Abstract
Control of gene expression involves the concerted action of multiple regulatory elements some of which can act over large genomic distances. Physical interaction among these elements can lead to looping of the chromatin fiber. Although posttranslational modifications of chromatin are thought to play a role in the conveyance of epigenetic information, it is largely unknown whether higher order chromatin organization such as looping contributes to epigenetic memory. A related unresolved question is whether chromatin loops are the cause or the effect of transcriptional regulation. Recent work from diverse organisms suggests a memory function for long-range chromatin interactions. It is proposed that higher order folding of the chromatin fiber can serve to maintain active and repressed states of gene expression.
Introduction
The eukaryotic genome is packaged in a highly organized fashion to fit within the spatial constraints of the cell nucleus and at the same time allow for access of regulatory factors to the underlying sequences. The first layer of packaging involves the winding of 147 base pairs of DNA around an octamer of core histone proteins to form a nucleosome, the crystal structure of which has been solved [1]. A string of nucleosomes without any further folding presents itself as an 11 nm fiber under the electron microscope. The next layer of packaging involves the helical stacking of nucleosomes to form a chromatin fiber with a diameter of ~30 nm of which the structural organization is beginning to be unraveled [2]. While the architecture of chromatin folding at the next higher level is much more obscure, it has become clear that the chromatin fiber is flexible and allows for movement inside of the nucleus [3].
In simple terms, chromatin fiber flexibility allows for physical interactions between distant regulatory sites in the genome in a manner involving contacts in cis and in trans (Figure 1). The term chromatin loop describes interactions in cis with the intervening sequence looped out. However, it is conceivable that trans interactions might turn out to be as important. Fluorescence in situ hybridization (FISH) and chromosome conformation capture (3C) and its derivatives have provided strong evidence that distant enhancers can loop to the promoters they control [4–7]. Importantly, chromatin loops can be found at both active and repressed genes and are not limited to enhancer-promoter interactions but can also involve insulator elements. Genetic loci at which chromatin loops have been found and the diverse roles loops might play have been extensively reviewed recently [8–12]. Moreover, with the advent of new technologies that detect a broader spectrum of chromatin contacts, interactions in trans are being increasingly appreciated, although their functions remain largely obscure [13]. A functionally relevant trans interaction is discussed below in the context of polycomb-mediated chromatin interactions. Here we limit our discussion to a specific aspect of chromatin looping, namely the possible role of long-range chromatin interactions in epigenetic memory.
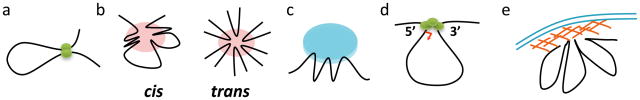
Figure 1.
Graphical representations of different arrangements of chromatin loops. a. In the simplest form of a chromatin loop two distant regions such as a promoter and enhancer are in close proximity to each other. The green ovals represent factor(s) that tie the loop. b. Multiple chromatin regions cluster together, creating three-dimensional active chromatin hubs (e.g. at the β-globin locus) or repressive clusters (e.g. the bithorax complex locus) by intra-chromosomal (cis) or inter-chromosomal (trans) interactions. Pink circles represent complexes of nuclear proteins that might contain gene-specific or general transcription factors. c. Loop formation might result from association of distal elements with shared sub-nuclear structures, such as the nucleolus, insulator bodies, transcription factories, or polycomb bodies. These associations might demarcate distinct genomic domains. Note that in this configuration, physical distances between juxtaposed regulatory elements can be much larger than those in gene specific intragenic loops (as in a), and may or may not influence each other. d. Interactions between the promoter and terminator sequences can result in the formation of intragenic gene loops. e. Formation of chromatin loops can be linked to targeting to the nuclear periphery such as the nuclear lamina or nuclear pores.
Our topic necessitates a brief description of the term epigenetics and how it will be used here. An epigenetic trait is defined as a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence [14]. The term “heritable” is most often used in the context of transgenerational inheritance or inheritance from mother to daughter cell, i.e. a form of memory that persists through mitosis or meiosis. However, frequently, the term epigenetic memory is employed when considering events that do not necessarily involve cell division, such as the memory of recent transcriptional activity. In the case of the inducible GAL1 gene in yeast, it has been proposed that changes in chromatin organization and subnuclear localization might mediate this form of transcriptional memory [15,16] (Figure 2, see below). However, additional work, including elegant heterokaryon experiments, indicates that this memory function can be imparted by information contained within the cytoplasm and can be maintained over several cell division cycles [17,18••].
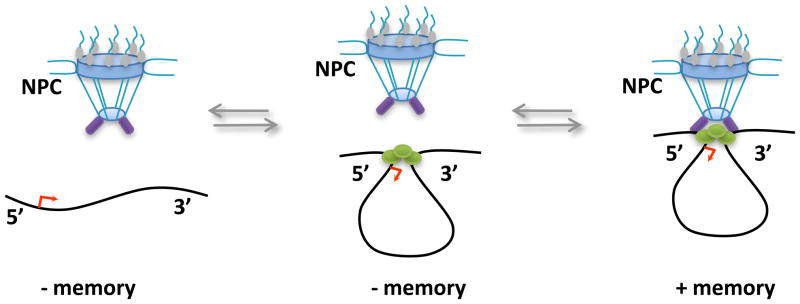
Figure 2.
Promoter-terminator loops as described for diverse genes in yeast and higher eukaryotes. In yeast, targeting of these structures to the nuclear envelope has been correlated with transcriptional memory. Disruption of the loop and/or its interaction with the nuclear envelope can lead to loss of transcriptional memory.
A form of transcriptional memory in mammalian cells, also referred to as transiently poised state or dynamic bookmarking has been linked to the persistence of general transcription factors. For example, a recent study in T cells showed that following repression, the prolonged presence of the histone acetyltransferase p300 and RNA polymerase II facilitates rapid re-induction of immediate early genes by stimuli that by themselves are ineffective without prior transcription [19•]. Thus, possible mechanisms that mark genes for rapid activation might include chromatin modifications, transcription factors and cytoplasmic proteins. In view of these possibilities, the above definition of epigenetics is somewhat arbitrarily confined, although it follows conventional and perhaps pragmatic thinking of epigenetics in the context of proteins associated with chromatin. In the following sections we specifically consider some recent but also older work from different systems suggesting that higher order chromatin organization might play a role epigenetic memory.
Stability and dynamics of chromatin loops
A memory function for chromatin loops might require that they are stable even in the absence of transcription or throughout the mitotic phase of the cell cycle when transcription is silenced globally [20] (Figure 3). Indeed, chromatin loops at the mammalian β-globin locus persist when transcription is silenced pharmacologically [21,22]. In addition, in yeast, promoter-terminator gene loops are maintained for extended periods of time even after transcription has been repressed [23••, 24•• ] (Figure 2). This suggests that higher order chromatin organization might contribute to a memory mechanism that facilitates rapid re-expression of a gene following a period of silencing or upon re-entry into the G1 phase. The chromatin fiber at imprinted gene loci such as the Igf2-H19 locus is folded in complex patterns involving interactions between enhancers and promoters, among insulators, and between insulators and promoters (for review see [25]). These looped structures are configured in an allele-specific manner and are maintained throughout many rounds of cell division. Since imprinting is regarded as a truly epigenetic phenomenon and involves DNA methylation, it is tempting to speculate that the looped structures might be stable throughout the cell cycle and contribute to epigenetic memory. Indeed, it has been reported that chromatin binding of the insulator protein CTCF and associated chromatin loops at the Igf2-H19 locus can persist throughout the mitotic phase of the cell cycle. This suggests that a degree of domain organization is preserved through the cell division cycle [26]. However, CTCF does not remain bound at all sites during mitosis [27], indicating that the observations at the Igf2-H19 locus might not be generalizable. Other examples for generationally stable allele-specific expression are the epialleles of the maize b1 gene that are involved in paramutation. Depending on the expression level, the alleles fold into distinct looped conformations, leading to speculation that these structures might be part of a meiotically stable memory machinery [28].
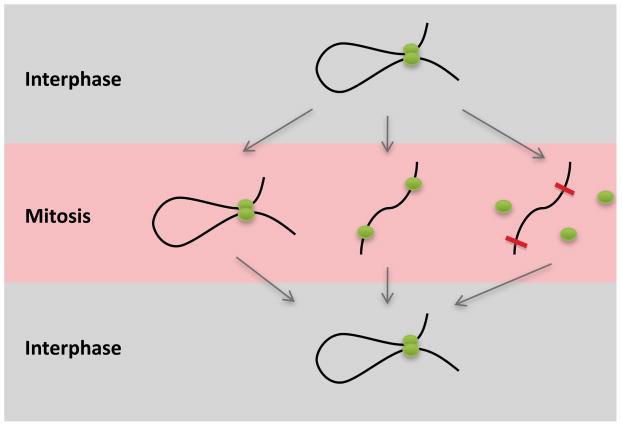
Figure 3.
Potential modes by which chromatin loops are transmitted through mitosis. The looped chromatin structure could persist throughout the cell division cycle. Alternatively, it could dissolve during mitosis but the looping factors might remain bound to chromatin to direct the reformation of the loops upon entry into G1. Finally, chromatin loops could dissolve along with removal of the looping factors. Epigenetic bookmarks (red bars) might facilitate reassembly of transcription factor complexes at the appropriate sites following cell division.
On the other hand, loops are clearly dynamic and can rapidly transition from one state into another during transcriptional activation or repression as revealed by experiments using conditional looping factors [29–31•].
Recent studies have begun to identify nuclear factors that establish chromatin loops in a gene-specific manner. Combinations of enhancer- and promoter-binding proteins cooperate to set up gene-specific chromatin loops as exemplified by studies of the tissue-specific transcription factors EKLF, GATA-1 and Ldb1 at the murine β-globin locus [29,30,32]. In addition, CTCF contributes to a subset of the chromatin loops at this locus [33,34]. It is unknown whether the looped configuration of the β-globin locus serves a transcriptional memory function throughout the cell division cycle. However, GATA-1 is globally removed from chromatin during mitosis [35], suggesting that the chromatin loops that depend on GATA-1 might also be lost. Nevertheless, it remains to be fully explored whether some higher order structures persist throughout the cell cycle at this locus, perhaps those dependent on CTCF.
Varying degrees of looped chromatin organization have been described at certain gene loci before the onset of gene transcription and even in non-expressing tissues [36–38]. However, either the frequencies of these interactions or their constellation are insufficient for full transcription. Whether these chromatin contacts underlie transcriptional competence in a manner that facilitates the onset of transcription upon a stimulus is an attractive possibility that remains to be explored further.
Collectively, these studies suggest that loops display properties of being both dynamic and relatively static in different contexts, indicating that their formation can be independently regulated. At present, the available evidence does not establish with certainty any causality between looped-chromatin and heritable gene expression states.
Polycomb mediated long-range interactions
A well studied model of epigenetic maintenance of transcription states involves the Polycomb group (PcG) and Trithorax group (trxG) regulatory systems. PcG complexes are typically associated with gene repression. Originally discovered in Drosophila, they are required to maintain repressed states of target genes throughout multiple rounds of cell division during development. Since PcG proteins can maintain repression in a manner independent of the initiating repressive event, they are thought to function via epigenetic chromatin-mediated mechanisms. Polycomb response elements (PRE) have been described as cellular memory modules and as such represent strong candidates for true epigenetic regulators. The observation that PcG bound elements can engage in long-range chromosomal interactions in cis and in trans [39–42] (for review see [43]) raised the possibility that higher order chromatin folding might be part of the memory mechanism.
An example is the PRE-containing Fab7 element that is involved in the developmental control of the Drosophila bithorax gene cluster. Fab7 can physically contact the promoters it represses. Importantly, transgenic Fab7 elements, regardless of their chromosomal locations, can interact with the endogenous Fab7 element to promote gene silencing [40••]. Both the physical pairing of Fab7 elements and repression require PcG proteins. This suggests that long-range interactions in cis or in trans are required for transcriptional silencing. Disruption of the endogenous Fab7 element led to derepression of a transgenic Fab7 -linked reporter gene in a manner that was maintained through multiple generations. Remarkably, even when the Fab7 element was bred back into the fly, the derepressed (i.e. “un-looped”) state of the transgene was generationally maintained [40••]. From this finding it was extrapolated that the PcG-dependent repressed state along with the associated long-range interactions is also part of an epigenetic memory mechanism. Likewise, the looped clustering of diverse PREs at the bithorax locus has been proposed to constitute part of a mitotically stable memory mechanism [39].
While these are tantalizing propositions, it has not been determined for any of the PcG mediated chromosomal interactions whether they are causally involved in transcriptional memory, or whether they are simply a reflection of the repressed state and its associated transcription factors or chromatin marks. It is also unclear whether the looped structures persist throughout the cell cycle or whether they dissolve and subsequently re-form after cell division (Figure 3). In the latter case, other bookmarking factors or epigenetic marks would have to exist to correctly reproduce these interactions.
It is possible that PcG proteins and the associated H3K27 tri-methylation preserve the looped chromatin configuration throughout mitosis. Alternatively, the higher order structures might be resolved. In that case, PcG proteins or histone modifications might provide marks to direct the correct re-organization of the chromatin fiber upon entry into G1. While these questions are unresolved, it is instructive to consider the fate of PcG proteins during S phase. PcG proteins have been shown to remain associated with replicating DNA templates in vitro [44], and mechanisms have been proposed by which the corresponding histone marks and associated reader proteins are propagated during DNA replication [45,46]. It is worth investigating whether higher order structures are propagated throughout S-phase and how they might be re-established on the newly replicated DNA templates.
Another consideration is that 3C-based methods examine interactions that reflect averages of a large number of alleles. In contrast, FISH based methods allow studies at the single cell level. For example, FISH experiments showed that the pairing of Fab7 elements occurs in only a fraction of nuclei [40••], raising the question as to how a relatively infrequent interaction can contribute to efficient gene silencing. Perhaps silencing is initiated by transient contacts and is subsequently maintained by other mechanisms. Alternatively, long-range contacts might be dynamic thus allowing capture by FISH only at a subset of alleles at any given time. If these interactions are indeed dynamic and short-lived, a role in epigenetic memory would seem unlikely.
As is the case for Drosophila, mammalian PcG proteins can spread across broad domains, which in turn are folded into complex looped structures, indicating that higher-order folding is a conserved feature of PcG regulation in metazoans [47•]. It remains to be elucidated how such broadly PcG-covered regions can fold into discrete loops. Moreover, future studies will have to assess whether the looped structures per se serve any memory or bookmarking functions.
Initiator-terminator loops and transcriptional memory
Transcriptional re-activation of a gene for the second time following an intervening period of silencing might require a shorter stimulus than activation for the first time. This phenomenon is referred as transcriptional memory. Many factors are implicated in the transcriptional memory of the inducible yeast GAL genes. Moreover, depending on whether short term (~ 1 or 2 cell divisions) or long term (5 or 6 cell divisions) memory are considered, factor requirements might vary. The molecular mechanism and the nature of “heritable” components are still controversial and might vary between strains, genes, and growth condition [48]. An early report suggested that transcriptional memory correlated with histone H3K4 methylation, thus potentially linking a chromatin modification with an epigenetic function [49]. However, later studies showed that H3K4 methylation as well as some additional histone marks are dispensable for this function [15,23••]. Instead the memory phenomenon was linked to the deposition of the histone variant H2A.Z, the association with the nuclear pore complex via Nup2 [16] (Figure 2) and the SWI/SNF chromatin remodeling complex [15]. As mentioned above, even cytoplasmic components are capable of transducing a transcriptional memory function [18••], indicating that multiple pathways that function in a linear or parallel fashion might maintain certain genes in a state poised for rapid activation.
Two recent reports added to the debate by suggesting that chromatin loops may underlie transcriptional memory in yeast [23••,24••]. Their findings were extensions of previous studies demonstrating that promoter and terminator sequences are in physical proximity at actively transcribed genes [50]. While these loops appear dispensable for steady state transcription [51], their sustained presence correlates with rapid transcriptional reactivation [23••,24••]. Specifically, promoter-terminator loops at several genes persisted following transcriptional repression, and rapid reactivation only occurred within the time window in which the loops where maintained. Once the loops were lost, transcriptional memory was erased, and genes reactivated with the same slow kinetics as during the initial activation. The correlation between loops and transcriptional memory was further supported by the use of mutants that are impaired for loop formation, for example, through mutation of TF-IIB (sua 7-1 allele) or loss of Mlp1 that is required for the targeting of the gene to the nuclear periphery.
Based on these correlations, the loops were termed memory gene loops (MGL). Indeed, one report claimed that these loops are both necessary and sufficient for transcriptional memory [24••]. However, we suggest withholding the term MGL until evidence is provided to support an essential or causative role of these loops in transcriptional memory. It is possible that TFIIB and Mlp1 (among others) might contribute to the memory independently of loop formation, in which case looping would simply be a consequence of a function enabling memory. Moreover, it is unclear how sufficiency of loop formation for memory can be established based on these data. Indeed, loss of Snf2 leads to loss of transcriptional memory without impairing loop formation or steady state transcription [23••]. This shows that the promoter-terminator loop is insufficient for transcriptional memory even under conditions that permit normal steady state transcription. Thus, the role of promoter-terminator loops during transcriptional memory, if any, deserves further investigation.
Promoter-terminator interactions have also been observed in mammalian cells at BRCA1, CD68, and proviral HIV-1 integrants [52–54]. In the case of BRCA1 there does not seem to be a strict correlation between transcription and loop formation as these loops are dissolved during high levels of transcription [52,53]. At the CD68 gene, 5′-3′ loops are also observed in the inactive state. At the HIV-1 proviral integrants, loops are strictly dependent on ongoing transcription and are lost upon blocking transcriptional elongation. Therefore, in these cases a role for looping in transcriptional memory appears unlikely.
Conclusions and Perspective
It is difficult to establish with certainty that a given chromatin modification exerts a truly epigenetic function. Chromatin marks are established or erased by nuclear factors that recruit the modifying enzymes. As such, chromatin modifications might simply be downstream mediators in a pathway rather than initiators of a given function [55]. The same holds for chromatin loops. If a loop persists during mitosis for example, is it the loop or the factors that tie the loop that specify epigenetic functions (Figure 3)? It is implicit that just because a histone mark, a nuclear factor, or a three-dimensional structure persists beyond the duration of transcription, this does not automatically imply a memory function.
Principally, proof of a direct epigenetic function for higher order chromatin organization will be equally as challenging as that for covalent histone modification. Nevertheless, experiments testing the cause-effect relationships of chromatin loops and memory can be conceived. In its simplest form, loops could be disrupted in a target manner using point mutants that impair contacts required for association between looping factors. Complementing “rescue” mutations in the binding partners could be engineered to restore binding and tested for their ability to restore looping.
Alternatively, short DNA sequences could be introduced at relevant sites into yeast genes followed by conditional expression of binding factors fused to dimerization domains. In this way, loop formation could be promoted or disrupted in a manner independent of endogenous nuclear factors, similar to what has been done in episomal gene constructs in mammalian cells [56].
If it could be established that a higher order chromatin structure underlies transcriptional memory, then what would be the mechanism? Perhaps a stable structure would increase the likelihood that an enhancer interacts with the appropriate promoter and prevent it from contacting inappropriate genes upon reassembly of the nuclear factors after mitosis. This would not only accelerate gene activation but would reduce unwanted transcription. For example, it has been observed that adventitious transcription of an irrelevant gene can occur when an enhancer from an unrelated locus loops to its promoter [57•]. Such “bystander” effects could be minimized by a stable intragenic loop.
A related open question is the role of looping in partitioning of genes to subnuclear compartments such as the nuclear envelope, the nucleolus, insulator bodies, polycomb bodies, or the nuclear pore complex (Figure 1, for review see [3]). It can be imagined how such partitioning might contribute to a memory function since at sites enriched for regulatory factors the assembly of transcription factor complexes would be accelerated. There are many examples of genes that move to the nuclear periphery upon activation in yeast [58]. Experiments in which genes were tethered to subnuclear sites have addressed cause-effect relationships between nuclear positioning and transcriptional memory. For example, tethering of the INO1 gene to the nuclear envelope in yeast has been shown to convey rapid re-activation kinetics [16] (Figure 2). Similar gain-of-function experiments can be envisioned in which long-range interactions are created under controlled conditions to explore whether higher order structures are drivers or passengers of mitotic memory.
Acknowledgments
We apologize to those whose work could not be discussed due to space limitations. We thank Ann Dean, Job Dekker, Christopher Vakoc, Jumin Zhao, and members of the laboratory for critical comments on the manuscript. GAB is supported by NIH Grants DK58044 and DK54937.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wulan Deng, Email: wuland@sas.upenn.edu.
Gerd A. Blobel, Email: blobel@email.chop.edu.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 4.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 5.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 6.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 7.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 8.Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Nunez E, Fu XD, Rosenfeld MG. Nuclear organization in the 3D space of the nucleus - cause or consequence? Curr Opin Genet Dev. 2009;19:424–436. doi: 10.1016/j.gde.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams A, Spilianakis CG, Flavell RA. Interchromosomal association and gene regulation in trans. Trends Genet. 2010;26:188–197. doi: 10.1016/j.tig.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 18••.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. Conjugation of haploid yeast cells with a mutation that renders them defective for nuclear fusion allowed the authors to generate heterokaryons that maintain separate nuclei. It was thus shown that long term galactose transcriptional memory can be imparted by cytoplasmic components including the Gall galactokinase. [DOI] [PubMed] [Google Scholar]
- 19•.Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, Bilke S, Haggerty CM, Player A, Wang YH, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci US A. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. It is shown in T-cells that occupancy at certain immediate early genes of p300, acetylated histones, and RNA polymerase 2 persist beyond the duration of transcription. This permits re-activation these genes by stimuli that on their own are incapable of activating transcription in the absence of prior conditioning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of Long-Range DNA Interactions after Inhibition of Ongoing RNA Polymerase II Transcription. PLoS ONE. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. Two reports showing that in yeast promoter-terminator gene loops persists following a cycle of transriptional activation and repression. Time course experiments and the use of various mutants establish a strong correlation between loop formation and the ability to rapidly re-activate gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. Embo J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komura J, Ikehata H, Ono T. Chromatin fine structure of the c-MYC insulator element/DNase I-hypersensitive site I is not preserved during mitosis. Proc Natl Acad Sci U S A. 2007;104:15741–15746. doi: 10.1073/pnas.0702363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell. 2009;21:832–842. doi: 10.1105/tpc.108.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among Distant Regulatory Elements at the beta-Globin Locus Requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 31•.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. During erythroid differentiation, transcription factor GATA-1 triggers the loss of enhancer-promoter proximity at the repressed Kit gene along with the de-novo formation of a loop within the transcribed portion within the body of the gene. This illustrates that loops can be associated with gene repression and highlights the dynamic nature of long range chromatin interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xin L, Zhou GL, Song W, Wu XS, Wei GH, Hao DL, Lv X, Liu DP, Liang CC. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol Biol. 2007;8:31. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 37.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 38.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 40••.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. A very provocative study raising the possibility that features of nuclear architecture involving the cellular memory module Fab7 and PcG proteins can be stably inherited throughout meiosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 43.Mateos-Langerak J, Cavalli G. Polycomb group proteins and long-range gene regulation. Adv Genet. 2008;61:45–66. doi: 10.1016/S0065-2660(07)00002-8. [DOI] [PubMed] [Google Scholar]
- 44.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 46.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. The authors show that polycomb dependent gene loops occur at the repressed GATA-4 locus in embryonic carcinoma cells. Looped structures are diminshed upon their differentiation and GATA-4 activation. Notably, colon cancer cells in which the GATA-4 gene is repressed by promoter methylation display a similar pattern of looped structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kundu S, Peterson CL. Dominant role for signal transduction in transcriptional memory of yeast GAL genes. Mol Cell Biol. 2010 doi: 10.1128/MCB.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 50.O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 51.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Reilly D, Greaves DR. Cell-type-specific expression of the human CD68 gene is associated with changes in Pol II phosphorylation and short-range intrachromosomal gene looping. Genomics. 2007;90:407–415. doi: 10.1016/j.ygeno.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 56.Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. Embo J. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Lower KM, Hughes JR, De Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci U S A. 2009;106:21771–21776. doi: 10.1073/pnas.0909331106. It is shown that a distal regulatory element of the human α-globin locus can act via looping on seemingly unrelated genes even when they reside at great distances from the α-globin genes and are interspersed by potential enhancer blocking insulators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21:127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]