Abstract
Flap endonuclease 1 (FEN1), a structure-specific endo- and exo- nuclease, exhibits multiple functions that determine essential biological processes, such as cell proliferation and cell death. As such, the enzyme must be precisely regulated in order to execute each of its functions with the right timing and in a specific subcellular location. Here, we report that FEN1 is methylated at arginine residues, primarily at R192. The methylation suppresses FEN1 phosphorylation at S187. The methylated form, but not the phosphorylated form of FEN1, strongly interacts with Proliferating Cell Nuclear Antigen (PCNA), ensuring the on and off timing of its reaction. Mutations of FEN1 disrupting arginine methylation and PCNA interaction result in unscheduled phosphorylation and cause failure of its localization to DNA replication or repair foci. This consequently leads to a defect in Okazaki fragment maturation, a delay of cell cycle progression, impairment of DNA repair, and high frequency of genome-wide mutations.
Structure specific flap endonuclease 1 (FEN1) is a multi-functional enzyme participating in DNA replication, DNA repair and apoptotic DNA fragmentation. Three mechanisms have been proposed to regulate how FEN1 executes its functions in DNA replication versus DNA repair 1: i) subcellular compartmentalization, ii) protein-protein interaction, and iii) post-translational modifications (PTMs). FEN1 is localized to the nucleus during the S phase of the cell cycle or in response to DNA damage 2,3. Recently, we demonstrated that FEN1 accumulates in the nucleoli and migrates to the nuclear plasma upon UV irradiation and phosphorylation 4. In addition to its nuclear localization, FEN1 also resides in the mitochondria and cooperates with DNA2 nuclease to process DNA intermediate structures during mitochondrial DNA replication and repair 5-7.
Protein-protein interaction plays a critical role in guiding FEN1 to regulate different biochemical pathways 1,8. To date, FEN1 has been reported to interact with at least 30 proteins 1. We have recently mapped the amino acid residues interacting with these proteins 9. FEN1 forms distinct protein complexes for DNA replication and repair. By interacting with PCNA, FEN1 is recruited to the replication foci for RNA primer removal and repair sites for DNA base excision repair 10. PCNA also stimulates flap endonuclease and exonuclease activities of FEN1 robustly for cleavage of RNA primers 8,11. Recently, the FEN1/PCNA interaction has been implicated in coordinating the sequential action of polymerase δ (Pol δ), FEN1, and DNA ligase 1 (Lig1) during Okazaki fragment maturation 12,13. Disruption of FEN1/PCNA interaction impairs Okazaki fragment ligation12. On the other hand, in response to stalled replication forks, the Werner syndrome protein (WRN) forms a complex with FEN1 and activates its gap-dependent endonuclease activity to initiate break-induced recombination 14. In addition to its roles in DNA replication and repair, during the process of apoptosis, FEN1 interacts with Endo G nuclease to degrade genomic DNA 15.
Posttranslational modifications (PTMs) are important in regulating the activity of FEN1. The protein is acetylated at its C-terminus by p300 histone acetylase in cellular response to UV irradiation 16. Cdk2/Cyclin E can phosphorylate FEN1 at Ser187 in late S phase of the cell cycle 17. Phosphorylation results in dissociation of FEN1 from PCNA. In addition to acetylation and phosphorylation, little is known about whether FEN1 undergoes other forms of PTMs and what are the functional impacts of these PTMs. The discovery of protein arginine methyltransferases (PRMTs) has lead to protein methylation coming out of the shadow of other forms of PTMs 18-20. In humans, PRMTs represent a family of several enzymes that utilize S-adenosyl methionine (SAM) as a methyl donor19. PRMTs are classified into type I or type II enzymes. Type I PRMTs (PRMT1, PRMT3, PRMT4, and PRMT6) produce asymmetrically di-methylated arginine (ADMA), whereas type II PRMTs (PRMT5 and PRMT7) catalyze the formation of symmetrically di-methylated arginine (SDMA)19. Arginine methylation has been shown to occur in proteins involved in signaling transduction 21, DNA damage response 22-24, and DNA repair 25-28. The diversity of reported substrates suggests that methylation may parallel other PTMs in levels of complexity for functional regulation of specific proteins. However, the mechanisms by which arginine methylation regulates protein functions remain largely unknown. Here, we report the discovery of FEN1 methylation at arginine residues which suppresses the nearby phosphorylation and facilities PCNA binding. We demonstrate that the cross-talk between methylation and phosphorylation plays important roles in regulation of FEN1 functions in DNA replication and repair.
Results
FEN1 methylation at the cellular level
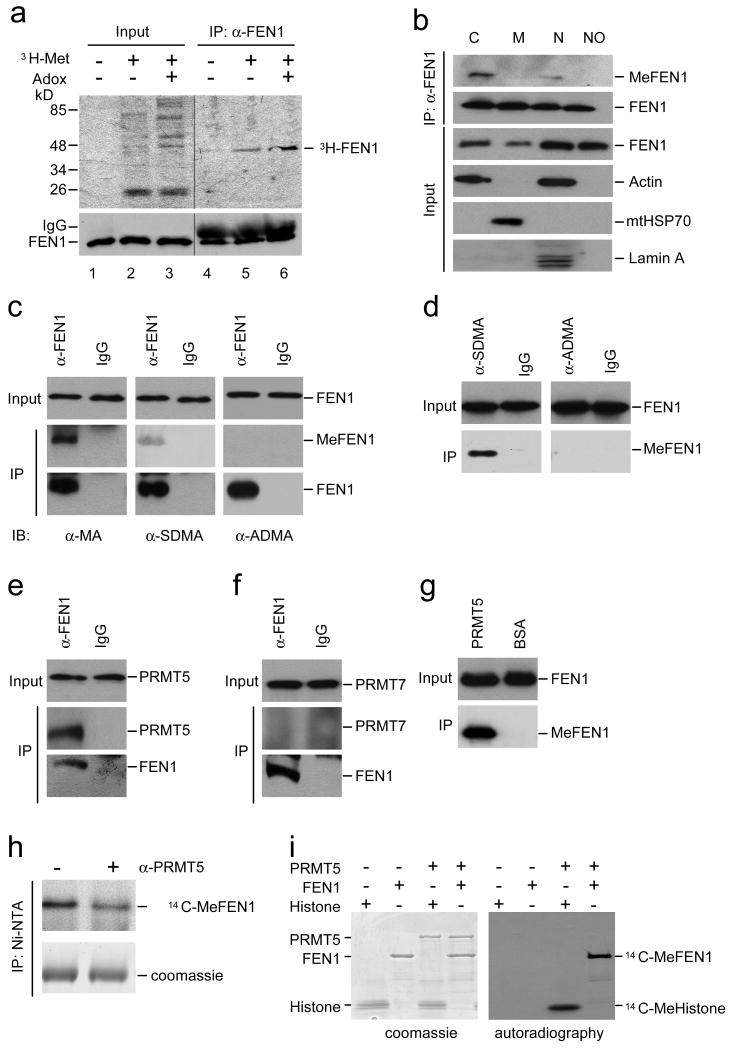
In cells, arginine methyltransferases catalyze methylation of substrate proteins using methionine as a methyl donor29. To address whether or not FEN1 protein can be methylated, HeLa cells were subjected to methyl group donor L-[methyl-3H] methionine (3H-Met) 29. 3H-labeled FEN1 was detected in the protein complex pulled down with an anti-FEN1 antibody (Fig. 1a). Pre-treating cells with Adox, which accumulates the levels of methyl-acceptable proteins 30, caused an increase in 3H-labeled FEN1 (Fig. 1a). Since FEN1 has been found in four different subcellular compartments i.e. the cytoplasm, mitochondria, nucleus and nucleoli (Fig. 1b and Ref 2,4,5), we determined the methylation status of FEN1 with regard to its subcellular localization. Although also localized in the nuclei, methylated FEN1 was mainly detected in the cytoplasm.
Figure 1. In vivo and in vitro methylation of FEN1 by PRMT5.
(a) In vivo methylation of FEN1. HeLa cells were metabolically labeled as described in Methods. The gel was loaded with 1% of the whole-cell lysate (Input) or the immunoprecipitate obtained with a FEN1-specific antibody (IP) (see Supplementary Methods for details). (b) Subcellular distribution of methylated FEN1. C: cytoplasm; M: mitochondria; N: nucleus; NO: nucleoli. (c) Immunoprecipitate from HeLa cell extract by FEN1-specific antibody was detected using arginine methylation specific antibodies. (d) Immunoprecipitate from HeLa cell extract by symmetrical and asymmetrical di-methylated arginine antibody was detected by using FEN1 antibody. (e-f) FEN1 interacts with PRMT5 in HeLa cell extract. Immunoprecipitate from HeLa cell extract by FEN1-specific antibody was detected using (e) PRMT5 or (f) PRMT7 antibodies. (g) PRMT5 interacts with FEN1 in vitro. PRMT5 or BSA was coated on CNBr-Sepharose beads then incubated with purified FEN1. Beads-bound FEN1 was detected by Western Blotting. (h) In vitro methylation assay by using HeLa cell extract as the enzyme resource. HeLa cell extract (with or without pre-depletion by anti-PRMT5 antibody) was incubated with His-tagged FEN1 and S-adenosyl-l-(methyl-14C) methionine (14C-SAM). His-tagged FEN1 was then purified by Ni-NTA beads, followed by SDS-PAGE and autoradiography. (i) In vitro methylation by PRMT5. Proteins were resolved by SDS-PAGE, stained with Coomassie blue (top panel), dried, and analyzed by autofluorography (bottom panel). For the full image, see Supplementary Figure 8.
We determined if FEN1 was indeed methylated at arginine and whether the modification was mono- or di-methylation. Immunoprecipitated FEN1 from HeLa cells was recognized by antibodies against mono-methylated arginine (MA) and symmetrical di-methyl arginine (SDMA) but not asymmetrical di-methyl arginine (ASDMA) (Fig. 1c). This result was confirmed by reciprocal immunoprecipitation using antibodies against SDMA and ASDMA, and immunoblotting with an anti-FEN1 antibody (Fig. 1d). These data suggest that FEN1 indeed contains SDMAs.
SDMA is the product of type II PRMTs, including two enzymes, PRMT5 and PRMT7. Utilizing a co-immunoprecipitation assay with stringent washing conditions (900 mM NaCl and 1% NP-40), we investigated whether FEN1 interacts with PRMT5 or PRMT7. Considerable amounts of PRMT5, but not PRMT7, were detected in the complex with FEN1 (Fig. 1e,f). The interaction between FEN1 and PRMT5 was confirmed by an in vitro pull-down assay using purified recombinant FEN1 and PRMT5 (Fig. 1g). To demonstrate FEN1 is a substrate of PRMT5, an in vitro methylation assay was performed using S-adenosyl-l-(methyl-14C) methionine (14C-SAM) as the methyl group donor and the crude extract of HeLa cells as the enzyme resource. When PRMT5 was pre-depleted from the extract by anti-PRMT5 antibodies, methylation of FEN1 was reduced dramatically, compared with untreated HeLa extract (Fig. 1h). Pre-depletion of PRMT7 did not affect FEN1 methylation under the same assay conditions, suggesting the specificity of PRMT5 in methylating FEN1. The ability of PRMT5 to methylate FEN1 was further confirmed by an in vitro methylation reaction where the purified recombinant FEN1 was incubated with PRMT5 recombinant protein (Fig. 1i). Quantification of the methylation efficiency revealed that more than 88% of FEN1 molecules were methylated in an overnight reaction in the presence of a 20-fold molar excess of 14 C-SAM and PRMT5, assuming that only one methylation reaction occurs in each FEN1 molecule.
Identification and validation of methylation sites
FEN1 protein contains 22 arginine residues. To identify which arginine residue is methylated, FEN1 was in vitro methylated by PRMT5 and subjected to ESI-LC-MS/MS analysis. GluC digestion lead to the detection of Arg19, Arg100, Arg104 and Arg192 as methylation sites (Fig. 2a and Supplementary Fig.1). A series of FEN1 mutants were made, each carrying a single mutation at R19, R100, R104 or R192, or combined mutations i.e. 3RK (R19K/R100K/R104K) or 4RK(Arg19K/Arg100K/Arg104K/Arg192K). When assessed for methylation by PRMT5, single mutation at any of these four residues resulted in defects in methylation (Fig. 2b). Mutation at R192 showed the greatest reduction in FEN1 methylation levels, compared to the remaining residues individually or in combination (3RK), suggesting that R192 is the major FEN1 methylation site (Fig. 2b). As expected, quadruple mutant 4RK displayed a similar degree of reduction of arginine methylation by PRMT5 as the R192 mutant (Fig. 2b,c). To demonstrate that these arginine residues are the main methylation sites in vivo, a c-myc tagged FEN1 4RK mutant was expressed in HeLa cells. Consistent with in vitro methylation data, the methylation of c-myc tagged FEN1 4RK was dramatically reduced at the cellular level, compared to the wild type (WT) control (Fig. 2d).
Figure 2. Identification and validation of FEN1 methylation sites.
(a) Mass spectrometry profile of FEN1. The sequence of FEN1 covering residue 180–198 is shown. Analysis by LC-MS/MS revealed the presence of a modified MDCLTFGSPVLMRHLTAS peptide containing methylated Arg 192. The mass spectrometry profiles used to identify the other three methylation sites, R19, R100, R104 are shown in Supplementary Figure 1. (b) Loss of the methylation sites individually or in combination results in failure of the PRMT5 methylation of FEN1. FEN1 proteins (WT and mutants) were in vitro methylated using 3H-SAM as the methyl donor, separated on SDS-PAGE, and stained by Coomassie blue (top panel). FEN1 bands were cut off for liquid scintillation assay (bottom panel). (c) In vitro methylation of the wild-type and mutant FEN1 by PRMT5, using 14C-SAM as the methyl donor. Proteins were resolved using SDS-PAGE gel, stained with Coomassie blue (top panel), and analyzed by autoradiography (bottom panel). (d) C-myc tagged WT or FEN1 mutants harboring mutations at the methylation sites of four arginine residues in combination were transfected into HeLa cells. Immunoprecipitate of HeLa cell lysate by anti-C-myc antibody was immunodetected by α-MA antibody. The total loading amount was examined by anti-FEN1 antibody. For the full image, see Supplementary Figure 8.
Methylation of FEN1 inhibits its phosphorylation
To investigate the functional role of arginine methylation of FEN1, we tested whether such PTMs had any impact on substrate binding, flap nuclease activity or the ability to interact with PCNA. No obvious effects were observed relative to FEN1 binding to DNA substrate, flap nuclease activity or the ability to interact with PCNA in vitro (Supplementary Fig. 2). We therefore considered an indirect effect via interplay with other PTMs, by examining sequential methylation and phosphorylation with purified recombinant FEN1 (Fig. 3a). Non-methylated FEN1 was readily phosphorylated by Cdk2/Cyclin E, but methylated FEN1 was resistant to phosphorylation (Fig. 3a). We excluded the possibility that PRMT5 inactivates Cdk2/Cyclin E by incubating histones with Cdk2/Cyclin E in the absence or presence of PRMT5, which showed no significant effects of PRMT5 on Cdk2/Cyclin E activity (Supplementary Fig. 3). In contrast to the findings that methylation of FEN1 inhibits its phosphorylation, FEN1 phosphorylation did not affect its methylation (Supplementary Fig. 4). To demonstrate that methylation prohibits FEN1 phosphorylation in vivo, R19, R100, R104 or R192 residues were replaced with phenylalanine to mimic arginine methylation for transfection into HeLa cells 31,32. R192F and 4RF but none of the other methylation-mimicking mutants suppressed FEN1 phosphorylation in vitro (Fig. 3b), supporting the idea that R192 methylation suppresses FEN1 protein phosphorylation.
Figure 3. Methylation of FEN1 at R192 inhibits its phosphorylation at S187.
(a) In vitro sequential methylation and phosphorylation assays. FEN1 was in vitro methylated with unlabeled SAM, followed by phosphorylation with Cdk2/Cyclin E and 32P-ATP. Reaction products were analyzed by SDS-PAGE (lower panel) and autoradiography (upper panel). (b) Methylation-mimicking mutations (arginine to phenylalanine) inhibit FEN1 phosphorylation. In vitro phosphorylation was performed using 32P-ATP. Labeled FEN1 proteins were separated on SDS-PAGE (upper panel). The protein bands were cut off from the gel for liquid scintillation quantification (middle panel). Autoradiography of labeled FEN1 proteins separated on a second SDS-PAGE (bottom panel). (c) Knockdown of PRMT5 in HeLa cells decreases FEN1 methylation while enhancing FEN1 phosphorylation. HeLa cells transfected either with control siRNA or siPRMT5 were lysed then immunoprecipitated with anti-FEN1 antibodies, followed by immunoblotting with anti-PhosSer or anti-SDMA antibodies. Whole-cell extracts were analyzed with anti-FEN1 and anti-PRMT5 antibodies. Semi-quantification of phosphorylated or methylated FEN1 is shown (right). (d) R192F blocks FEN1 phosphorylation in cells. HeLa cells with exogenous expression of WT or mutant FEN1 were lysed, followed by immunoprecipitation with anti-c-myc antibody. Immunoprecipitate was detected by anti-FEN1 and anti-PhosSer antibodies. For the full image, see Supplementary Figure 9.
PRMT5 methylation of FEN1, resulting in the suppression of FEN1 phosphorylation, was further validated in vivo using siRNA to knock down PRMT5. Reduction of PRMT5 protein level resulted in a clear decrease in arginine-methylated FEN1 and a concomitant increase in phosphorylated FEN1 (Fig. 3c). When methylation-mimicking mutant R192F or methylation-deficient mutant R192K was expressed in HeLa cells, R192F failed to be phosphorylated, whereas R192K was hyper-phosphorylated at both G1 and S cell cycle phases (Fig. 3d). Taken together, our data suggest that R192 methylation of FEN1 suppresses its phosphorylation.
Methylation of FEN1 facilitates its interaction with PCNA
Phosphorylation of FEN1 has been shown to abolish its PCNA interaction 17. FEN1 does not bind to PCNA upon phosphorylation in vitro (Fig. 4a). By methylation, FEN1 becomes resistant to phosphorylation and keeps its affinity for PCNA (Fig. 4a). At the cellular level, methylation-mimic mutation R192F or non-methylated FEN1 displayed similar affinity with PCNA (Fig. 4a and Supplementary Fig. 2). While phospho-mimic mutant S187D failed to interact with PCNA, FEN1 methylation could not restore the PCNA binding of S187D (Supplementary Fig. 5). On the other hand, methylation-mimic mutant R192F, which could not be phosphorylated by Cdk2/Cyclin E, strongly interacted with PCNA. Similar to the S187A mutant, R192F FEN1 constitutively bound to PCNA (Fig. 4b). In contrast, methylation-defective mutant R192K was readily phosphorylated and failed to bind to PCNA (Fig. 4b). 4RK FEN1 is non-methylated but highly phosphorylated in the cell S phase. The interaction between FEN1 and PCNA was also interrupted (Fig. 4b). These data suggested that arginine methylation preserves the FEN1 interaction with PCNA by inhibiting FEN1 phosphorylation.
Figure 4. Methylation of FEN1 ensures its interaction with PCNA via suppression of its phosphorylation.
(a) Methylated FEN1 is resistant to phosphorylation and forms a complex with PCNA. Methylated or non-methylated FEN1 were incubated with Cdk2/Cyclin E followed by pull-down with PCNA-beads. Bound proteins were analyzed by anti-FEN1 or anti-PCNA antibodies. (b) In vitro phosphorylation and PCNA binding assay of WT and FEN1 mutants. WT or mutant FEN1 proteins, with or without phosphorylation, were subjected to PCNA beads. Phosphorylation of FEN1 was analyzed by autoradiography and the pulled-down proteins were immunodetected by anti-FEN1 and anti-PCNA antibodies. (c) Co-localization of FEN-1 and PCNA in S-phase cells. Cells were fixed and co-stained with anti-FEN-1 and anti-PCNA antibodies. (d) Co-localization of FEN-1 and BrdU in cells in S phase. Cells were labeled (1 h) with BrdU, followed by fixation and co-staining with anti-FEN-1 and anti-BrdU antibodies. Scale bars, 10 μm. For the full image, see Supplementary Figure 10.
At the S phase of the cell cycle, FEN1 is recruited to DNA replication loci via interaction with PCNA. Disruption of the FEN1/PCNA interaction impairs such localization 12. If methylation is important for FEN1 to remain under-phosphorylated and, therefore, interact with PCNA, failure in methylation should lead to a defect in localization of FEN1 to the replication foci. WT FEN1 co-localized with PCNA and BrdU positive signal, a marker of DNA replication foci, while phospho-mimic mutant S187D failed to form discrete foci with PCNA and BrdU (Fig.4c,d). The methylation-defective mutant R192K was unable to localize to the foci, but methylation mimic mutant R192F co-localized with PCNA and BrdU (Fig. 4c,d). Similar to R192K, the 4RK FEN1 mutant failed to co-localize to the replication site. These data suggested that FEN1 methylation and phosphorylation were important for regulation of its subnuclear localization and that methylation and phosphorylation are at two opposite sides of an equilibrium regulating FEN1 localization to replication foci.
FEN1 methylation enhances DNA replication and repair
As described above, 4RK mutant FEN1 is non-methylated, highly phosphorylated, does not interact with PCNA and fails to localize to DNA replication foci (Fig. 5a,b), which is important for Okazaki fragment maturation12. Okazaki fragment maturation assay was performed using a dNTP mixture containing radio-labeled dCTP and a model substrate containing RNA-DNA flap mimicking the Okazaki fragment maturation intermediate. The assay simulates the sequential reactions of gap filling, RNA primer removal and DNA ligation during the process of Okazaki fragment maturation. When the assay was performed in vitro, the nuclear extracts of 4RK transfected cells showed a clear decrease in the efficiency to remove RNA primer flaps and were defective in DNA ligation (Fig. 5c). As a result, the un-ligated product was accumulated in 4RK cells (Fig. 5c). To exclude the possibility that the 80 nt product observed in Figure 5c resulted from the synthesis to the end of the 80 nt template by strand displacement, we performed ligation assay, in which the DNA-RNA flap was labeled at the 3′-end and the assay was similar to what was done in Figure 5c, except for the use of a cold dNTP mixture (Fig. 5d). As a result, the 80 nt would only be able to be observed if the extended upstream primer was ligated with the 5′-phospho group-containing downstream labeled oligonucleotides. The 5′-phospho group would result from the removal of RNA-DNA flap. If DNA synthesis utilized upstream oligos as a primer, and without ligation, the product would not be visible on the gel. The observed 80 nt product on Figure 5d indicates the involvement of ligation in this reaction.
Figure 5. Methylation defect retards Okazaki fragment maturation, DNA replication and induces DNA damage.
(a) Detection and quantification of exogenous FEN1 (Cymc-tagged) in stable cell lines. (b) Cmyc-FEN1 was immunoprecipitated from HeLa cell extracts, followed by detection with indicated antibodies. (c and d) Okazaki fragment maturation assay. A gap substrate with a 20 nt RNA-DNA flap (with or without a label at the 3′end, top) was incubated with indicated amounts of cell lysates, followed by autoradiography (middle panel) and quantification (bottom). Panel (c) and (d) are similar except unlabeled substrate and [α-32P]dCTP were used in (c) while 3′-end labeled substrate and cold dCTP were used in (d). (e) Double strand DNA breaks (DSBs) were detected with immunoassays using an antibody against γH2AX. Nuclei were stained with DAPI. Pink spots were scored as positive nuclei with DNA strand breaks (left panel). Right panel, quantification of γH2AX positive stained nuclei. Scale bar, 40 μm. (f) Thymidine incorporation assay. For full image, see Supplementary Figure 11.
Previous reports have indicated that defects in the Okazaki fragment maturation process during DNA replication or ligation during DNA repair could lead to accumulation of DNA double strand breaks33,34. Therefore, 4RK cells are expected to display a higher level of DNA damage. To verify this, WT and 4RK cells were cultured to determine the levels of the phosphorylated form of H2AX histone variant (γH2AX), an early marker of the cellular response to DNA breaks. In the absence of any exogenous source of DNA damage, basal levels of phosphorylated γH2AX in 4RK cells were higher than that in WT (Fig. 5e), indicating the accumulation of DNA double strand breaks due to 4RK expression. The lower efficiency of Okazaki fragment maturation and the higher DNA damage may affect DNA replication of 4RK cells. We evaluated the DNA replication rate in WT and 4RK cells using a 3H-thymidine incorporation assay. The data show that 3H-thymidine incorporation in 4RK cells was approximately 50% of that in WT cells (Fig. 5f).
Cell cycle progression was measured to demonstrate that inability of FEN1 methylation is sufficient to cause spontaneous DNA damage and cell cycle arrest. Our data showed that 4RK cells have a slower cell cycle progression rate than WT cells (Supplementary Fig. 6a). When cells were arrested at G1/S border by double thymidine treatment 35 and released for 20 h, WT cells progressed from the first G1 phase to the next G1 phase, while 4RK cells were distributed in G2/M phase (Supplementary Fig. 6a). It took more than 30 h for 4RK cells to finish one cell cycle. In addition to the slow cell cycle progression, the majority of 4RK cells were arrested at the prophase of mitosis, while WT cells were evenly distributed in pro-, meta-, and ana- phase of mitosis (Supplementary Fig. 6b,c). 4RK FEN1 mutant cells also displayed a low overall proliferation rate (Supplementary Fig. 6d). These data suggest that defects in Okazaki fragment maturation in the S-phase result in prophase mitotic arrest in 4RK cells.
Previous studies indicate that protein arginine methyltransferases are involved in cellular responses to oxidative stresses36. We determined whether oxidants alter FEN1 methylation status. Indeed, methylation of FEN1 was induced by H2O2 in HeLa cells (Fig. 6a). H2O2 also facilitated the translocation of FEN1 from the cytoplasm to the nucleus where DNA damage occurs (Fig. 6b). These data suggest a possible role of FEN1 methylation in DNA repair. Long-patch base excision repair (LP-BER) was measured using a 40 nt DNA substrate containing an abasic tetrahydrofuran (THF) residue, an analogue of LP-BER substrate, and radio-labeled dCTP in a dNTP mixture (Fig. 6c). Again, successful repair would require three sequential steps: (1) polymerization and displacement of THF-contained oligonucleotide to form a flap; (2) flap removal and (3) ligation of up- and down-stream oligos. We found both WT and 4RK cell nuclear extracts could generate ligation products of the expected size (40 nt). However, the rate of ligation of 4RK nuclear extracts was approximately 70% lower than WT (Fig. 6c). To confirm that ligase activity was indeed involved in this reaction, we employed an alternative assay using a substrate with 3′-end label on the THF-containing oligo and with a nick at the 5′ to the THF site (Fig 6d). The 40 nt product shown in Fig. 6d was only possible when THF was removed, the gap was filled and upstream and downstream oligos were ligated. Our observation that 4RK caused defective LP-BER led us to propose that cells expressing the 4RK variant would be sensitive to DNA base damaging agents. To test this, 4RK and WT HeLa cells were treated with H2O2 at various concentrations for determining the survival rate (Fig.6e). Consistent with the biochemical data, 4RK cells were more sensitive to H2O2 treatment (Fig. 6e). The HPRT assay also demonstrated that 4RK cells displayed an approximate 32-fold greater spontaneous mutation rate than that of the WT HeLa cells (Fig. 6f and Supplementary Table 1).
Figure 6. Methylation of FEN1 is induced by H2O2 and important for DNA repair.
H2O2 treatment induces FEN1 methylation (a) and drives FEN1 to the nucleus from the cytoplasm (b). (c-d) LP-BER assay using cell extracts. Tetrahydrofuran-containing substrates (THF, top panel, circle), were incubated with indicated amounts of cell extract, followed by autoradiography (middle panel) and quantification (bottom panel). Unlabeled substrate and hot dCTP were used in (c) while nicked substrate with 3′-end label on the TFH-containing oligo and cold dNTP mixture were used in (d). (e) 4RK cells are sensitive to H2O2 stress. HeLa cells (WT or 4RK) were treated (1 h) at indicated concentrations and cellular sensitivity was determined by growth inhibition experiments; mean ± s.d., n=4. (f) Spontaneous mutation frequency was evaluated with an Hprt mutant assay. (g) Model for FEN1 dynamic interaction with PCNA and DNA substrates regulated by FEN1 methylation and phosphorylation. Upon formation of flap structure in Okazaki fragment mutation or LP-BER, methylated FEN1 is recruited to the replication fork by interacting with PCNA, following the dissociation of Pol δ/β. Methylation of FEN1 ensures its interaction with and stimulation by PCNA to remove the flap structure. After DNA flap cleavage, FEN1 undergoes de-methylation and subsequent phosphorylation by cell cycle-dependent kinases, leading to disruption of the FEN1/PCNA interaction and dissociation of the nuclease from DNA substrates. Lig I is then recruited by interaction with PCNA and seals the nicks between two DNA fragments. For the full image, see Supplementary Figure 12.
Discussion
Here we report that methylation serves as a primary event intersecting with other regulatory circuitry, such as protein-protein interactions and subcellular localization. We identified arginine methylation as a novel PTM of FEN1 and demonstrated its roles in regulation of FEN1 activities in relationship to DNA replication and repair. FEN1 can be methylated in vitro and in vivo with R192 as the major methylation site. Methylation of FEN1 at R192 precludes its phosphorylation at S187, ensuring its interaction with PCNA and hence recruitment to the DNA replication or repair foci. Methylation-defective mutants display impaired DNA replication and repair efficiency, resulting in accumulation of spontaneous double strand DNA breaks, retarded cell cycle and higher sensitivity to DNA damage agents. Our data provide strong evidence for the regulation of DNA replication and repair by methylation events that indirectly control other posttranslational events, in this case phosphorylation.
Methylation at the arginine residue serves as a mechanism for functional regulation of non-histone proteins. To date, approximately 130 non-histone proteins have been reported containing methylated arginine (Supplementary Table 2). Several proteins involved in DNA replication and repair have been examined for arginine-methylation as a switch for “loss-of-function” or “gain-of-function” 37,38. Among these are Mre11, 53BP1 and Polβ 22,23,27,28,39. In the case of Polβ, there are two methylation events at two different arginine residues mediated by PRMT6 and PRMT1. PRMT1-derived methylation enhance polymerase activity due to exaggerated DNA binding and processivity25. It is worth noting that methylation of R137 of Polβ by PRMT1 impairs its interaction with PCNA 26. This is contrary to FEN1, in which methylation prevents phosphorylation but ensures PCNA binding. This opposite effect leads to the postulation that methylation may allow Polβ and FEN1 to sequentially access PCNA and the DNA substrate during LP-BER.
It has long been accepted that the dynamic interaction between FEN1 and PCNA is critical for faithful and efficient DNA flap removal during Okazaki fragment maturation and LP-BER12,40. Our findings suggest a molecular model to describe how FEN1 binds to and dissociates from PCNA, thus dynamically accessing and departing from DNA substrates (Fig. 6g). In early S phase, FEN1 methylation at the R192 residue prevents FEN1 phosphorylation at the S187 residue and ensures the FEN1/PCNA interaction, which allows the nuclease to replace the polymerase (Polδ or Polβ) and access flap DNA substrates. When flap cleavage is done and FEN1 is de-methylated, possibly by an unknown demethylase. The non-methylated FEN1 is then phosphorylated by the Cdk/cyclin complex, forcing FEN1 to dissociate from PCNA and the DNA substrate. This step is critical in that it avoids constant binding of FEN1 to PCNA and DNA substrates and blocking Ligase I action. Supporting this model, we observed a decrease in methylated FEN1 (Supplementary Fig. 7) and an increase in phosphorylated FEN1 in later S phase (Fig.3d and Supplementary Fig.7). The phenomenon of dynamic methylation and demethylation has also been reported in the estrogen receptor α (ERα), in which the methylation level increases significantly after 5 min of exposure to estrogen 2 (E2), then decreases rapidly at 15 min, but with little change observed in overall protein levels 41. Although these data suggest the existence of enzymes that reverse arginine methylation, it is unclear how the methyl groups in FEN1, ERα, and other proteins are removed. Several studies have described the process of de-amination 42,43, by which the methyl group from an arginine is removed, converting monomethylated arginine into citrulline. Recently, JMJD6 has been reported to remove methyl groups from methylated arginine residues of histones, however it has not been determined whether its activity is limited to histones or it is also able to demethylate nonhistone proteins 44. The mechanism by which Fen1 is demethylated remains to be investigated.
Posttranslational modifications may play a critical role in the specificity and timing of FEN1-protein interaction. In addition to methylation and phosphorylation, acetylation has been reported in FEN1 protein. The physiological significance of FEN1 acetylation is not yet understood. Unlike phosphorylation, acetylation did not affect FEN1/PCNA interaction.16 Whether there is a cross-talk between acetylation and phosphorylation or methylation remains to be addressed. To date, FEN1 has been reported to interact with more than 30 proteins that can be categorized into at least five different pathways1. One may postulate that posttranslational modifications play a role in switching FEN1 between different pathways by controlling its interaction with specific partner proteins under a particular condition. Mapping modifications sites of FEN1 enables the study of cross-talk among different forms of posttranslational modification and of their physiological consequences in cell life or death.
Our findings have demonstrated the biological significance of the methylation-mediated dynamics of the FEN1/PCNA interaction. Deficiency in FEN1 methylation, due to mutations such as R192K and 4RK, increases the potential for it to be constitutively phosphorylated, leading to a failure to interact with PCNA. As a consequence, mutant cells expressing 4RK have defects in DNA replication, DNA repair and a high incidence of spontaneous mutations. These observations are consistent with those observed in a mouse model in which point mutations F343A/F344A (FFAA mutation) were used to disrupt FEN1/PCNA interaction, resulting in defects in DNA replication and LP-BER12. FFAA mutant cells have an aberrant cell cycle progression and display chromosomal instabilities. Heterozygous mice for the FFAA mutation develop a wide spectrum of cancer. These studies illustrate the potential prevalent consequences of impaired FEN1 arginine methylation in cancer etiology. The imbalance in PRMT activity has been implicated in many diseases, including cancer45,46. It is also worth noting that a single nucleotide polymorphisms (SNPs) has been identified on arginine 192 of FEN1 (R192Q, rs4989586) in the human population47. Study of this polymorphism might be helpful to link the genetic variation and protein arginine methylation with the population susceptibility to certain types of cancers.
Methods
Antibodies
The following antibodies were used: anti-FEN1 and anti-PCNA (GeneTex), anti-γH2AX and anti-mono methyl arginine (Abcam), anti-PhosSer, anti-asymmetric dimethyl-arginine and anti-symmetric dimethyl-arginine antibodies (Millipore), anti-Myc (sc-33) (Santa Cruz Biotechnologies), anti-PRMT5 (IMG 505) and anti-PRMT7 (IMG 512A) (Imgenex), anti-phosphor-Histone H3 (S10) and anti-α/β tubulin (Cell Signaling Technology), and anti-BrdU (Becton Dickinson).
In vitro methylation assay
Purified recombinant proteins were incubated (1 h, 30°C, cold methylation) with 1 μg of recombinant PRMT5 in 30 μl of methylation buffer (50 mM HEPES (pH 8.0), 0.01% (v/v) Nonidet P-40, 10 mM NaCl, 1 mM DTT, and 1 mM PMSF) supplemented with 2 μl of S-adenosyl-l-(methyl-3H or methyl-14C) methionine (3H-SAM or 14C-SAM, Amersham Biosciences; radioactive methylation) and 20 nmol of S-adenosyl-l-methionine sulfate p-toluenesulfonate (SAMe-PTS, Sigma, St. Louis, MO). Reactions were stopped either by addition of 2× SDS-PAGE sample buffer, followed by heating (5 min, 95°C) or by addition of 100 mM NaCl and incubation (10 min, 4 °C). Samples were analyzed by SDS-PAGE followed by autoradiography or liquid scintillation assay.
In vitro kinase assays
In vitro phosphorylation assays were performed with 100 ng of purified Cdk–Cyclin complexes to a final volume of 18 μl, containing 40 mM HEPES-NaOH (pH 7.5), 8 mM MgCl2, 33.3 mm ATP, 10 mCi of [γ-32P]ATP (3000 Ci/mmol; GE Health) and substrates to be tested. Reactions were stopped after 45 min (30°C) by adding SDS sample buffer. Samples were analyzed by SDS-PAGE followed by autoradiography or liquid scintillation assay.
DNA replication efficiency assay
DNA replication efficiency of cells was measured by 3H incorporation assay48. Briefly, 5 × 104 cells were seeded onto a 6-cm dish in DMEM for 12 h. 3H-thymidine was added to a final concentration of 1 μCi/ml. Cells were incubated in 3H-thymidine containing DMEM medium for a specific time period and washed with ice-cold PBS buffer. DNA was precipitated by treating cells with 10% ice-cold trichloroacetic acid and 10 mM thymidine (15 min, 4 °C). After extensive washes with PBS buffer, DNA was solubilized in 0.5 M NaOH. Radioactivity levels in the sample were measured using a liquid scintillation counter.
Okazaki fragment maturation assay and LP-BER assay using cell extract
Gap filling, RNA/DNA primer removal, and DNA ligation reactions were assayed with gapped substrates, following a modified version of published protocols6,12. Briefly, cell extracts were incubated with specific gapped substrates in a reaction buffer containing 5 μCi [α-32P]dCTP and 50 μM each of dATP, dGTP, dTTP and ATP. Reactions were carried out (60 min, 37°C) and products were separated on a 15% denaturing PAGE, followed by autoradiography. LP-BER reconstitution was performed using a protocol similar to the Okazaki fragment maturation assay, but with a Tetrahydrofuran-containing substrate6,49.
H2O2 sensitivity assay
Sensitivity to DNA damage reagent H2O2 was determined by cell growth inhibition assay. HeLa cells were seeded (1,500/well), incubated (overnight, 37°C), treated (1 h, 37°C) with multiple dilutions of H2O2, washed in a fresh medium (DMEM containing 10% fetal bovine serum), and incubated (72 h) under normal growth conditions (37°C, 5% CO2). The number of viable cells was determined by the CellTiter 96 AQueous one-solution cell proliferation assay (Promega). At least four replications for each clone were averaged. Data are expressed as the percentage of growth relative to untreated controls.
Hprt mutation frequency assay
The spontaneous Hprt mutation frequency was detected as previously described50. HeLa cells (1 × 105, WT or 4RK) were plated (10-cm plates) and incubated (1 d). The medium was then changed to HAT (Hypoxanthine Aminopterin Thymidine) for 24 h and HT (Hypoxanthine and Thymidine) medium for another 48 h to remove pre-existing Hprt cells in the cell population. Cells were subcultured every 3 days, for 20 days, and density maintained at 106 cells. Cells (200 per plate) were then seeded on five plates for 10 days to count the relative cloning frequency. After cell attachment (18–24 h), the medium was replaced with fresh medium containing 5 μg/ml 6-thioguanine (6-TG). Cells were maintained in 6-TG-containing medium for 10 days. The plates were then washed with PBS, fixed and stained with 1% methylene blue.
Supplementary Material
Acknowledgments
We thank George Tsaprailis in the Proteomics Core facility of University of Arizona for technical assistance in determining the methylation sites on FEN1. We thank Dr. S.R. da Costa (City of Hope) for editorial assistance. This work was supported by NIH grants RO1 CA073764 and R01CA085344 to B.H.S.
Footnotes
Author contributions: Z.G., L.Z., and B.H.S. designed the experiments, analyzed the data and wrote the manuscript. H.X. purified recombinant proteins. H.D., M.Z. and M.R.P. performed cell biological experiments. Q.M. C. performed the experiments with mass spectrometry and analyzed the data.
Competing financial interests: The authors declare no competing financial interests.
Contributor Information
Zhigang Guo, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Li Zheng, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Hong Xu, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Huifang Dai, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Mian Zhou, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Mary Rose Pascua, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
Qin M. Chen, Department of Pharmacology, University of Arizona, Tucson, AZ 85724
Binghui Shen, Department of Cancer Biology, City of Hope National Medical Center and Beckman Research Institute, 1500 East Duarte Road, Duarte, CA 91010.
References
- 1.Shen B, et al. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–29. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 2.Qiu J, Li X, Frank G, Shen B. Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J Biol Chem. 2001;276:4901–8. doi: 10.1074/jbc.M007825200. [DOI] [PubMed] [Google Scholar]
- 3.Shibata Y, Nakamura T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J Biol Chem. 2002;277:746–54. doi: 10.1074/jbc.M109461200. [DOI] [PubMed] [Google Scholar]
- 4.Guo Z, et al. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol Cell Biol. 2008;28:4310–9. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–87. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–36. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8:1242–9. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, et al. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–43. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, et al. Comprehensive mapping of the C-terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J Mol Biol. 2008;377:679–90. doi: 10.1016/j.jmb.2007.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen U, Chen S, Saha P, Dutta A. p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex. Proc Natl Acad Sci U S A. 1996;93:11597–602. doi: 10.1073/pnas.93.21.11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–12. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 12.Zheng L, Dai H, Qiu J, Huang Q, Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol Cell Biol. 2007;27:3176–86. doi: 10.1128/MCB.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–22. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 14.Zheng L, et al. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–9. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. Embo J. 2003;22:3451–60. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasan S, et al. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell. 2001;7:1221–31. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 17.Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 2003;22:4301–13. doi: 10.1038/sj.onc.1206606. [DOI] [PubMed] [Google Scholar]
- 18.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 19.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber S, Bauer UM. Arginine methylation in interferon signaling: new light on an old story. Cell Cycle. 2009;8:1464–5. [PubMed] [Google Scholar]
- 22.Boisvert FM, Hendzel MJ, Masson JY, Richard S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle. 2005;4:981–9. doi: 10.4161/cc.4.7.1830. [DOI] [PubMed] [Google Scholar]
- 23.Adams MM, et al. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 2005;4:1854–61. doi: 10.4161/cc.4.12.2282. [DOI] [PubMed] [Google Scholar]
- 24.Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–9. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 25.El-Andaloussi N, et al. Arginine methylation regulates DNA polymerase beta. Mol Cell. 2006;22:51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 26.El-Andaloussi N, et al. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007;21:26–34. doi: 10.1096/fj.06-6194com. [DOI] [PubMed] [Google Scholar]
- 27.Boisvert FM, Dery U, Masson JY, Richard S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005;19:671–6. doi: 10.1101/gad.1279805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boisvert FM, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005;4:1834–41. doi: 10.4161/cc.4.12.2250. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–8. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen DH, Wu KT, Hung CJ, Hsieh M, Li C. Effects of adenosine dialdehyde treatment on in vitro and in vivo stable protein methylation in HeLa cells. J Biochem. 2004;136:371–6. doi: 10.1093/jb/mvh131. [DOI] [PubMed] [Google Scholar]
- 31.Mostaqul Huq MD, et al. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber S, et al. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes Dev. 2009;23:118–32. doi: 10.1101/gad.489409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–63. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 34.Soza S, et al. DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol Cell Biol. 2009;29:2032–41. doi: 10.1128/MCB.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–98. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–31. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–62. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 39.Dery U, et al. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol Cell Biol. 2008;28:3058–69. doi: 10.1128/MCB.02025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J Biol Chem. 2000;275:10498–505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 41.Le Romancer M, et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–21. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 43.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 45.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–6. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 46.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–15. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 47.Mitra AK, et al. Association of polymorphisms in base excision repair genes with the risk of breast cancer: a case-control study in North Indian women. Oncol Res. 2008;17:127–35. doi: 10.3727/096504008785055567. [DOI] [PubMed] [Google Scholar]
- 48.Lu R, Serrero G. Inhibition of PC cell-derived growth factor (PCDGF, epithelin/granulin precursor) expression by antisense PCDGF cDNA transfection inhibits tumorigenicity of the human breast carcinoma cell line MDA-MB-468. Proc Natl Acad Sci U S A. 2000;97:3993–8. doi: 10.1073/pnas.97.8.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–56. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian Y, et al. Molecular events after antisense inhibition of hMSH2 in a HeLa cell line. Mutat Res. 1998;418:61–71. doi: 10.1016/s1383-5718(98)00108-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.