brown ED. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J Bacteriol. 2004;186:1381–1387. doi: 10.1128/JB.186.5.1381-1387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, et al. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem. 2006;281:24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 16.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 17.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 18.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 19.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An WG, Chuman Y, Fojo T, Blagosklonny MV. Inhibitors of transcription, proteasome inhibitors and DNA-damaging drugs differentially affect feedback of p53 degradation. Exp Cell Res. 1998;244:54–60. doi: 10.1006/excr.1998.4193. [DOI] [PubMed] [Google Scholar]
- 21.Blagosklonny MV, Demidenko ZN, Fojo T. Inhibition of transcription results in accumulation of Wt p53 followed by delayed outburst of p53-inducible proteins: p53 as a sensor of transcriptional integrity. Cell Cycle. 2002;1:67–74. [PubMed] [Google Scholar]
- 22.Demidenko ZN, Blagosklonny MV. Flavopiridol induces p53 via initial inhibition of Mdm2 and p21 and, independently of p53, sensitizes apoptosis-reluctant cells to tumor necrosis factor. Cancer Res. 2004;64:3653–3660. doi: 10.1158/0008-5472.CAN-04-0204. [DOI] [PubMed] [Google Scholar]
- 23.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Y, Liu Z, Zhao S, Lou F, Nilsson S, Ekman P, et al. Nucleostemin mRNA is expressed in both normal and malignant renal tissues. Br J Cancer. 2006;94:1658–1662. doi: 10.1038/sj.bjc.6603145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kafienah W, Mistry S, Williams C, Hollander AP. Nucleostemin is a marker of proliferating stromal stem cells in adult human bone marrow. Stem Cells. 2006;24:1113–1120. doi: 10.1634/stemcells.2005-0416. [DOI] [PubMed] [Google Scholar]
- 27.Liu SJ, Cai ZW, Liu YJ, Dong MY, Sun LQ, Hu GF, et al. Role of nucleostemin in growth regulation of gastric cancer, liver cancer and other malignancies. World J Gastroenterol. 2004;10:1246–1249. doi: 10.3748/wjg.v10.i9.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beekman C, Nichane M, De Clercq S, Maetens M, Floss T, Wurst W, et al. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol Cell Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohrum MA, et al. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 35.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S, et al. Critical role of nucleostemin in pre-rRNA processing. J Biol Chem. 2009;284:4968–4977. doi: 10.1074/jbc.M804594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60S pre-ribosomal particles. J Biol Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- 39.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, et al. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, et al. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 2005;3:21. doi: 10.1186/1741-7007-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Zhou J, Ochs RL, Henning D, Jin R, Valdez BC. Downregulation of RNA helicase II/Gu results in the depletion of 18 and 28 S rRNAs in Xenopus oocyte. J Biol Chem. 2003;278:38847–38859. doi: 10.1074/jbc.M302258200. [DOI] [PubMed] [Google Scholar]
- 42.Holmstrom TH, Mialon A, Kallio M, Nymalm Y, Mannermaa L, Holm T, et al. c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J Biol Chem. 2008;283:7046–7053. doi: 10.1074/jbc.M709613200. [DOI] [PubMed] [Google Scholar]
- 43.Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Wade C, Shea KA, Jensen RV, McAlear MA. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol Cell Biol. 2001;21:8638–8650. doi: 10.1128/MCB.21.24.8638-8650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang M, Itahana K, Zhang Y, Mitchell BS. Depletion of guanine nucleotides leads to the Mdm2-dependent proteasomal degradation of nucleostemin. Cancer Res. 2009;69:3004–3012. doi: 10.1158/0008-5472.CAN-08-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002;21:1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinyamu HK, Archer TK. Estrogen receptor-dependent proteasomal degradation of the glucocorticoid receptor is coupled to an increase in mdm2 protein expression. Mol Cell Biol. 2003;23:5867–5881. doi: 10.1128/MCB.23.16.5867-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 49.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Wang H, et al. MDM2 Is a Negative Regulator of p21WAF1/CIP1, Independent of p53. J Biol Chem. 2004;279:16000–16006. doi: 10.1074/jbc.M312264200. [DOI] [PubMed] [Google Scholar]
- 51.Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosby R, Cui Z, Rogers E, deLivron MA, Robinson VL, DiMario PJ. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth and midgut precursor cell maintenance. Mol Biol Cell. 2009;20:4424–4434. doi: 10.1091/mbc.E08-06-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lunardi A, Di Minin G, Provero P, Dal Ferro M, Carotti M, Del Sal G, et al. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc Natl Acad Sci USA. 107:6322–6327. doi: 10.1073/pnas.1002447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye F, Zhou C, Cheng Q, Shen J, Chen H. Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cada Z, Boucek J, Dvorankova B, Chovanec M, Plzak J, Kodets R, et al. Nucleostemin expression in squamous cell carcinoma of the head and neck. Anticancer Res. 2007;27:3279–3284. [PubMed] [Google Scholar]
- 56.Tamase A, Muraguchi T, Naka K, Tanaka S, Kinoshita M, Hoshii T, et al. Identification of tumor-initiating cells in a highly aggressive brain tumor using promoter activity of nucleostemin. Proc Natl Acad Sci USA. 2009;106:17163–17168. doi: 10.1073/pnas.0905016106. [DOI] [PMC free article] [PubMed] [Google Scholar]