Abstract
Background
The heterogeneous nuclear ribonucleoprotein (hnRNP) K is an essential RNA and DNA binding protein involved in gene expression and signal transduction. The role of hnRNP K in cancer is relatively understudied. However, several cellular functions strongly indicate that hnRNP K is involved in tumorigenesis. Oncogenes c-Src, c-myc, and eIF4E are regulated by hnRNP K. We have shown an increased cytoplasmic hnRNP K in pancreatic cancer. In the present study, we investigated the altered expression of hnRNP K protein and its correlation with p-ERK in melanoma using human melanoma cell lines and tissue microarray.
Materials and Methods
The protein levels of hnRNP K and p-ERK in 8 human melanoma cell lines and a melanoma progression tissue microarray containing 80 melanoma, 23 dysplastic nevi, and 14 benign nevi specimens were analyzed using Western blot and immunohistochemistry analysis. hnRNP K was knocked down by siRNA, and its effect on melanoma cells was assessed.
Results
We showed a higher hnRNP K protein level in both melanoma cell lines and melanoma tissue specimens, which correlated with a higher c-myc expression. An increase in the cytoplasmic hnRNP K and eIF4E protein levels in melanoma cells is also seen. p-ERK level was also higher in dysplastic nevi and melanoma tissues, but did not correlate with hnRNP K protein level. We then demonstrated that knocking down of hnRNP K by siRNA inhibited melanoma cell growth and colony formation, as well as c-myc expression.
Conclusions
hnRNP K expression correlated with melanoma and may play a role in melanoma tumorigenesis.
The heterogeneous nuclear ribonucleoprotein (hnRNP) K, a component of the hnRNP complex, is an essential RNA and DNA binding protein. Genetic studies in C. elegans and metazoans reveal that hnRNP K deletion is embryonic lethal.1 The three KH domains of hnRNP K protein are almost completely conserved between X. Laevis and mammals and are well conserved in fly, nematode, and yeast.1 hnRNP K protein is found in several subcellular compartments where it is thought to be involved in multiple processes that comprise gene expression.2 The hnRNP K protein interacts with many different molecules involved in gene expression and signal transduction including chromosome remodeling, DNA transcription, RNA processing, RNA splicing, RNA stability, and translation.3
The role of hnRNP K in cancer is relatively understudied. However, several cellular functions strongly indicate that hnRNP K is involved in tumorigenesis. Firstly, the expression of many genes involved in tumorigenesis is regulated by hnRNP K, such as oncogenes c-Src, eIF4E, and c-myc.4,5 hnRNP K was shown to bind a core polypyrimidine element in the eIF4E promoter and positively regulate its transcription.4 Overexpression of hnRNP K increases translation initiation, cell division, and neoplastic transformation in an eIF4E-dependent manner.4 Oncogene c-myc is an important transcription factor that regulates the expression of many essential genes involved in tumorigenesis. hnRNP K protein binds to the CT element present within the c-myc P1 promoter.6 Overexpression of hnRNP K protein increases the activity of the c-myc gene promoter, an effect that is enhanced when hnRNP K is coexpressed with the TFIID TATA box-binding protein (TBP).7 In addition, hnRNP K binds and activates the c-src promoter in cooperation with the transcription factor Sp1.8 Secondly, hnRNP K was identified as a member of the p53/HDM2 pathway.9 hnRNP K is a HDM2 target and plays key roles in coordinating transcriptional responses to DNA damage by serving as a cofactor for p53. Other important observations implicating the tumorigenic role of hnRNP K include that hnRNP K can be regulated by many oncogenic kinases. Phosphorylation of hnRNP K by proto-oncogene c-Src leads to the removal of hnRNP K from the differentiation control element (DICE) and the relief of the translational silencing.1 Interestingly, the activation of the Ras/Raf/MEK/ERK pathway results in cytoplasmic accumulation of hnRNP K and regulation of translation.10 Therefore, hnRNP K plays an essential role in tumorigenesis. However, no one has studied the direct important role of hnRNP K in human melanoma.
Recently, hnRNP K was found to be upregulated in many cancers including colorectal, prostate, hepatic, esophageal, breast cancer, and leukemia.11–16 Our group found that the cytoplasmic hnRNP K is increased in pancreatic cancer and knockdown of hnRNP K inhibited pancreatic cancer cell growth and colony formation.17 Interestingly, the increased cytoplasmic localization is also observed in colorectal cancer.18 Overexpression of hnRNP K can transform cells, enhance passage through G1 phase, and increase global translation.4 These findings further support that hnRNP K has oncogenic potential.
In the present studies, we have investigated the altered expression of hnRNP K protein and its correlation with p-ERK in melanoma. We also examined the effect of knocking down of hnRNP K on melanoma cell growth and colony formation.
Materials and Methods
Cell Culture and Tissue Specimens
Mel-STV cell line is an immortalized normal melanocyte cell line. It is a gift from Dr. Robert Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA) and was cultured in DMEM supplemented with 10% fetal bovine serum (Omega Scientific, Inc., Tarzana, CA).19 A375, UACC647, UACC903, UACC1460, UACC2565, UACC1227, UACC827, and UACC612 human melanoma cell lines, primary human fibroblasts, and normal primary melanocytes were obtained from American Type Culture Collection (ATCC, Manassas, VA). These cells were cultured at 37°C with 5% CO2 in RPMI 1640 medium (Mediatech, Inc., Herndon, VA), supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin (Invitrogen). All transfections were carried out using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. Collection of melanoma specimens was approved by the Institutional Review Board of Human Subjects of University of Arizona.
Tissue Microarray and Immunohistochemistry Analysis
Human melanoma progression tissue microarray (TMA) was obtained from Dr. Galen Hostetter (Translational Genomics Research Institute (TGen), Phoenix, AZ). Immunohistochemistry (IHC) was performed using the hnRNP K antibody (1:1000) and p-ERK antibody (1:1000) as described previously except that we used NovaRED substrate (Vector Lab., Burlingame, CA) instead of DAB substrate for p-ERK IHC.20 The TMA IHC was evaluated by light microscopic examination, and the intensity of immunostaining in each core was assessed by 2 pathologists (AB and FL) using the scoring system described. The intensity of immunostaining in each core was graded as negative = zero, weak = 1, moderate = 2, or strong = 3.21 The proportion of cells staining positively was also assessed as percentage. The score was then calculated as the numbers representing intensity times the percentage of cells stained (Score = Intensity × % of positive cells).20
Cell Fractionation
Cytoplasm, nucleus, membrane, and cytoskeleton fractionations were performed using the Compartment Protein Extraction Kit (Millipore, Billerica, MA) following the manufacturer's instruction. Cytoplasm and nucleus fractionation were performed as previously described by our group.22
Knockdown of Endogenous hnRNP K by siRNA, Cell Growth and Colony Formation Assay
Predesigned hnRNP K siRNA (Eurogentec, San Diego, CA) or negative control siRNA (Mission siRNA Universal Negative Control No. 1, Sigma–Aldrich, St. Louis, MO) were transfected into A375 or primary human fibroblast cells using Lipofectamine 2000 (Invitrogen). For hnRNP K siRNA, two predesigned oligonucleotides 5′ ggaacaagcauuuaaaaga 3′ and 5′ ucuuuuaaaugcuuguucc 3′were synthesized and annealed according to the manufacturer's protocol. Cells were harvested 48 h after transfection. Cells were lysed and Western blot was performed as previously described.23 For cell growth assay, 24 h after transfection, 2 × 104 cells were seeded into 100-mm plates in triplicate, and cell growth was monitored by counting total cell numbers every 2 or 3 days as described previously by our group.23 For colony formation assay, 24 h after transfection, 1000 cells were seeded into 100-mm plates in triplicate and incubated for 2 weeks to allow colony formation. Then the media were removed and colonies were stained with methylene blue solution (50% methanol and 0.5% methylene blue) at room temperature for 5 min. The plates were rinsed with water, and colony number was counted.
Western Blot Analysis
Western blot was performed as previously described.24 α-tubulin monoclonal antibody was purchased from Oncogene Research Product (Gibbstown, NJ). Monoclonal hnRNP K antibody was purchased from Sigma–Aldrich (St. Louis, MO). c-myc and eIF4E antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). p-ERK and ERK1/2 antibodies were purchased from Cell Signaling (Danvers, MA). The intensities of the bands were quantified using ImageJ software (NIH) and normalized to α-tubulin.
Statistical Analysis
All data are reported as mean ± standard deviation (SD). When appropriate, differences between two groups were compared using the t test. Correlation assay was used to correlate hnRNP K and p-ERK level. Differences were considered significant at P < 0.05.
Results
Higher hnRNP K Expression in Human Melanoma Cells
To evaluate hnRNP K protein expression in melanoma cell lines, we performed Western blot analysis in 8 melanoma cell lines using hnRNP K antibody. Compared with normal primary melanocytes and fibroblasts, hnRNP K protein expression is significantly higher in all of the melanoma cell lines (Fig. 1). To examine if higher hnRNP K protein level correlates with higher protein levels of 2 of the important oncogenes regulated by hnRNP K, the membrane was stripped and immunoblotted with eIF4E and c-myc antibodies. The intensities of the bands were quantified and normalized to tubulin. As expected, c-myc expression was higher in all of the melanoma cell lines (Fig. 1). However, eIF4E expressions were lower in most of the melanoma cell lines compared to normal melanocytes (Fig. 1). Because the MAPK pathway is often constitutively activated in melanoma, we examined p-ERK and total ERK level in these cell lines to see if they correlate with hnRNP K expression. To our surprise, p-ERK level is lower in 7 of 8 melanoma cell lines compared with normal melanocytes (Fig. 1). Total ERK is higher in 4 of 8 and lower in 3 of 8 melanoma cell lines compared with normal melanocytes (Fig. 1). Interestingly, p-ERK levels in 2 melanoma cell lines (A375 and UACC903) that were known to have B-Raf mutation were not higher than normal melanocytes. Therefore, we did not see a correlation between hnRNP K expression and MAPK activation in these melanoma cell lines.
FIG. 1.
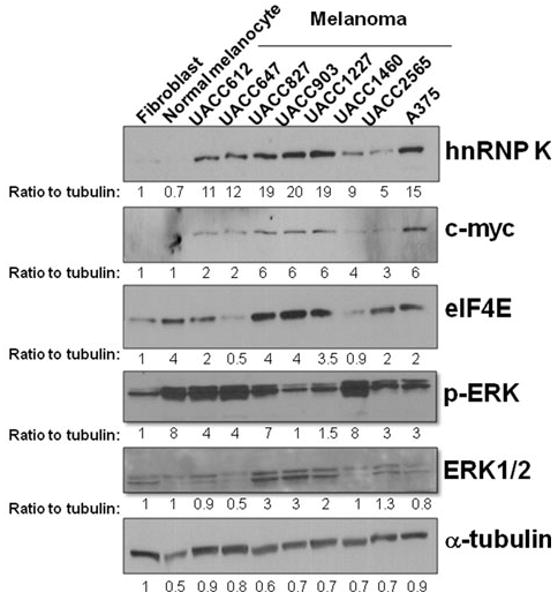
Higher hnRNP K protein level in human melanoma cell lines. Primary fibroblast, primary normal melanocytes, and 8 melanoma cell lines were grown at the log phase and harvested. Western blot analysis was performed using hnRNP K, c-myc, eIF4E, p-ERK, ERK1/2, and α-tubulin antibodies. The intensities of the bands were quantified using ImageJ software and normalized to α-tubulin. The ratio to tubulin was indicated below each band. ERK1/2 was measured as 1 band because of the close distance between these 2 bands
To confirm the higher hnRNP K protein level in human melanoma tissues, we obtained human melanoma progression tissue microarrays (TMA) from Dr. Galen Hostetter (Translational Genomics Research Institute (TGen), Phoenix, AZ). The detailed tissue information can be found in the Supplemental Table 1. hnRNP K protein expression was analyzed by immunohistochemistry (IHC) using hnRNP K antibody. The mean score of the hnRNP K protein expression in dysplastic nevi and melanoma tissues was significantly higher compared with benign nevi tissues (P < 0.05, Table 1; Fig. 2a).
TABLE 1.
Mean scores of hnRNP K protein expression and p-ERK level in benign nevi, dysplastic nevi, and melanoma tissue specimens
| No. of cases | hnRNP K score | P value | pERK Score | P value | Correlation | |
|---|---|---|---|---|---|---|
| Nevus | 14 | 1.79 ± 0.56 | 0.02 ± 0.08 | −0.139 | ||
| Dysp nevus | 23 | 2.30 ± 0.48 | .009a | 0.21 ± 0.43 | .029a | 0.033 |
| Malignant melanoma | 80 | 2.58 ± 0.55 | .025b | 0.71 ± 0.90 | .0004b | 0.097 |
Compared to nevus
Compared to dysplastic nevus
FIG. 2.
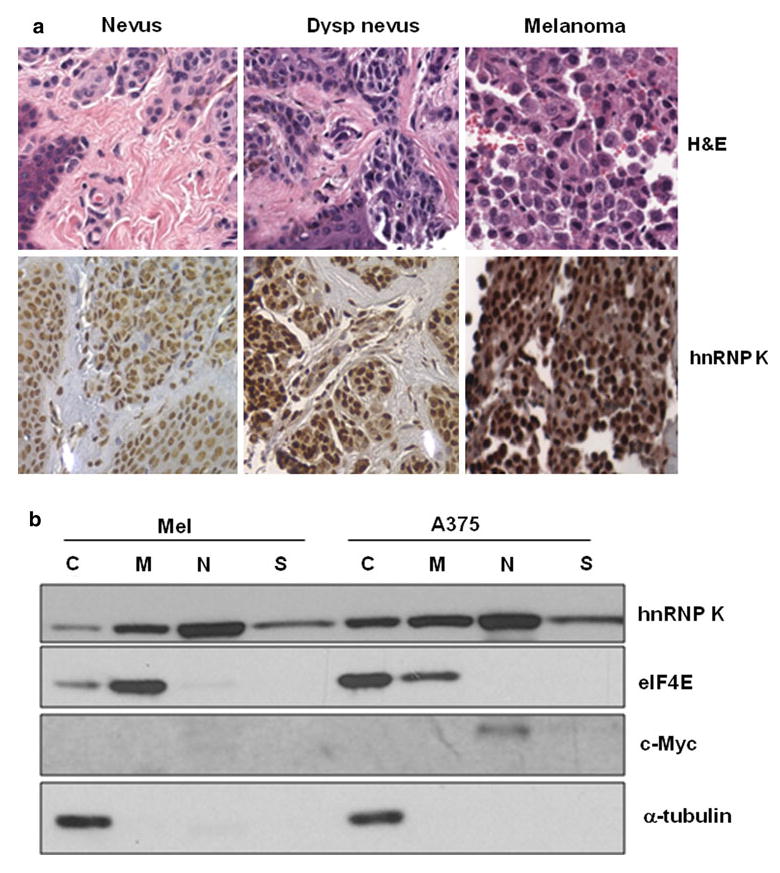
a hnRNP K protein is increased in human dysplastic nevus and melanoma tissues. H&E staining and immunohistochemistry was performed on a melanoma progression tissue microarray using hnRNP K antibody. Representative H&E and immunohistochemistry analysis of the hnRNP K protein in benign nevus, dysplastic nevus, and melanoma tissues were shown. Magnification: 400 ×. b Increased cytoplasmic hnRNP K protein in melanoma cells. Mel-STV (Mel) and A375 cells were fractionated, and Western blot analysis was performed using hnRNP K, eIF4E, c-myc and α-tubulin antibodies. C cytoplasm, M membrane, N nucleus, S cytoskeleton
We have reported that there is a higher cytoplasmic fraction of hnRNP K in pancreatic cancer.25 To investigate whether there is also a higher protein level of hnRNP K in the cytoplasms of melanoma cell lines, subcellular fractionation followed by immunoblot analysis was performed using hnRNP K antibody (Fig. 2b). A significant amount of hnRNP K protein was localized to the cytoplasmic fraction of the A375 melanoma cells. In contrast, in the immortalized “normal” human melanocytes (Mel-STV), hnRNP K protein level was very low in the cytoplasmic fraction (Fig. 2b).19 This higher cytoplasmic distribution of hnRNP K in melanoma cells is similar to our previous observation in pancreatic cancer cells.25 We also assessed the subcellular localizations of eIF4E and c-myc. Interestingly, eIF4E was mostly translocated to cytoplasm in melanoma cells (Fig. 2b, second panel). eIF4E was suggested to promote translation in the cytoplasm of tumor cells. c-myc was only detected in the nucleus of A375 melanoma cells (Fig. 2b).
Because MAPK pathway is often constitutively activated in melanoma, we examined p-ERK level in the same TMA tissue specimens and performed a correlation analysis with the hnRNP K protein level. The p-ERK levels in dysplastic nevi and melanoma tissues are significantly higher compared with benign nevi tissues (P < 0.05, Table 1; Fig. 3a). However, the hnRNP K protein level does not correlate with p-ERK level in these tissues (Table 1; Fig. 3b). To further investigate whether activated MAPK contributes to the cytoplasmic accumulation of hnRNP K or eIF4E, we treated A375 melanoma cells with MEK inhibitor U0126 and ask if this will decrease the accumulation of hnRNP K or eIF4E in the cytoplasm.10 Consistent with our previous data, inhibition of p-ERK did not block the cytoplasmic accumulation of hnRNP K and eIF4E (Fig. 3c). Our data suggested that MAPK pathway did not contribute to the cytoplasmic translocation of hnRNP K and eIF4E in this melanoma cell line.
FIG. 3.
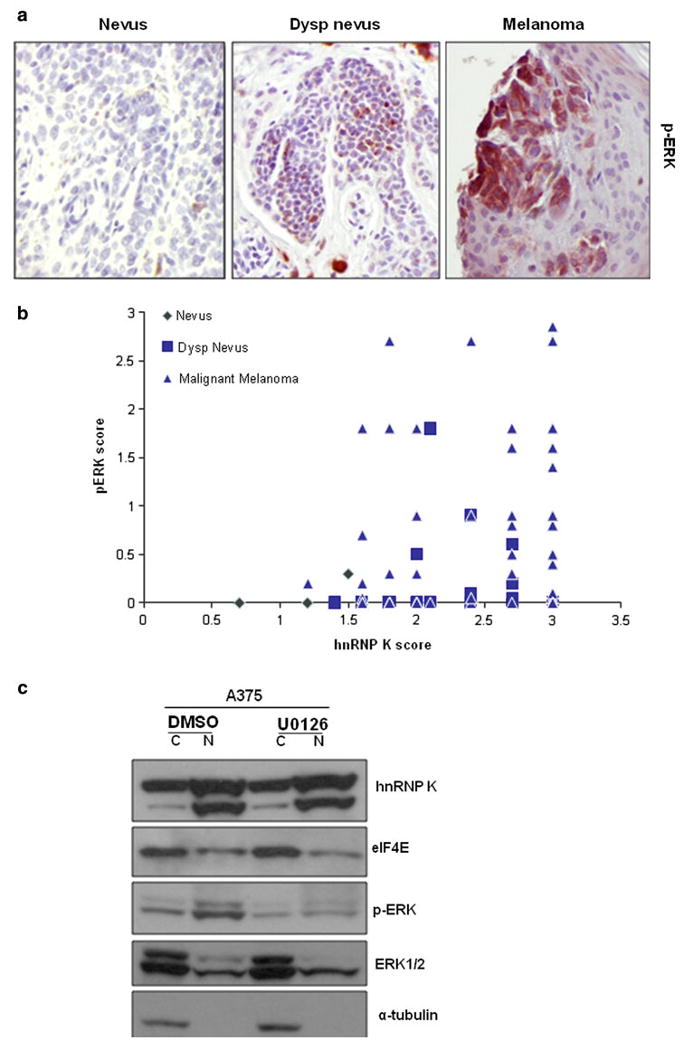
a p-ERK level is higher in human dysplastic nevus and melanoma tissues. Immunohistochemistry was performed on the same melanoma progression tissue microarray using p-ERK antibody. Representative immunohistochemistry analysis of p-ERK in benign nevus, dysplastic nevus, and melanoma tissues were shown. Magnification: 200 ×. b p-ERK level does not correlate with hnRNP K protein level. The IHC scores of the hnRNP K protein level and p-ERK level of the same tissue specimens were plotted against each other. c MEK inhibitor did not decrease the accumulation of hnRNP K and eIF4E in the cytoplasm. A375 cells were treated with MEK inhibitor U0126 (10 μM) or vehicle control DMSO for 1 h and fractionated. Western blot analysis was performed using hnRNP K, eIF4E, p-ERK, ERK1/2, and α-tubulin antibodies. C cytoplasm, N nucleus
To evaluate if hnRNP K expression correlates with known prognostic variables in melanoma, we gathered pathological information of the melanoma tissues on the TMA and performed statistical analysis. hnRNP K expression level seems to correlate with satellitosis (P < 0.05) (Table 2). Tumors that have satellitosis appear to have higher hnRNP K expression. There is a trend that tumors with ulceration, tumor-infiltrating lymphocytes (TIL), regression, and high mitotic rate (> 6/mm2) have higher hnRNP K expression (Table 2). However, it is not statistically significant (P > 0.05). There is no correlation between hnRNP K expression and tumor thickness or TNM stage (Supplemental Fig. 1).
TABLE 2.
Correlation of hnRNP K expression scores with known prognostic variables in melanoma
| Prognostic variables | No. of cases | hnRNP K score | P value | |
|---|---|---|---|---|
| + | − | |||
| Ulceration | 21 | 2.59 ± 0.50 | 2.55 ± 0.52 | 0.88 |
| High mitotic rate | 18 | 2.80 ± 0.45 | 2.45 ± 0.51 | 0.19 |
| Regression | 20 | 2.57 ± 0.51 | 2.54 ± 0.52 | 0.94 |
| Satellitosis | 18 | 2.90 ± 0.17 | 2.41 ± 0.51 | 0.014 |
| TIL | 22 | 2.55 ± 0.50 | 2.53 ± 0.55 | 0.93 |
TIL tumor-infiltrating lymphocytes
Knockdown of Endogenous hnRNP K by siRNA Inhibited Melanoma Cell Growth and Colony Formation
To determine the role of the endogenous hnRNP K protein in melanoma cell growth and colony formation, predesigned hnRNP K siRNA or negative control siRNAs were transfected into A375 melanoma cells. Endogenous hnRNP K protein levels were efficiently and specifically reduced at 72 h after transfection as was determined by Western blot analysis (Fig. 4a). c-myc protein was decreased in hnRNP K knockdown cells. Interestingly, eIF4E protein was reversely increased in hnRNP K knockdown cells (Fig. 4a). To examine the effect of hnRNP K silencing on tumor cell growth, we seeded the same number of indicated cells in the plates and counted total cell numbers every 2 or 3 days. Knockdown of hnRNP K significantly inhibited melanoma cell growth compared with control RNAi cells in A375 cells after day 5 (Fig. 4b). We further compared the ability of this cell line to form colonies. Consistent with our cell growth data, hnRNP K-knockdown cells formed significantly fewer colonies than control RNAi cells (Fig. 4c). To examine if this effect is also true in normal cells, we performed the same experiments in primary human fibroblasts. These cells are primary normal cells and can be effectively transfected and grow compared with primary normal melanocytes. hnRNP K silencing slightly and transiently reduced cell growth, but cell growth was recovered to the control level by day 7 (Fig. 4d). The total colony formation was much lower than tumor cells, but hnRNP K silencing reduced colony number compared with control RNAi cells (Fig. 4e). These data suggested that hnRNP K plays an important role in cell growth and proliferation.
FIG. 4.
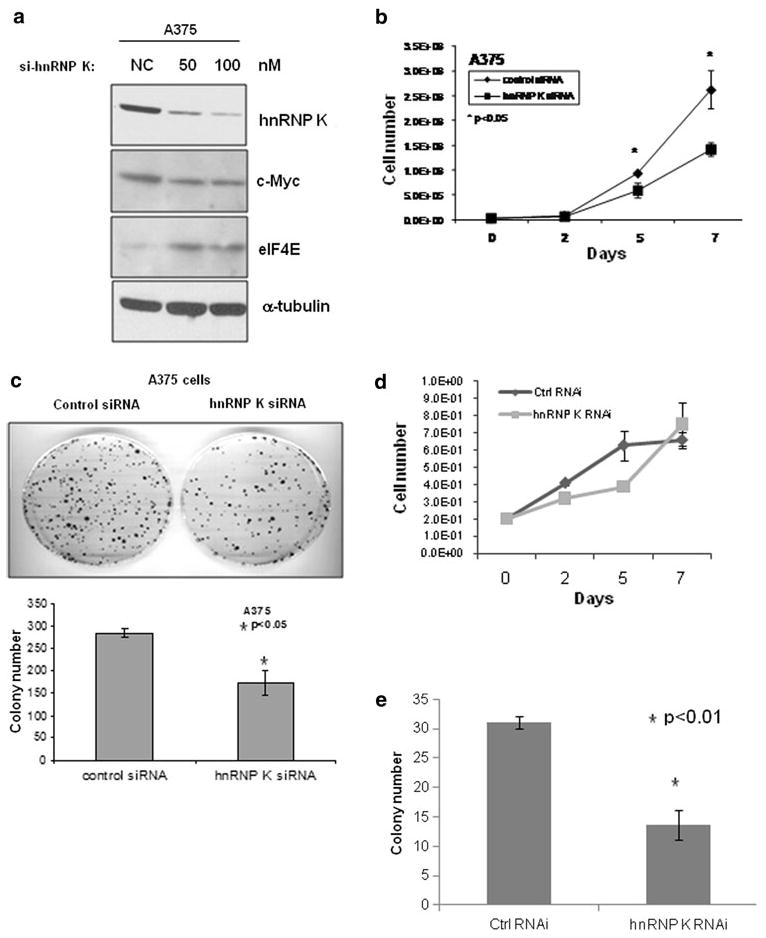
siRNA knock down of hnRNP K inhibited melanoma cell growth and colony formation. a A375 melanoma cells were transfected with negative control (NC) or 50 nM or 100 nM of hnRNP K siRNAs. Cells were harvested and lysed 48 h after transfection, followed by Western blot analysis using hnRNP K, c-myc, eIF4E, and α-tubulin antibodies. b A375 cells were transfected with 50 nM of negative control or hnRNP K siRNA. Then, 24 h after transfection, 2 × 104 cells were seeded into 100-mm plates in triplicate, and cell growth was monitored by counting total cell numbers every 2 or 3 days; or c 1000 cells were seeded into 100-mm plates in triplicate and incubated for 2 weeks to allow colony formation. Then media were removed, and colonies were stained with methylene blue solution. The plates were rinsed with water, and the colony number was counted. d, e Same experiments as described in b and c were repeated in primary human fibroblasts
Discussion
The hnRNP family members play important roles in DNA repair, chromosome remodeling, telomere biogenesis, cell signaling, and gene expression at both transcriptional and translational levels. Aberrant expression of individual hnRNP members in different tumor tissues supports the hypothesis that the hnRNPs are involved in tumorigenesis. However, their essential roles in tumor development and progression are largely underexplored. hnRNP K protein expression has been shown to increase in lung and liver cancers.13,26 Nagano et al. showed that hnRNP K was upregulated in an immortalized prostate cancer cell line; however, it was not reported whether the same was true in cancer tissues of the prostate.11 Our group has shown that hnRNP K was upregulated in pancreatic cancer.25 Here we reported for the first time that hnRNP K protein expression is significantly higher in melanoma cell lines and tissues. We also demonstrated that the silencing of hnRNP K resulted in decreased tumor cell proliferation and colony formation. Cellular localization of hnRNPs has been shown to be altered in cancer tissues, as is the case with hnRNP D in mouse neoplastic lung tissue.27 This is especially relevant because certain hnRNPs have important roles in mRNA nuclear-cytoplasmic shuttling. An increased cytoplasmic localization of hnRNP K has been observed in colorectal cancer and pancreatic cancer.18,25 Herein we have shown for the first time that an increased cytoplasmic localization of hnRNP K in melanoma cell lines.
hnRNP members have been identified as transcription factors with hnRNP K being the most extensively studied. Transcription factors can either be activators or repressors of gene expression. hnRNP K has been shown to bind double- or single-stranded DNA in the promoter regions of human proto-oncogenes c-Src, eIF4E, and c-myc in a sequence-specific manner.4,7,8 Expression of egr-1 is associated with the transient recruitment of hnRNP K to the promoter and transcribed regions.28 Furthermore, in vitro, hnRNP K was able to stimulate transcription and interact with the TATA binding protein (TBP).7 Therefore, hnRNP K appears to be a transcription activator, but it has also been documented as a functional repressor. hnRNP K was shown to interact with C/EBP-β, which repressed C/EBP-β transcriptional activation.29 hnRNP K has also been shown to mediate translation of c-myc mRNA by activating the IRES.30 Mutated c-myc IRES found prevalently in patients with multiple myeloma bound hnRNP K more efficiently in vitro and was translated to a greater extent by hnRNP K in vivo. These observations suggest a possible mechanism by which hnRNP K contributes to multiple myeloma by increasing translation of the oncogene c-myc. In the present study, we demonstrated that an increased c-myc expression is correlated with the increased hnRNP K expression in melanoma cell lines (Fig. 1).
eIF4E has an important role in cellular proliferation and is overexpressed in several tumors including head and neck, breast cancers, colon, lung, lymphoma, and bladder.4 hnRNP K was shown to bind to a promoter element in the eIF4E gene, resulting in increased transcription. eIF4E transcription was augmented if both c-myc and hnRNP K was overexpressed.4 However, this is not the case in our melanoma cell line. We saw an almost reverse correlation between hnRNP K/c-myc expression and eIF4E expression. This controversial observation may be caused by the different tumor type we study and regulation by other undetermined signaling pathways that are unique to melanoma. However, we observed a consistent cytoplasmic translocation of eIF4E in both melanoma and pancreatic cancer cells, suggesting an increased cytoplasmic translation in tumor cells.25 Interestingly, there is also an increased hnRNP K in the cytoplasm of tumor cells. The MAPK pathway has been shown to phosphorylate eIF4E. However, the MAPK pathway does not seem to play a role in the cytoplasmic translocation of eIF4E.
N-Ras/B-Raf is activated in most melanoma cells. Activation of MAPK pathway can lead to phosphorylation and cytoplasmic accumulation of hnRNP K in Hela cells and leukemia cells.10,31 Whether this is also true in melanoma is unknown. Here we showed that the activation of MAPK pathway does not seem to be the reason for higher expression and cytoplasmic accumulation of hnRNP K protein in melanoma, because p-ERK level did not correlate with hnRNP K level in melanoma cell lines and tissues and blocking MAPK activity did not decrease the cytoplasmic accumulation of hnRNP K. Whether MAPK pathway regulates hnRNP K expression is controversial. The only report by Notari et al. showed that inhibiting MAPK, but not PI3 K, PKC, or PLCγ, decreased hnRNP K expression in leukemia cells.31 However, overexpression of ERK1/2 did not increase hnRNP K expression.31 Exactly how hnRNP K expression is regulated in other human cancers is not known. Our data suggested that MAPK may not regulate hnRNP K expression or subcellular localization in melanoma cells. Further investigation of alterations of the downstream transcription factors of ERK (Ets, Elk-1, Stat1/3, MITF) in melanoma cells may explain the potential mechanisms. Besides ERK, hnRNP K can be phosphorylated by JNK, PKC, Src, Lck, Lyn, and Fyn.1 It can also be regulated by methylation and ubiquitination.32 Whether these signaling pathways regulate hnRNP K localization or protein level needs further investigation.
Melanoma is the most aggressive form of skin cancer. Many genetic alterations in melanoma have been identified. Chromosome region 9q21.32 was reported to be linked to familial melanoma.33 hnRNP K is exactly localized to 9q21.32. This coincidence together with our findings may point to an important oncogenic function of hnRNP K in melanoma. Besides Ras/Raf/MEK/ERK and PI3 K pathways, CDKN2A (INK4A, ARF) deletion or mutation will in turn impact on RB and p53 tumor suppressors.34 Interestingly, hnRNP K was identified as a member of the p53/HDM2 pathway.9 We have also showed that hnRNP K interacts with mutant p53.25 Therefore, altered p53 pathway in melanoma cells might explain hnRNP K alterations. Other important alterations in melanoma are receptor tyrosine kinases including EGFR, c-MET, and c-KIT.34 Upregulation of these tyrosine kinases can also activate the MAPK or PI3 K signaling pathways. Therefore, many genetic alterations in melanoma seem to interfere with these 2 signaling cascades, which may complicate the unique features of melanoma. Another important gene amplified in melanoma is microphthalmia-associated transcription factor (MITF), which is also downstream of ERK.35 Therefore, one reason for uncorrelated pERK and hnRNP K expression may be the alteration of the downstream transcription factors of ERK in melanoma. Melanoma has high heterogeneity. Identifying the key regulators of melanoma is necessary for better understanding of this disease.
Supplementary Material
Acknowledgments
This work was supported by Arizona Biomedical Research Commission grant (0007), G.I. SPORE CA95060, NCI/NIH grant CA133449, and NIH Arizona Cancer Center Support Grant CA023074. We thank Dr. Mark Nelson and Dr. Ronald Heimark for their discussion and support.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1245/s10434-010-1121-1) contains supplementary material, which is available to authorized users.
References
- 1.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 2.Klimek-Tomczak K, Wyrwicz LS, Jain S, Bomsztyk K, Ostrowski J. Characterization of hnRNP K protein-RNA interactions. J Mol Biol. 2004;342:1131–41. doi: 10.1016/j.jmb.2004.07.099. [DOI] [PubMed] [Google Scholar]
- 3.Mikula M, Dzwonek A, Karczmarski J, Rubel T, Dadlez M, Wyrwicz LS, et al. Landscape of the hnRNP K protein-protein interactome. Proteomics. 2006;6:2395–406. doi: 10.1002/pmic.200500632. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25:6436–53. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baber JL, Libutti D, Levens D, Tjandra N. High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J Mol Biol. 1999;289:949–62. doi: 10.1006/jmbi.1999.2818. [DOI] [PubMed] [Google Scholar]
- 6.Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, et al. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J Biol Chem. 1993;268:18249–58. [PubMed] [Google Scholar]
- 7.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–60. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie SA, Pasha MK, Batten DJ, Sharma RK, Olson DJ, Ross AR, et al. Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 2003;31:1502–13. doi: 10.1093/nar/gkg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 10.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3:325–30. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 11.Nagano K, Masters JR, Akpan A, Yang A, Corless S, Wood C, et al. Differential protein synthesis and expression levels in normal and neoplastic human prostate cells and their regulation by type I and II interferons. Oncogene. 2004;23:1693–703. doi: 10.1038/sj.onc.1207297. [DOI] [PubMed] [Google Scholar]
- 12.Klimek-Tomczak K, Mikula M, Dzwonek A, Paziewska A, Karczmarski J, Hennig E, et al. Editing of hnRNP K protein mRNA in colorectal adenocarcinoma and surrounding mucosa. Br J Cancer. 2006;94:586–92. doi: 10.1038/sj.bjc.6602938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostrowski J, Bomsztyk K. Nuclear shift of hnRNP K protein in neoplasms and other states of enhanced cell proliferation. Br J Cancer. 2003;89:1493–501. doi: 10.1038/sj.bjc.6601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatakeyama H, Kondo T, Fujii K, Nakanishi Y, Kato H, Fukuda S, et al. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics. 2006;6:6300–16. doi: 10.1002/pmic.200600488. [DOI] [PubMed] [Google Scholar]
- 15.Mandal M, Vadlamudi R, Nguyen D, Wang RA, Costa L, Bagheri-Yarmand R, et al. Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J Biol Chem. 2001;276:9699–704. doi: 10.1074/jbc.M008514200. [DOI] [PubMed] [Google Scholar]
- 16.Perrotti D, Neviani P. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res. 2007;13:1638–42. doi: 10.1158/1078-0432.CCR-06-2320. [DOI] [PubMed] [Google Scholar]
- 17.Zhou R, Shanas R, Nelson MA, Bhattacharyya A, Shi J. Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int J Cancer. 2009;126:395–404. doi: 10.1002/ijc.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carpenter B, McKay M, Dundas SR, Lawrie LC, Telfer C, Murray GI. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br J Cancer. 2006;95:921–7. doi: 10.1038/sj.bjc.6603349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–54. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelson MA, et al. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog. 2008;47:235–44. doi: 10.1002/mc.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dundas SR, Lawrie LC, Rooney PH, Murray GI. Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J Pathol. 2005;205:74–81. doi: 10.1002/path.1672. [DOI] [PubMed] [Google Scholar]
- 22.Shi J, Hershey JW, Nelson MA. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett. 2009;583:971–7. doi: 10.1016/j.febslet.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, et al. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–36. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- 24.Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW, et al. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem. 2003;278:5062–71. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- 25.Zhou R, Shanas R, Nelson MA, Bhattacharyya A, Shi J. Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int J Cancer. 2010;126:395–404. doi: 10.1002/ijc.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pino I, Pio R, Toledo G, Zabalegui N, Vicent S, Rey N, et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–43. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 27.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 28.Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Bomsztyk K. Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res. 2003;31:3954–62. doi: 10.1093/nar/gkg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miau LH, Chang CJ, Shen BJ, Tsai WH, Lee SC. Identification of heterogeneous nuclear ribonucleoprotein K (hnRNP K) as a repressor of C/EBPbeta-mediated gene activation. J Biol Chem. 1998;273:10784–91. doi: 10.1074/jbc.273.17.10784. [DOI] [PubMed] [Google Scholar]
- 30.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, et al. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–20. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 31.Notari M, Neviani P, Santhanam R, Blaser BW, Chang JS, Galietta A, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107:2507–16. doi: 10.1182/blood-2005-09-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem Biophys Res Commun. 2009;380:431–6. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonsson G, Bendahl PO, Sandberg T, Kurbasic A, Staaf J, Sunde L, et al. Mapping of a novel ocular and cutaneous malignant melanoma susceptibility locus to chromosome 9q21.32. J Natl Cancer Inst. 2005;97:1377–82. doi: 10.1093/jnci/dji280. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh P, Chin L. Genetics and genomics of melanoma. Expert Rev Dermatol. 2009;4:131–43. doi: 10.1586/edm.09.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


