Abstract
Protein Arginine Deiminase (PAD) activity is dysregulated in numerous diseases, e.g., Rheumatoid Arthritis. Herein we describe the development of a fluorescence polarization-Activity Based Protein Profiling (fluopol-ABPP) based high throughput screening assay that can be used to identify PAD-selective inhibitors. Using this assay, streptonigrin was identified as a potent, selective, and irreversible PAD4 inactivator.
Dysregulated Protein Arginine Deiminase (PAD) activity contributes to a number of human pathologies, including Rheumatoid Arthritis (RA), colitis, and cancer, thus highlighting the importance of designing inhibitors for these enzymes.1 In humans, there are five PAD isozymes (PADs 1-4 and 6) that catalyze the posttranslational conversion of peptidyl-arginine to peptidyl-citrulline in numerous protein substrates.2 While these enzymes are highly related at the amino acid level, they do exhibit different tissue expression patterns and, to a lesser extent, substrate specificities.1, 3 Additionally, their individual contributions to human disease are unknown, although PADs 2 and 4 are strongly associated with these pathologies.1b
We recently described the design and synthesis of two highly potent PAD inhibitors, i.e., F- and Cl-amidine,4 which have been important chemical tools in discerning the physiological role of PAD4.4b, c, 5 These compounds are mechanism-based inactivators that covalently modify an active site Cys in PAD4 that is critical for catalysis.6 However, these compounds are pan PAD inhibitors because Cl-amidine, and to a lesser extent F-amidine, inhibit all of the active PAD isozymes with similar potency.3 Thus, the identification of PAD-selective inhibitors remains of paramount importance; such compounds will allow us to discern the individual contributions of PAD isozymes to both normal human physiology and disease.
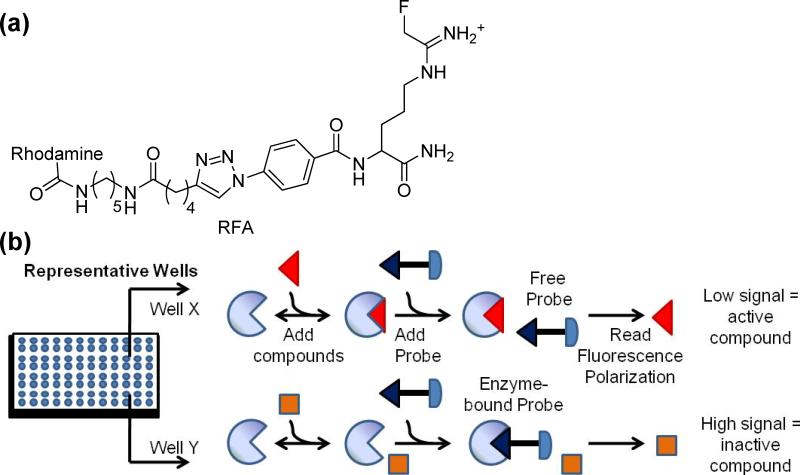
To identify PAD-selective inhibitors, we set out to develop an efficient high-throughput screening (HTS) assay. Previously, we described a gel-based assay that relies on competition between compounds from a library and a PAD specific Activity Based Protein Profiling (ABPP) reagent termed rhodamine-conjugated F-amidine (RFA).7 RFA is composed of a mechanism-based PAD inhibitor (i.e., F-amidine) and a fluorophore (Fig. 1).8 This gel-based assay can visibly track the enzyme's activity and overcomes many of the limitations of preceding PAD assays (i.e., the use of toxic compounds, high temperatures, and strong acids). Although useful for screening small compound libraries, this assay requires 1-D SDS-PAGE gels and is consequently not compatible with large compound libraries (i.e., >1000).
Fig 1.
(a) Structure of Rhodamine-conjugated Fluoro-amidine (RFA). (b) The fluopol-ABPP assay. The compound is either an inhibitor of the enzyme (top) or is inactive (bottom).
Based on our recent work with ABPP probes targeting serine hydrolases,9 we surmised that our gel-based assay could be converted into an HTS-amenable format that monitors the changes in fluorescence polarization (fluopol) that occur when RFA binds to PAD4. As fluorescent molecules are excited with plane polarized light, the RFA-PAD4 complex, will rotate slowly and therefore emit highly polarized light. In contrast, free RFA rotates more quickly and emits depolarized light. If an inhibitor is bound to PAD4, it will prevent RFA from binding, resulting in a lower fluopol signal (Fig. 1).
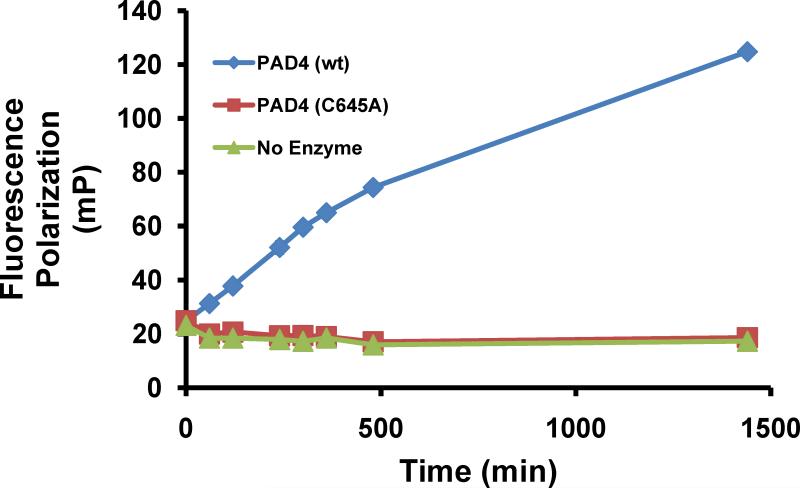
To develop this HTS assay, titrations of the ABPP probe RFA and PAD4 were initially performed to identify the conditions needed for a strong, time-dependent increase in fluopol (Fig. 2). The fluopol of the reaction between RFA and either wtPAD4, the PAD4C645A mutant (a catalytically deficient PAD4 mutant in which the active site Cys is replaced by an Ala), or no enzyme, was measured over a range of time (0-1440 min) to identify the proper assay conditions that would yield an adequate Z' factor (> 0.5). After 300 min of incubation under the assay conditions (i.e., 2 μM PAD4, 75 nM RFA), we identified a robust Z' value of 0.73. These kinetically controlled conditions allow for the identification of both reversible and irreversible PAD4 inhibitors. As expected, no labeling of the mutant PAD4 enzyme was observed, and gratifyingly the fluopol values were comparable to the wells without enzyme. Therefore, we used no enzyme as a positive control.
Fig. 2.
RFA reacts with PAD4 in a time dependent manner. A strong, time-dependent increase in fluopol was observed at 300 min.
Using these assay conditions, we screened 2,000 compounds (5 μM final) from the NIH Validation Set at The Scripps Research Institute in La Jolla, CA (Pubchem AID 463073). To determine the high and low boundaries for FP signals, we included negative control reactions (no compounds) and positive control reactions (without PAD4) on each plate, respectively.
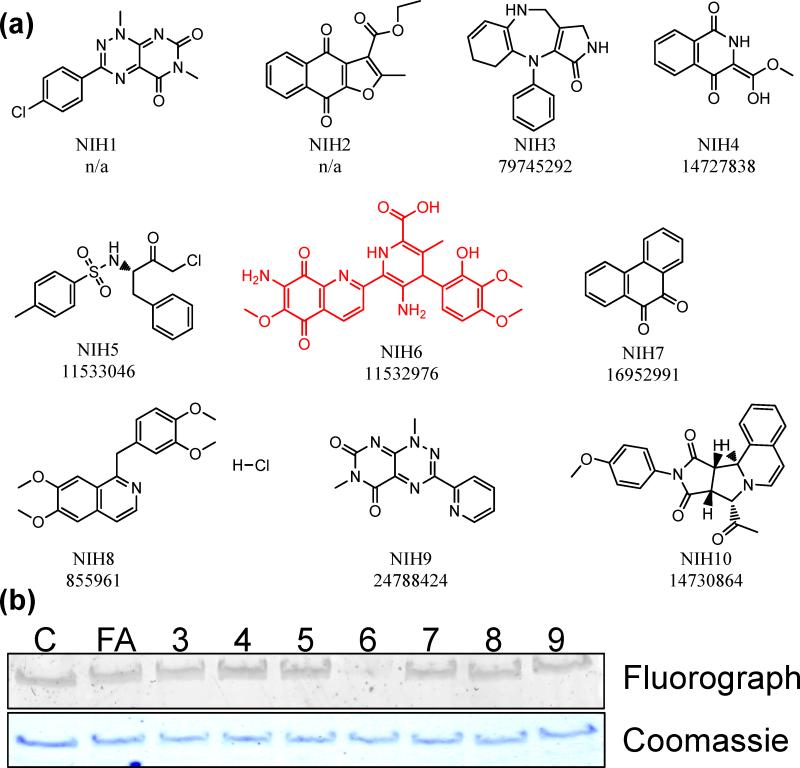
This HTS identified 10 compounds that demonstrated greater than 30% inhibition of PAD4 (Table 1; Fig. 3a and S1). Unfortunately, three of these are no longer commercially available, i.e., NIH1, 2, and 10. Of the remainder, their ability to inhibit PAD4 was validated using the gel-based ABPP assay described above (Fig. 3b and Table 1). It is noteworthy that one of the compounds identified in the screen was tosyl phenylalanyl chloromethyl ketone (TPCK; NIH5), which is an inhibitor of cysteine proteases. Cysteine proteases, like the PADs, possess a Cys-His dyad in their active site that is critical for catalysis and this compound likely inactivates PAD4 via a similar mechanism. However, this compound proved to be one of the weakest inhibitors in the secondary gel-based ABBP-based screen.
Table 1.
The ‘hits’ identified by the Fluopol-ABPP HTS
| Hit ID | % Inhibition (library screen) | % Inhibition (Gel based ABPP) | IC50 for PAD4 (μM) |
|---|---|---|---|
| NIH1 | 90.7 | n/da | n/da |
| NIH2 | 61.9 | n/da | n/da |
| NIH3 | 58.3 | 64.4 | ≥ 100 |
| NIH4 | 66.5 | 13.3 | 42 ± 13 |
| NIH5 | 39.7 | 12.9 | 13 ± 2 |
| NIH6 | 92.5 | 93.9 | 2.5 ± 0.4 |
| NIH7 | 94.4 | 22.3 | 16 ± 4.0 |
| NIH8 | 74.4 | 12.2 | 39 ± 11 |
| NIH9 | 98.2 | 22.2 | 21 ± 5.0 |
| NIH10 | 37.2 | n/da | n/da |
n/d = not determined because the compound was not available.
Fig. 3.
Structures of ‘hits’ and gel-based secondary screen. (a) The numbers below the structure are fluoPol-ABPP primary hit number (top) and NIH validation collection compound number (bottom). n/a = not available. (b) Inhibition of RFA labeling of PAD4 by the primary ‘hits’. Lane 1 is a DMSO control (C), lane 2 is a positive control, i.e. F-amidine (FA), and lanes 3-9 are compounds NIH3-NIH9 (10 μM final).
In contrast, the largest decrease in fluorescence intensity was observed with the antitumor antibiotic streptonigrin (compound NIH6; NIH #11532976), thus quickly focusing our efforts on characterizing the inhibitory properties of this compound. Streptonigrin is a metabolite produced by Streptomyces flocculus, and belongs to a group of aminoquinone-containing antitumor agents that inhibit DNA replication, interfere with cellular respiration, and disrupt cell replication.10 The antitumor activity of this compound has been described for numerous types of cancers, including breast, melanoma, lung, and lymphoma.11
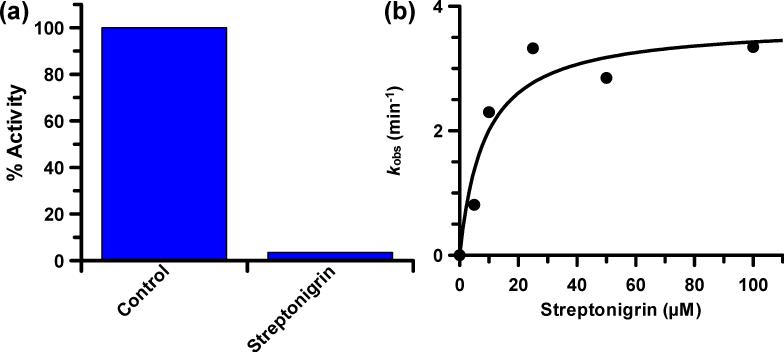
Further examination of the inhibitory properties of streptonigrin demonstrated that this compound exhibits an IC50 value of 1.87 ± 0.24 μM for PAD4. This led us to determine the mechanism by which streptonigrin inhibits PAD4. For these experiments, streptonigrin was incubated with PAD4 for 30 min, after which the reaction was dialyzed overnight (~20 h) and residual PAD4 activity was then measured. The fact that no PAD4 activity was recovered indicates that streptonigrin is an irreversible PAD4 inhibitor (Fig. 4a). Notably, substrate could protect against inactivation (Fig. S2), thereby indicating that inactivation is due to the preferential modification of an active site residue in PAD4, most likely, Cys645.
Fig. 4.
Streptonigrin is an irreversible PAD4 inactivator. (a) The preformed PAD4•streptonigrin complex was dialyzed for 20 h and the % activity remaining determined. (b) Plot of the pseudo-first order rate constants of inactivation, i.e. kobs, versus streptonigrin concentration.
Given that MS experiments could not identify the specific residue modified by this compound, it is difficult to speculate on the specific mechanism of inactivation. Nonetheless, streptonigrin is known to generate reactive oxygen species in the presence of Zn2+ or Cu2+, as well as a suitable reductant (e.g., NADH);10a thus, an oxidative mechanism is possible. However, NADH is not present in the assay buffer and both metal ions are at trace levels in our assays.2 Furthermore, little to no inter-isozyme selectivity would be expected for the oxidative inactivation of PAD4. Thus, inactivation most likely proceeds via nucleophilic attack on the quininoid ring system of streptonigrin; the two carbons ortho to the carbonyls are potential Michael acceptors. Collapse of the intermediate, and subsequent protonation of the ring, would undoubtedly lead to the formation of a stable adduct.
To further characterize the mechanism of inhibition, we next examined the time and concentration dependence of streptonigrin-induced PAD4 inactivation. The results of the experiments are consistent with a two-step mechanism of inactivation (Fig. 4b). The kinact and KI values for PAD4 inhibition are 3.7 ± 0.5 min-1 and 8.5 ± 4.3 μM, respectively, and yield a kinact/KI value of 4.4 × 105 M-1 min-1, which is more than 33-fold higher than Cl-amidine, a previously described PAD inhibitor that had been established as the most potent PAD inactivator to date.
We then examined inter-isozyme selectivity trends (Table 2). Based on the IC50 values for PADs 1, 2, and 3 (Table 2) these data suggest that streptonigrin inhibits PADs 3 and 4 over PADs 1 and 2 with >10-fold selectivity. However, IC50 values are dependent on the substrate concentration and the relative affinity of an enzyme for that substrate. Therefore, we determined kinact/KI values to more accurately assess the selectivity of this compound (Table 2). The kinact/KI values for PADs 1, 2, and 3 are 3.7 × 103 M-1 min-1, 12 × 103 M-1 min-1, and 3.5 × 103 M-1 min-1, respectively (Table 2). These values are at least 35-fold lower than the value observed for PAD4; this data indicates that streptonigrin is a PAD4-selective inhibitor. The discrepancy between the IC50 value obtained for PAD3 and the kinact/KI is attributable to the fact that BAEE is a very poor PAD3 substrate.3
Table 2.
Inactivation of PADs 1, 2, 3, and 4a
| PAD1 | PAD2 | PAD3 | PAD4c | |
|---|---|---|---|---|
| IC50 (μM) | 48.3 ± 34.2 | 26.1 ± 0.3 | 0.43 ± 0.03 | 1.87 ± 0.24 |
| KI (μM) | 11 ± 7.0 | 17 ± 5.8 | 49 ± 34 | 8.5 ± 4.3 |
| kinact (min-1) | 0.04 ± 0.004 | 0.2 ± 0.02 | 0.2 ± 0.1 | 3.7 ± 0.5 |
| kinact/KI (min-1M-1) | 3.7 × 103 | 12 × 103 | 3.5 × 103 | 4.4 × 105 |
| Fold selectivityb | 119 | 37 | 126 | 1 |
Kinetic parameters were determined by incubating enzyme with varying concentrations of streptonigrin at 37 °C for various periods of time. Then residual PAD activity was measured with BAEE.
Fold selectivity is based on the relative values of kinact/KI.
Given that the PADs are highly homologous, we reasoned that one or more of the compounds identified in the screen might also preferentially inhibit a different PAD isozyme. Therefore, we determined whether NIH3 to NIH9 could inhibit PADs 1, 2, and 3 (Fig. S3). For these studies, we used our standard PAD substrate assay to determine the percent inhibition at a single fixed concentration of inhibitor. Interestingly, most of these compounds are relatively weak inhibitors, and some even actually activated these PAD isozymes by ~2-fold. However, as this effect was relatively small, we felt that further investigations of this phenomenon were unwarranted. Note that in this assay streptonigrin showed a trend similar to the IC50 values stated above for PADs 1, 2, and 3, thereby validating the results of this method to examine inter-isozyme selectivity trends.
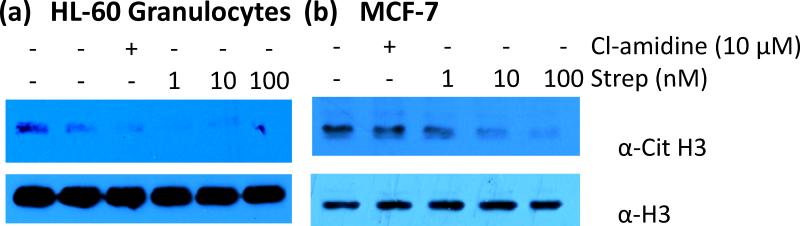
Finally, the ability of streptonigrin to inhibit in cellulo PAD4 activity was evaluated by determining whether streptonigrin could decrease histone H3 citrullination in MCF-7 and HL-60 granulocytes. PAD4 is known to deiminate histone H3 in these cell lines in response to estrogen and LPS, respectively.12 Western blots using an anti-citrulline histone H3 antibody showed that as little as 1 nM of streptonigrin reduced the levels of citrullinated histone H3 in both HL-60 granulocytes and MCF7 cells in a dose dependent manner (Figure 5). As a control, the levels of citrullinated histone H3 were measured in the presence of Cl-amidine (10 μM final). These data indicate that streptonigrin inhibits PAD4 activity in cellulo. Relative to Cl-amidine, the high in cellulo potency likely reflects the fact that streptonigrin (i) is 33-fold more potent than Cl-amidine; and (ii) likely has greater bioavailability due to the fact Cl-amidine is positively charged and highly hydrophilic, which would be expected to limit its cell permeability.
Fig. 5.
Bioavailability of streptonigrin in (a) HL-60 granulocytes and (b) MCF-7 cells. Cell lines were treated with either Cl-amidine (10 μM) or streptonigrin (1, 10, or 100 nM) and then probed with anti-citrulline H3 antibody (top). The blot was stripped and re-probed with anti-H3 antibody to show equal loading of protein (bottom).
In summary, we have developed a fluopol-ABPP HTS assay to identify PAD inhibitors. This screen quickly identified 10 PAD inhibitors from a library of 2,000 compounds. This list was reduced to only one compound by using a number of secondary screens, including our previously described gel-based ABPP assay.7a These studies also demonstrated that streptonigrin is a potent and selective PAD4 inhibitor. Given the known antitumor activity of this compound, it is interesting to speculate that the pharmacological effects of streptonigrin are due, at least in part, to its ability to inhibit PAD4 activity. This seems plausible when one considers the fact that PAD4 is overexpressed in numerous tumor types13 and we have shown that PAD4 inhibition triggers apoptosis of several different cancer cell lines.5a However, streptonigrin is unlikely to represent either a useful PAD4 probe or a therapeutic, as this compound is known to exhibit pleitropic effects on a number of cellular processes, including DNA replication, cellular respiration, and cellular replication. Additionally, this compound possesses a number of less than desirable side effects, including nausea, diarrhea, and hair loss. Further, bone marrow depression and high toxicity has limited its use as a cancer therapy.14 Nonetheless, these studies demonstrate the power of fluopol-ABPP to identify PAD-selective inhibitors, the discovery of which provides valuable information (i.e., novel chemical scaffolds) for the design of more potent PAD4 inhibitors. In the future, we plan to utilize this assay to screen PAD4 against larger compound libraries, and to adapt this assay to identify inhibitors of PADs 1, 2, and 3.
Funding was provided in part by the University Of South Carolina, TSRI, Scripps Florida, and by National Institutes of Health grant GM079357 to P.R.T.
Supplementary Material
Footnotes
This article is part of the ‘Enzyme and Proteins’ web-theme issue for ChemComm.
Electronic Supplementary Information (ESI) available: Detailed experimental procedures and results. See DOI: 10.1039/b000000x/
Notes and references
- 1.a Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]; b Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Kearney PL, Bhatia M, Jones NG, Luo Y, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 3.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Chembiochem. 2010;11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. J Am Chem Soc. 2006;128:1092–1093. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Mol Cell Biol. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. J Biol Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knuckley B, Bhatia M, Thompson PR. Biochemistry. 2007;46:6578–6587. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Knuckley B, Luo Y, Thompson PR. Bioorg Med Chem. 2008;16:739–745. doi: 10.1016/j.bmc.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 8.Luo Y, Knuckley B, Bhatia M, Thompson PR. J Am Chem Soc. 2006;128:14468–14469. doi: 10.1021/ja0656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Bolzan AD, Bianchi MS. Mutat Res. 2001;488:25–37. doi: 10.1016/s1383-5742(00)00062-4. [DOI] [PubMed] [Google Scholar]; b Knipp M, Vasak M. Anal. Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 11.a Oleson JJ, Calderella LA, Mjos KJ, Reith AR, Thie RS, Toplin I. Antibiot Chemother. 1961;11:158–164. [PubMed] [Google Scholar]; b Teller MN, Wagshul SF, Woolley GW. Antibiot Chemother. 1961;11:165–173. [PubMed] [Google Scholar]; c Reilly HC, Sugiura K. Antibiot Chemother. 1961;11:174–177. [PubMed] [Google Scholar]; d Beall HD, Mulcahy RT, Siegel D, Traver RD, Gibson NW, Ross D. Cancer Res. 1994;54:3196–3201. [PubMed] [Google Scholar]; e Beall HD, Murphy AM, Siegel D, Hargreaves RH, Butler J, Ross D. Mol Pharmacol. 1995;48:499–504. [PubMed] [Google Scholar]
- 12.a Neeli I, Khan SN, Radic M. J Immunol. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]; b Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson WL, Labra C, Barrist E. Antibiot Chemother. 1961;11:147–150. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.