Abstract
Spectacular increases in the quantity of sequence data genome have facilitated major advances in eukaryotic comparative genomics. By exploiting homology with classical model organisms, this makes possible predictions of pathways and cellular functions currently impossible to address in intractable organisms. Echoing realization that core metabolic processes were established very early following evolution of life on earth, it is now emerging that many eukaryotic cellular features, including the endomembrane system, are ancient and organized around near-universal principles. Rab proteins are key mediators of vesicle transport and specificity, and via the presence of multiple paralogues, alterations in interaction specificity and modification of pathways, contribute greatly to the evolution of complexity of membrane transport. Understanding system-level contributions of Rab proteins to evolutionary history provides insight into the multiple processes sculpting cellular transport pathways and the exciting challenges that we face in delving further into the origins of membrane trafficking specificity.
Keywords: Trafficking, Evolution, Eukaryogenesis, Rab protein, Systems biology
Introduction
One of the major evolutionary transitioning events was eukaryogenesis, the origin of the nucleated cell [1]. Internal compartments are rare in Archaea and Eubacteria, when present at all [2]. The increased complexity of cellular compartmentalization that occurred in nascent eukaryotic cells facilitated separation of major biochemical processes and allowed much finer control and sophistication. For example, the nuclear envelope separates transcription from translation, allowing increased complexity, in both regulation of protein biosynthesis/turnover and control of gene expression, to arise independently [3]. While the nucleus serves as both an evocative and essentially definitive example, eukaryotic cells possess numerous other autogenously derived membrane compartments that are diverse in structure and function. With the exception of the peroxisome, these compartments are principally involved in the biosynthesis, targeting, and turnover of surface molecular components, together with uptake and degradation of molecules and particles from the cellular environment, via endocytosis and phagocytosis respectively (Fig. 1). Collectively, this is the endomembrane system, and its origins and subsequent evolutionary history, distinct from the endosymbiotic mitochondrion and chloroplast organelles, may well be critical to our understanding of the emergence of the eukaryotic state. In this article we will review the evolution of the Rab GTPases, key regulators of organelle specificity, as a potent example from which to infer general mechanisms for the generation of increased organelle complexity and of regulatory control in autogenously derived eukaryotic compartments.
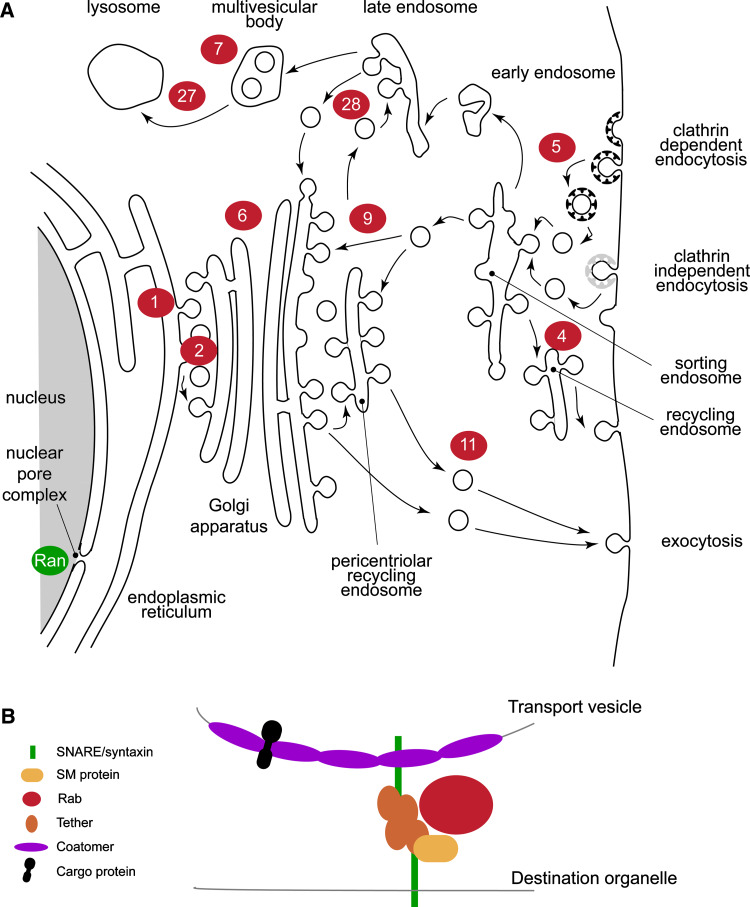
Fig. 1.
General features of the eukaryotic endomembrane system and specificity components. a Organelles and transport routes for major conserved pathways are shown, with Rab proteins known to mediate each step show as red lozenges. The related GTPase Ran is also shown as a green lozenge at the nuclear pore complex. Transport routes are indicated by arrows. Note that many Rabs, for example Rab11, participate extensively in transport routes and hence the localization of this protein is quite extensive. The figure is highly schematic. Endocytic coats are shown as black and gray ‘T’s. b Fusogenic apparatus of vesicle transport. The transport vesicle and destination organelle membranes must be brought into close proximity and fuse; this represents a considerable free energy barrier, which is surmounted by the fusogenic apparatus. Depicted are the minimal components, and many that are vital for targeting and fusion are omitted. However, at the center of this network sits the Rab protein that coordinates SNARE binding, cytoskeletal interactions, and other functions
Pathways and functions of the endomembrane system
Functionally, the endomembrane system can be subdivided into exocytic and endocytic systems. Exocytosis is the export of biosynthetic products to the surface, for secretion, or for population of endomembrane organelles. In all extant eukaryotes, the pathway initiates at the endoplasmic reticulum where the nascent polypeptide is imported into the ER [4]. Following a period of folding, post-translational modification, and quality control, mature proteins are exported from the ER in coated vesicles that assemble at specialized membrane subdomains or ER exit sites. Vesicles are delivered to the cis-face of the Golgi complex and cargo migrates through the multiple stacks of this organelle, to emerge at the trans-face. Transport between the ER and Golgi complex is mediated by several GTPases, Rab1 and Rab2 (Figs. 1, 2) which function to co-ordinate cytoskeletal interactions and specificity in fusion with membranes at the cis-Golgi [5].
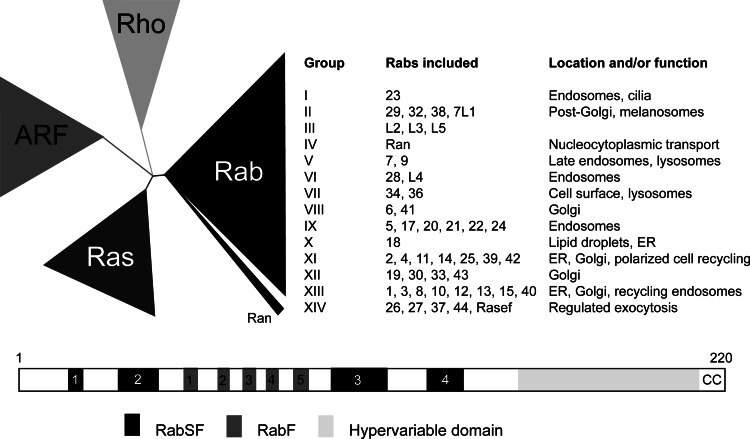
Fig. 2.
GTPase structure, evolution, and function. a Eukaryotic Ras-like GTPases are generally defined by membership to four superfamilies, Ras, Arf, Rho, and Rab/Ran. The latter family is the largest in most organisms, and is itself divisible into a number of subfamilies, ranging from 8 [41] to 14 [43]; phylogenetic reconstruction of this family is problematic. To some extent, the Rab subfamilies demonstrate related function between the members, but this is not always the case, and is also confounded by Rab protein participation in multiple interactions and pathways (figure adapted from [103] and [43]). b Ribbon representation of Rab GTPase emphasizing the positions of Rab family (RabF) and subfamily (RabSF) motifs. The protein is drawn with the N-terminus to the left. Most Rab GTPases are approximately 24 kDa in molecular weight, and comprise ~220 amino acids, as indicated. A total of five RabSF regions and four RabF regions have been described, and these correspond to surface loops involved in protein–protein interactions. The hypervariable region at the C-terminus is indicated as is the dicysteinyl prenylation signal, by ‘CC’
Exit from the Golgi apparatus can take one of several routes. Delivery to the cell surface requires incorporation into exocytic vesicles, which then fuse with the plasma membrane, releasing soluble contents to the exterior or membrane-bound proteins into the plasma membrane, involving Rab11 and related Rab proteins. A second regulated secretory route is mediated by stalling the progress of exocytic carriers; these vesicles will then fuse with the plasma membrane upon stimulation of the cell by a secretagogue. Multiple additional routes also originate at the trans-Golgi face, with destinations including sorting endosomes and the lysosome. Finally, there is also retrograde transport through the Golgi complex, which facilitates recovery and retrieval of proteins, both of which are mediated by Rab6 [6].
Similarly to post-Golgi transport, endocytosis includes several distinct pathways. The precise mechanism employed depends on the size of the object being taken up. Classically, endocytosis refers to small objects <~500 nm in diameter, while phagocytosis is reserved for larger cargo. Uptake from the surface depends on one of several coats, the best characterized of which are clathrin and caveolin. Endocytic cargo is rapidly delivered to Rab5-containing early endosomes [7], which rapidly maturate into sorting endosomes. Here material is either recycled, by a Rab4/Rab11-mediated pathway that intersects with the exocytic system, or is delivered via Rab7- and Rab9-mediated pathways to the lysosome via late endosomes and multivesicular bodies (MVBs). Excellent reviews describing the molecular machinery of endocytosis and exocytosis are available, to which the reader is referred for more details (e.g., [8–10]).
While we have described these transport pathways on a framework of Rab proteins, the various steps require participation of many additional proteins. Indeed, a general model of vesicle formation involving the Sar/Arf GTPases, cargo adaptors, and membrane-deformation complexes has been elegantly deduced, as has a robust model of vesicle fusion involving tethering proteins, coiled-coil SNAREs, and Rabs. Importantly, these models hold for all trafficking events at the various endomembrane organelles in the cell and, with the probable exception of the tether factors [11], the dominant proteins are all members of either small (SM proteins) or extensive (Rabs, SNAREs) paralogous families [12].
It is significant that intracellular trafficking pathways represent a considerable level of both complexity and diversity in terms of mechanism, function, and morphology. This diversity is evident both between individual species and lineages as well as cell types in multicellular organisms. Hence, sculpting the membrane trafficking system is likely most important for adaptation of unicellular organisms and division of labor in higher animals and plants. Together the endomembrane system accounts for ~10% of protein coding sequence of the eukaryotic genome, and while encompassing the paralogous Rab, ARF and SNARE families also extends to GTPase-activating proteins (GAPs), nucleotide exchange factors, and many others. Despite very clear structural and functional discrimination between exocytic and endocytic transport and the associated molecular machinery, considerable progress has been made in documenting the origins of these trafficking system components, how they interact to provide specificity, and the deeper origins of trafficking pathways.
Overview of evolution of the endomembrane system
It is now generally accepted that the major eukaryotic kingdoms or supergroups originated at least a billion years ago, and that a rapid radiation of lineages occurred at some point following this initial eukaryogenesis event. In the 1980s and 1990s, the prevailing paradigm [13] was that a small number of lineages (Archezoa) had evolved early and prior to the invention of key cellular attributes (i.e., introns, Golgi bodies, peroxisomes, and mitochondria). Such organisms, like Giardia, Trichomonas, Trypanosoma, or Entamoeba, were thus presumed to be cytologically simple due to retention of this primitive state. However, through a combination of more sophisticated molecular evolutionary models, improved taxonomic sampling and elegant molecular and cellular biology, this paradigm has been overturned [14]. Hypotheses identifying the earliest eukaryotes have come and gone and there is currently no robustly supported placement of the eukaryotic root [15], essentially dispending, for the time being, deductions of early/primitive versus advanced/higher/crown group eukaryotes.
What has emerged, directly from the huge increase in genome sequence data, is a robust picture of eukaryotic diversity, now subdivided into five or six supergroups [16, 17]. Animals and fungi are sister taxa within a single supergroup, a surprising relationship for many cell biologists. Significantly, the previously “primitive” Archezoa are spread throughout at least three supergroups. Despite the lack of a clear eukaryotic root, however, the relationships between the supergroups are increasingly well established [18, 19]. The topology makes reconstruction of the last eukaryotic common ancestor (LECA) conceptually clear; analysis of the presence of individual genes across eukaryotic lineages, or of entire paralogous families of genes, should reveal the point in evolution at which such genes arose. For example, if a gene is present in only a single supergroup, then the most likely interpretation is that the gene has arisen after that group diverged from the remaining supergroups. If, on the other hand, the gene is found in all supergroups, then the gene is inferred to have been present in the LECA. The intermediate case, i.e., where the gene of interest is only found in a subset of supergroups, may suggest either very early emergence post-LECA or secondary loss (Fig. 3).
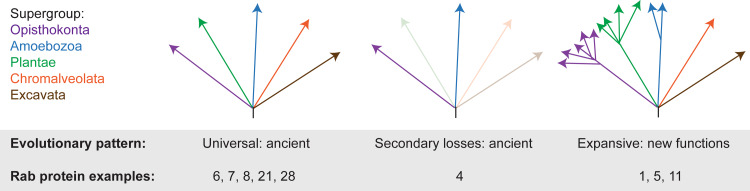
Fig. 3.
Evolutionary modes as inferred by comparative genomics and phylogeny. Five hypothetical extant eukaryotic supergroups are shown as arrows emanating from an unresolved node representing the last eukaryotic common ancestor (LECA). Left: universal representation of a gene in modern taxa is most consistent with an ancient origin at or before the LECA. Good examples of these types of genes are the core Rab groups, which are found in the vast majority of modern genomes and populate the basic exocytic and endocytic pathways. Center: when representation in taxa is sparse this may still be consistent with an ancient origin for the gene, but accompanied by frequent secondary loss from multiple taxa. Rab4 is a good example of this mode of evolution. The pattern may also indicate lineage-specific innovation, depending on the specific supergroups represented. Right: expansion of paralogues, leading to new functions. Importantly, this mode also implicates ancient origin for the primordial paralogue. A good example here is Rab5, which is clearly ancient due to near-universal representation, but is expanded in many lineages to a small paralogous family. This mode appears to be a dominant one in the generation of highly complex systems as seen in metazoa, plants, and several other taxa
Applying this logic to the extensive genomic data now available, it becomes clear that much of the basic machinery and the overall configuration of the endomembrane system is ancient. This encompasses the ER, Golgi complex, and major exocytic pathways, suggesting that the biosynthesis and exocytosis of secretory and surface molecules in the LECA was of similar, or even greater, complexity than many modern cells [12]. This paradigm is further supported by comparative genomics and biochemical studies, for example of N-glycosylation pathways comparing both the gene complements and enzymatic capacity of a wide range of organisms to build N-glycan precursors. As N-glycosylation plays a central role in protein folding and quality control, the finding that the core of the pathway is highly conserved across supergroups is excellent evidence for sophisticated ER function in LECA [20, 21]. Further, comparative genomic and reverse genetic studies of the proteins responsible for assisting in folding of polypeptides translocated into the ER lumen suggests that a complex repertoire of chaperones, protein disulphide isomerases, and quality-control factors are widely retained in modern lineages, once more reflecting a sophisticated system in the LECA [20, 22]. Coupling comparative gene representation analysis with phylogenetics has revealed that not only were SNAREs, many vesicle coats, SM proteins, and Rabs present in the LECA, but the major organelle or pathway specific family members were already differentiated ([12] inter alia). This provides further depth in understanding LECA, right down to the level of specific cellular compartments.
Additionally, while most analysis readily confirms evolutionary homology, direct experimental evidence has demonstrated that for most orthologues, similar roles are retained. For example, Rab proteins retain functions in the same intracellular transport steps in essentially all organisms where such definitions have been made; this has been done systematically with trypanosomes, members of the Excavata [23, 24] and extensively for higher plants, specifically Arabidopsis thaliana [25, 26]. Similar studies are underway for several other taxa.
Organellar paralogy; a model for novel compartment evolution
The last eukaryotic common ancestor appears to have possessed a significant diversity of organelles and transport pathways, defined by paralogous assemblies of proteins interacting to encode specificity [12]. Because many of these proteins are similarly ancient and diverse, we proposed that the complexity of endomembrane organelles could have arisen from a single internal membranous compartment by an interative process of gene duplication of specificity-encoding proteins, specialization of these duplicates, and co-evolution with additional members of the specificity module system [12, 27]. Significantly, the process of establishing the diverse compartments was likely achieved in two distinct stages. Prior to LECA, all major organelles were established together with the core gene families. Subsequently, many of these pathways became elaborated, specialized, or multiplexed, in a lineage-specific manner. Further, good examples of the evolution of (apparently) completely novel pathways, paralog expansions leading to multiple pathways, secondary losses and protein domain shuffling generating new functionality from pre-existing units are frequent.
For example, differential uptake of trans-membrane domain versus glycosylphosphatidylinositol (GPI)-anchored proteins by distinct routes may have differential aspects between metazoa and other taxa [24], and it is possible that metazoa are part way to evolution of genuinely distinct pathways. Specifically, GPI-anchored proteins associate with glycolipid/cholesterol-rich rafts, which is an ancient cellular feature [28–30]. In some cases, GPI-anchored proteins also associate with caveolae, flask-like plasma membrane structures requiring expression of caveolin; these latter are restricted to metazoa and are not essential for GPI-anchored protein endocytosis [31–33]. GPI-anchored proteins are endocytosed by multiple routes in mammalian cells, including conventional clathrin-dependant pathways, common with protozoa [31, 34], but in metazoa there are several novel endocytic pathways that are absent from most lineages. Therefore the multiple endocytic routes in metazoan cells offer a high level of complexity to GPI-endocytosis, and potential material for evolutionary exploitation.
A distinct example of lineage-specific evolution of new pathways by paralogous expansion is provided by Rab5. LECA probably possessed a single Rab5 gene but multiple Rab5 paralogs have arisen in a lineage-specific manner [27]. This has potentially dual significance. Firstly, division of labor between differentiated Rab5 pathways is not necessarily equivalent across the eukaryotes and the roles of distinct Rab5 paralogues cannot be predicted with precision. Secondly, the independent expansion of Rab5 on multiple occasions suggests a common response to evolutionary pressure and that, on multiple occasions since LECA, there has been selection for differentiation of early endocytic pathways.
Evolution of Rab GTPase sequences; loops and framework regions
Rab proteins constitute the largest subfamily of the Ras-related GTPase superfamily (Fig. 2). Members of this supergroup regulate a vast array of processes, including responses to external stimuli, cytoskeletal remodeling, and intracellular transport. In this last aspect, three GTPase subfamilies are essential participants; Arf, Ran, and Rab. Ran has a clearly distinct mechanism and function, lacking a membrane targeting domain and operating in the context of nucleocytoplasmic transport [35]. Both Arfs and Rabs are membrane localized by virtue of post-translational modification, myristoylation, and prenylation, respectively, and function in membrane transport; while there are clear differences in their roles, the biological reasons for this are unclear.
Generating a clear view of Rab sequence diversity and evolutionary history has been far from trivial. An early attempt to characterize Rab sequence diversity, predating the availability of complete genome data, exploited sequence heterogeneity within two surface loops, and were labeled as “specificity domains” [36]. Chimeric proteins incorporating sequence derived from Saccharomyces cerevisiae Ypt1, a Rab involved in ER to Golgi trafficking, and Sec4, which functions in exocytosis at the plasma membrane, were used to map sequences responsible for targeting and function. A hybrid with both specificity domains of Ypt1 in a Sec4 framework produced a protein that complements Ypt1, confirming that these regions contain information sufficient to specify function. Similar experiments assessed targeting of various Rab5-Rab6 hybrids to function as Rab5 [37]. Substitution of the Rab5 C-terminus into Rab6, which normally localizes to the Golgi apparatus, targeted the hybrid to early endosomes and the plasma membrane, but did not stimulate endocytosis. However, the addition of further Rab5 sequence corresponding to RabSF1, RabF4, and RabSF2, respectively, ([38] see below), produced a chimera that apparently had Rab5 function. This clearly implicated these new regions in interactions between Rabs and their effectors. Further, Rabenosyn-5, a Rab5 effector protein, contains two distinct sites that interact with a variety of Rabs, offering an opportunity to map sequences required for interaction. The binding affinities of these two regions, RbsnA and RbsnB, for over 30 Rabs was assessed [39], demonstrating differentiation in recognition between distinct Rab-Rabenosyn5 complexes as RbsnA selectively bound Rab4 and Rab14 but RbsnB bound Rab5, Rab22 and Rab24. Significantly, RbsnA and RbsnB share very similar helical hairpin structures that bind equivalent sites on their respective Rab proteins, explaining the dual interaction sites. Finally, three residues are key to the discrimination between Rab subsets and alteration of these residues in RbsnA to the equivalent in RbsnB could reverse specificity.
By definition, those regions that change conformation between the GDP- and GTP-bound states, the “switch regions”, are key to mediating effector-binding and function. Since Rabs all have a similar protein fold, with a six-stranded beta-sheet and five alpha-helices making up the core structure, the effector specificity would be expected to be largely restricted to surface loops [40]. However, some residues significant in binding Rab3A and an effector, Rabphilin, are conserved throughout the Rab family, making an understanding of specificity rather difficult. Interestingly, orientation of these residues is incompatible with involvement in binding in some Rab proteins, e.g., Rab5C and hence variability in orientation likely contributes to effector interaction specificity. These primarily empirical approaches highlight some of the complexity with assigning sequence to function within the Rab family, a prerequisite to understanding the evolutionary history.
Pereira-Leal and Seabra [38] analyzed the primary structures of a collection of mammalian Rab proteins in order to formulate a comprehensive definition of the Rab family within the broader superfamily of Ras GTPases. Motifs involved in nucleotide and Mg2+ binding, labeled G1-3 and PM1-3, respectively, were conserved on a superfamily level. Following alignment of known Rabs, five short regions (five or six residues) of consensus sequence unique to the Rab family were identified. These regions, labeled RabF1-5, map to the switch regions, with the implication that these sequences contribute to interaction with regulators/effectors distinguishing between GDP- and GTP-bound conformations.
The same approach was taken to defining subfamilies of Rab proteins. Following phylogenetic analysis of known mammalian Rabs, the aim was to make a useful distinction between isoforms (proteins with unusually high sequence homology and probably similar functions) and proteins that are simply closely related. Four regions, RabSF1-4, were identified to have significantly higher conservation within, rather than between, subfamilies. When mapped onto the 3D structure of the Rab3A-Rabphilin3A complex RabSF1, 3 and 4 corresponded to complementary-determining regions that form a binding pocket for the Rab3A effector, while RabSF2 mapped to the opposite surface of the Rab3A within the putative “switch I” region. The presence of two separate subfamily specific surfaces suggests that effector-Rab interactions are highly variable and employ multiple binding faces. This feature underscores both the flexibility and complexity of Rab interaction pathways.
Rab family evolution at the whole-genome level
With the availability of increased sequence data, the possibility of more insightful analysis of Rab evolution arose. Revisiting this question, Pereira-Leal and Seabra expanded analysis to include new data from Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens and Arabidopsis thaliana [41]. Phylogenetic analysis resulted in co-segregation of known orthologues, and specific conservation within RabSF regions was suggested as a mechanism for assigning putative orthologues and hence classification. However, assignment of orthologues in A. thaliana, essentially the only non-opisthokont included, was problematic due to extensive expansion and divergence within subclasses, which resulted in an alternate nomenclature [26]. More significantly, comparisons between different species demonstrated a non-linear relationship between cell number and the number of Rabs encoded in the genome, suggesting that expansion of the Rab repertoire correlates with multicellularity and/or tissue differentiation. Significantly, as the multicellular state has arisen on multiple occasions, it has been suggested that plasticity within the Rab family may be a facilitator, or is at least permissive, for multicellularity [41]. Further, higher-order grouping within the Rab subfamily was detected, predicting eight clades. Most importantly, a degree of functional relatedness was apparent for members of the same clade. For example, clade V contains Rab5 and Rab22, both of which are involved in endosome transport, while members of clade II, Rab2, 4, 14, 11, and 25, tend to be associated with endosomal recycling. Using principle component analysis, an additional two clades were identified, which again confirmed the concept that Rabs sharing similar functions and/or locations are more closely related than Rab proteins with distinct functions [42]. A subsequent analysis, using dendrogram sequence clustering has suggested a total of 14 Rab clades [43].
Detailed investigations of Rab clade I, as defined by Periera-Leal and Seabra [38], also suggest functional flexibility, even within restricted subfamilies [44]. Rabs within this group function to regulate ER to Golgi transport, and include metazoan Rab1 and Rab35, Ypt1 in S. cerevisiae and the RabD subgroup in A. thaliana. Additional paralogues arise through duplication and divergence, for example Rab35 in metazoa appears to have acquired a new function in endosomal recycling. Large expansions of Rab1 are present in Excavata and Amoebozoa, but better sampling is required to determine the points of duplication within recent evolutionary history. Rab1A in Plasmodium falciparum represents a unique paralogue; the location is consistent with a role early in the secretory pathway but several novel, specific substitutions have been identified that map to effector-binding sites, and thus may confer additional function [45].
Analysis of Rab clade VII, characterized by Rab7 and Rab9, demonstrated that Rab9 branched within the choanoflagellate/metazoan clade of Rab7A proteins and is restricted to Metazoa and Monosiga brevicollis [44]. Significantly, there is also evidence for multiple expansions and then subsequent loss for Rab7 and Rab9 genes, essentially retaining a small gene copy number. Evolution in both Clade I and VII demonstrates that frequent expansions occur and that these are often rapidly eliminated. However, frequent expansion clearly can provide regular opportunities for the evolution of novel paralogues and hence pathways, selecting for roles specific to tissue type, environmental conditions or life-cycle stage.
Significantly, a full analysis of Rab evolution, encompassing the complete diversity of the Eukaryota, remains to be achieved as the studies above have focused rather heavily on Opisthokonta and plants, neglecting the majority of taxa. Our own analysis in progress suggests that the phylogenetic history of Rab proteins is in fact rather more complex than previously suspected, with LECA possessing an elaborate repertoire of Rab proteins (M. Elias, M.C. Field, and J.B. Dacks, unpublished data). This implicates secondary loss as a major mechanism for sculpting of the endomembrane system.
Rab evolution across the supergroups
Focusing on a limited range of taxa, and specifically the fungi, an indication of how whole-genome complements of Rabs evolve has been deduced. Generally, the number of Rabs is very low in fungi, with little variation in the total family size between species [46]. Indeed 24 of 26 species analyzed encode between eight and 12 Rabs, with no apparent correlation between lifestyle/lineage and the number of Rabs present. There are six subfamilies present in all free-living fungi: Ypt1, Sec4, Ypt3, Ypt5, Ypt6, and Ypt7, and indeed C. albicans has no additional Rabs, but is still able to exist in yeast and hyphal forms. This minimal set represents massive loss from LECA ([46], M. Elias, M.C. Field, and J.B. Dacks, unpublished data). In addition, there are expansions of Ypt5 and Ypt3 within the fungal kingdom that are independent from the mammalian expansions. Despite relatively common duplications, secondary loss leads to maintenance of this relatively small set of Rabs. While the driving force for expansion of Rabs is usually assumed to be to increased functionality or flexibility, this is not apparent; for example Ypt31 and Ypt32 appear to be functional redundant. In summary, fungi appear able to support multicellularity and a saprophytic lifestyle with a surprisingly small number of Rabs.
In higher plants, and specifically the angiosperm A. thaliana, 57 Rabs forming eight clades have been identified [26]. The degree of divergence within these clades prevented putative orthologues being ascribed, suggesting considerable selective pressures propelling novel Rab evolution. Analysis of the RabSF regions allowed these eight groups to be further divided into 18 subclasses, many of which contain multiple isoforms, suggesting significant expansion of specific Rab subsets. For example A. thaliana RabA, corresponding to mammalian Rab11, contains six members, suggesting the evolution of multiple and distinct recycling pathways.
Entamoeba histolytica is an enteric protozoan parasite within the Amoebozoa and has over 90 Rab genes [47]. This extremely large Rab family within a unicellular organism runs counter to the paradigm that major increases in Rab number accompany multicellularity, and is nearly an order of magnitude greater than S. cerevisiae, despite the genome being only 1.6 times larger. Over 20 Rabs showed >40% sequence similarity with opisthokont Rabs, but with an unusually large number of isoforms, e.g., nine Rab7 paralogues. A further 30 Rabs showed >40% sequence similarity to other Rabs within the same genome, clustering into nine subgroups, while 39 showed <40% sequence similarity to human/yeast Rabs or other amoebic Rabs; this suggests a large degree of unique or lineage-specific Rab evolution in E. histolytica. Interestingly, ~20% of the predicted Rabs had significant regions deleted, yet still appear to be transcribed, while several have either novel or no prenylation signal, suggesting novel functions or mechanisms. Two of the Rab7 paralogues, EhRab7A and EhRab7B, have been investigated experimentally and EhRab7A functions at an earlier stage in lysosomal delivery than EhRab7B [47]. As most organisms, including multicellular taxa, possess only one or two Rab7 paralogues, this suggests an unprecedented level of sophistication in late endosomal trafficking for E. histolytica.
On account of their importance as disease agents, considerable attention has been devoted to the Chromalveolata, and the Apicomplexa in particular. The malaria parasite, Plasmodium falciparum, has 11 Rabs, a remarkably compact family for an organism with two hosts and multiple life-cycle stages with radically differing morphologies [48]. Ten are expressed in the erythrocytic stage, and PfRab11B, the sole exception may represent a stage-specific Rab. Comparisons between P. falciparum and additional apicomplexan taxa indicate that, as a group, these organisms have small numbers of Rab genes, with Cryptosporidium parvum at eight and Toxoplasma gondii with 15 [49]. The difference in repertoire size between T. gondii and C. parvum may relate to the presence of the parasitophorous vacuole membrane for T. gondii. Interestingly, PfRab5B is unusual, lacking a C-terminal prenylation motif, and is coordinately transcribed with PfRab2, PfRab6 and PfRab11B rather than with the other PfRab5 isoforms, suggesting possible functional coupling [48]. Additional studies demonstrate minimization of other components of the trafficking machinery in apicomplexan parasites [11, 50, 51], almost certainly due to specific secondary loss and hence sculpting, as other chromalveolates do not share this feature. For example Tetrahymena thermophila, also an alveolate, encodes ~70 Rab proteins, with clear patterns of both lineage-specific expansions and innovation [52].
The possibility that small Rab gene families are associated with parasitism is contradicted by E. histolytica and, spectacularly by Trichomonas vaginalis [53, 54]. T. vaginalis is an excavate protozoal parasite transmitted by sexual contact. It exists in both free-swimming trophozoite and amoebic forms, and possesses a massive Rab complement approaching 300 members. Many of these are transcribed, but only 14 formed clades with previously characterized sequences from opisthokonts or plants, the remaining forming unique clades. The unexpectedly large number of Rabs in T. vaginalis begs the question of what is driving Rab expansion in T. vaginalis and E. histolytica; neither the number of life-cycle stages, complexity of the endomembrane system, nor multicellularity provide a satisfactory explanation. Common features of T. vaginalis and E. histolytica include parasitism and amoebic life-cycle stages. However, the latter can also be discounted as Naegleria gruberi, another amoebic member of the Excavata comparatively closely related to T. vaginalis, possesses around 30 Rab genes [55], while Nagleria’s even closer relatives, T. brucei, T. cruzi, and Leishmania major, all parasitic excavates, possess ~20 Rab genes, most of which are clearly orthologous with metazoan Rabs [23, 56].
Interestingly, if the Rab proteins have evolved by a combination of a large LECA repertoire and lineage-specific scultping, the paralogous GTPase family involved in trafficking, the Arfs, arose almost exclusively by paralogous expansion following the LECA [56, 57]. This latter observation has at least two implications. Firstly, the precise functions of Arf proteins are likely unique in the different supergroups, exemplified by the rather distinct phenotypes obtained with genetic analysis of Arf function in, for example, S. cerevisiae, plants and trypanosomes [58–61]. Secondly, it is unlikely that Arf proteins per se played a significant role in the initial laying out of the endomembrane system [58–61], potentially implying that this role was taken on by the Rab proteins.
Evolution of Rab membrane targeting specificity
The initial delivery of Rabs to membrane is accompanied by post-translational modifications, specifically the addition of a prenyl group or groups to the C-terminus, proteolytic processing, and carboxymethylation [62]. Prenylation takes place on the SH groups of cysteine residues in the carboxy-terminal prenylation signal, and is essential for membrane targeting [41, 63]. The prenylation signal (trivially CAAX box) for Rabs tends to include two cysteine residues, for example XXCC or XCXC, resulting in double prenylation, and given the hydrophobicity of prenyl-moieties, this results in rather stable association with lipid bilayers. Not all Rabs, however, are dually prenylated, and the functional basis behind these differential modifications is unclear. Rabs are modified by a specific Rab-type geranylgeranyltransferase (RGGTase) that catalyses addition of geranylgeranitol (C20), and which differs from the other prenyltransferases (farnesyltransferase (FTase) and geranylgeranyltransferase I (GGTase I)) by a requirement for Rab escort protein (REP) [41]. All prenyltransferases share structurally related α- and β-subunits, and the common origin is exemplified by the α-subunit of FTase and GGTase I, which is the same gene product. For modification to occur, a complex of Rab, REP and RabGGTase must form (Fig. 4). REP also acts as a chaperone for the prenylated Rab protein prior to membrane delivery, preventing it from aggregating in solution. It has proved difficult to determine the crystal structure of this complex, but it was possible to analyze independently the binary complexes of REP1-RabGGTase and Rab7-REP1 [64]. A second cycle concerns extraction of Rab proteins from the target membrane and their recycling; again this is made more complex due to the presence of the highly hydrophobic prenyl groups, but is facilitated by guanine dissociation inhibitor (GDI), which preferentially binds GDP-Rab, secludes the prenyl groups away from the aqueous environment, rendering the Rab soluble, and also inhibits GDP to GTP exchange.
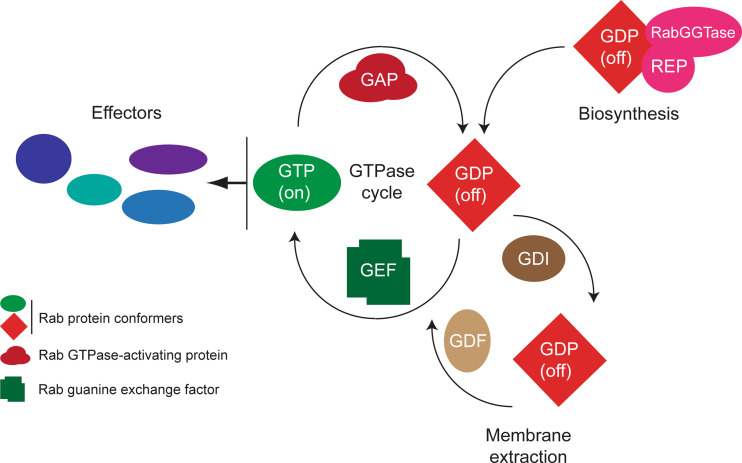
Fig. 4.
Basic functional cycles of Rab proteins. The core Rab function is the GTPase cycle; this process serves to elicit a conformational switch, which importantly regulates interaction with effector molecules (blue lozenges), principally in the GTP-bound (or active ‘on’ state). Switching between these states involves the action of GTPase-activating proteins (GAPs) or guanine nucleotide exchange factors (GEFs), which accelerate the intrinsic hydrolytic activity and nucleotide exchange reactions by several orders of magnitude. A second important cycle is the relocation of the Rab protein following vesicle fusion at the target membrane; usually the Rab is in the GDP, or ‘off’ conformation, which allows specific interactions with a soluble guanine-dissociation inhibitor (GDI) that is able to both sequester the C-terminal prenyl moiety and solubilize the Rab protein. Reintegration into the donor membrane compartment requires connection with a GDI-displacement factor (GDF) and also a GEF, to restore the molecule to the GTP-bound state. Finally, initial targeting following biosynthesis requires insertion utilizing Rab escort protein (REP). Significantly, there are a great number of effector molecules, facilitating a disseminative mode of action for Rab proteins. In most organisms, a similar number of GAPs, GEFs, and Rabs are found in the genome, suggesting an intimate relationship between the GTPase and the factors facilitating the GTPase cycle, while there are usually few GDIs and a single REP
The RabGGTase α-subunit interacts with domain II of REP1 to form the complex. Since the LRR and Ig-like domains of the α-subunit are not present in the structurally similar GGTaseI and FT, it was predicted that they would be significant in the interaction between RabGGTase and REP1—this, however, turned out not to be the case. Likewise, since GDI (similar in structure to REP1) has low affinity for RabGGTase, it was thought that structures which are absent from GDI but present in REP would be involved in the interaction—again, this was not seen. Rather it is the presence of key residues (F279 and R290) specific to REP that are responsible for the differing affinities [64–66].
The interface between Rab7 and REP1 has also been analyzed with reference to the structurally similar Ypt1-GDI complex [67]. REP and GDI perform similar roles in the broadest sense that both proteins deliver Rab to a target membrane. The crystal structure of the Ypt1-GDI complex reveals that the Rab switch regions are involved in interaction. Further, a lipid-binding pocket is present within domain II, composed of helices D, E, F, and H, and occupied by the geranylgeranyl group of Ypt1. A site in domain I had previously been identified as a putative lipid-binding site, but is now hypothesized to interact transiently during Rab retrieval. Structural analysis of the Rab7-REP1 complex revealed a strikingly similar interaction with a hydrophobic tunnel formed by α-helices structurally homologous to D, E, F, and H helices of GDI and occupied by the geranylgeranyl group. Further, this hydrophobic tunnel was only partially open when the structure of unprenylated-Rab7-REP was analyzed; the change in conformation to open the hydrophobic tunnel results in the displacement of the key residue F279, thus weakening the interaction with RabGGTase. Clearly, the similarity of the REP and GDP-binding interface suggests retention of an ancestral-binding mode.
Evolution of Rab prenylation has been investigated in some detail, and appears to be comparatively simple. The system is ancient, and RGGTase, FTase, and GGTase I, were probably present in LECA [68]. The potential for some flexibility within the prenyltransferase family has been noted as the α-subunits contain a crescent-shaped, double-layered, right-hand superhelix topology, which wraps around the β-subunit. Indels are present in some of the linkers between each helix, but there is some constraint. The similarity of this structure to the karyopherins, which also bind to small GTPases, may be more than coincidence [69]. The RGGTase of some lineages possesses an additional leucine-rich repeat (LRR) domain, which was probably present in LECA but has been lost on multiple occasions; neither the function nor the reason for this are known [68]. Only rarely are paralogous expansions of RGGTase subunits observed, suggesting that one prenyltransferase is sufficient for recognition of the entire repertoire of Rab proteins in most organisms.
Rab escort protein belongs to the same protein family as GDI, and therefore shares a common, possibly bi-functional, ancestor [70]. The conserved architecture is present as two domains, with the N-terminal domain containing the Rab-binding site. The REP C-terminal domain mediates binding to the α-subunit of RGGTase. Importantly, the mode of recognition of Rabs is common between REP and GDI and both retain sequence-conserved regions that mediate this interaction. Again, REP appears able to accommodate insertions in selected sites in different lineages, but the functional consequences are somewhat unclear. Interestingly, metazoa have two REP genes [68], and there appears to be some substrate specificity [71]. If this is simply to accommodate an expanded metazoan Rab repertoire, or has some more specific consequence, is still unclear. Overall, it is clear that the system for prenylation, delivery, and extraction of Rab proteins from membranes has both an ancient basis and is rather non-specific in terms of precise sequence recognition.
The mechanisms by which Rab proteins achieve their characteristic subcellular distributions are complex and incompletely understood. Clearly, this represents an important and exciting challenge to address for increasing understanding of both the function and evolution of membrane-trafficking specificity. Various studies highlighted the hypervariable C-terminal domain in targeting. For example, Rab5 is targeted to early endosomes and Rab7 to late endosomes, and these proteins contain CCSN and CSC prenylation motifs, respectively. Although prenylation is required for membrane association, the motif alone carries no targeting information as Rab5 containing a CSC Rab7 prenylation motif localized to its original site. However, a chimera where the 34 C-terminal residues of Rab7 replace the Rab5 C-terminus targets the chimera to late endosomes [72]. However, subsequent studies suggest that the targeting mechanism is more complex [73] and a panel of chimeras produced through reciprocal sequence exchange between Rabs from different parts of the endomembrane system indicate that the hypervariable regions is not always sufficient. For example, Rab27A with the Rab5A C-terminus retains the original Rab27A localization and function. Rab5 orthologues from divergent organisms (trypanosomes) with little C-terminus sequence conservation were directed to the correct endosomal location. Further, RabSF4 and RabSF3 contribute to Rab5A targeting and RabSF2 and RabSF3 to Rab27A targeting. As these regions are involved in effector binding, this implicates effector association as a targeting mechanism, and the possibility that localization depends on cooperative interaction with multiple partners. For example, there is evidence that TIP47, a Rab9 effector, is important in the localization of Rab9 [74]. The dominant sites may not be the same for all Rab subfamilies, such that simple analysis of sequence alone is, in some cases, insufficient for prediction of location or function. The Rab hypervariable C-terminus also contains a hydrophobic motif that varies in both position and composition. This region interacts with a hydrophobic region called the C-terminal binding region (GDI/REP domain I) while the “mobile effector loop” (domain II), which is required for generic targeting function, is also closely associated and may contribute to the specific interaction with receptor proteins (GDFs) at target membranes.
The task of unpicking the details of Rab targeting remains. Receptor molecules are thought to be involved and indeed certain membrane-bound factors have been identified. These are referred to as GDI-displacement factors (GDFs) and show distinct subcellular localization. However, there are currently no data regarding the mechanism of recognition or lipid transfer. Targeting is likely to involve such receptors but there may also be a role for GEFs, cascade-like mechanisms, luminal content, and a plethora of Rab-effector interactions. That specificity is likely not simply encoded in primary sequence, but is instead based on dynamic interactions with co-expressed binding partners, and has several implications for understanding the evolution of membrane-trafficking. Most simply, it means that we must integrate multiple proteins into evolutionary schemas [27]. It means that we need to start looking at transcriptomic and interaction data for trends of conservation, not merely comparative lists of components. These are challenges, but exciting ones to take on as we move forward. It also means that we can begin developing and incorporating ideas about how these interaction networks would have evolved and shaped the regulation and function of the membrane-trafficking system.
Evolution of pathways and connectivity; making connections
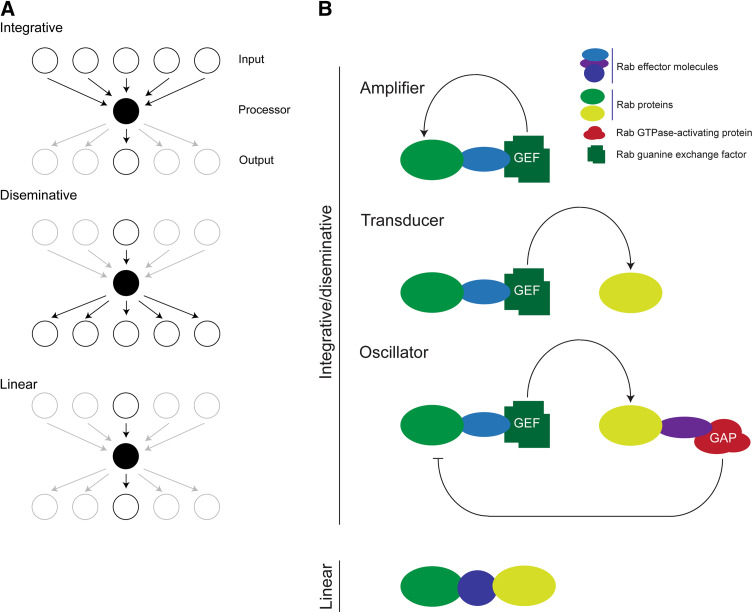
Very few proteins function in isolation, and most operate in the context of complexes. Such complexes may be stable, essentially representing the entire population of a given protein, or highly transient, so that a protein spends little of its time in such association. The interactions between proteins serve to control activity, location, stability, and specificity, and all represent points at which evolutionary selective pressure can operate. Depending on the topology of a pathway, i.e., the interaction network, information may be aggregated or disseminated (Fig. 5). For example, a protein that interacts with a large number of upstream factors, but a smaller population of downstream ones serves to aggregate biological information, but a protein where the opposite is the case disseminates. Changes in either the topology or the identity of interaction partners can greatly influence cellular behavior, and the remarkable flexibility of small GTPase pathways probably represents an evolutionary advance facilitating the emergence of multiple new functions (Fig. 5). All GTPases participate in a GTP cycle, and which, due to very slow intrinsic hydrolytic and GDP/GTP exchange rates, is normally assisted by several cofactors (Fig. 4). Principally, a GTPase can be considered in an active state in the GTP form (green oval in Fig. 4), and can interact specifically with effector proteins (blue ovals); for Rabs these include SNARE complexes, SM proteins and many others [37, 75, 76]. Indeed, Rab5 in mammals possesses up to 50 effectors, a bewildering array of potential downstream targets [77–81]. The GTPase is inactivated by a GTPase-activating protein, or GAP (dull red blob in Fig. 4), converting the protein to the GDP form. GAPs accelerate the rate of GTP hydrolysis by two or more orders of magnitude. Reactivation is achieved by nucleotide exchange, and again the rate here is vastly accelerated by guanine nucleotide exchange factors, or GEFs. Unlike Rab GAPs, this family is rather complex and they are not defined by possession of a single domain.
Fig. 5.
Functional changes by alterations to GTPase pathway topology. a Three hypothetical network topologies are presented, focusing on a central processor node (black), with input and output layers shown as open circles above and below the processor layer, respectively. Active interactions between factors are shown by arrows, inactive nodes and interactions are shown shaded. Top: an integrative network, where the central processor receives input from multiple nodes, but is tightly coupled to a single output node. Center: a disseminative network, where a single input interaction facilitates the processor interacting with multiple output nodes. Lower: a simple linear network where the processor is restricted to interactions with a single input and output node. b Examples of networks that involve Rab proteins. Top: integrative and disseminative networks. Uppermost scheme shows an amplification circuit where the Rab (green lozenge) interacts via an effector (blue lozenge) with a protein that has GEF activity, maintaining the Rab in the GTP-bound, or on, state. Middle scheme shows a transduction circuit, whereby the Rab effector interacts with a different G protein (light green lozenge) transferring information along a chain. Lower scheme shows a negative feedback loop or oscillator, whereby a second Rab protein recruits, via an effector, a GAP protein for the first Rab, thereby shutting off the pathway. Critically, the addition of further components facilitates increased complexity within the signaling pathway, and the circuits have been drawn to emphasize that additional complexity may be added incrementally. At the bottom, a linear pathway is created when two Rab proteins bind to the same effector. Depending on other criteria, this pathway topology may also function to integrate distinct Rab functions in a co-ordinate manner
Rab proteins represent a potent example of GTPase flexibility and adaptability and their range of potential interactions is very great indeed. The Rab-interacting proteins which are best characterized include their GAPs and GEFs, but also a range of proteins termed effectors specifically interact with the GTP-bound form; these encompass sorting adaptors, kinases, phosphatases, GAPs, GDIs, and cytoskeletal motors (Fig. 4). The interaction is conformation-specific; the vast majority of known interactions are with the GTP form, and these are defined as effectors as the GTPase is considered to be active, and transducing information, and hence act as sensors to GTPase activation. A minority of interaction partners have been described that are specific for the GDP conformer, although at present little is known about this class of Rab interaction molecules [82]. While central to Rab function, the range of effectors clearly provides extensive material for evolution of novel functions and specificity.
A number of Rab interactions have been identified that conceptually are analogous to electronic logic circuits, and provide interesting potential mechanisms for the evolution of specificity; empirical studies confirm that this view probably reflects in vivo mechanisms of action of Rab proteins and hence has functional relevance [83]. Examples include amplifiers, whereby a Rab protein activates a GEF, which then acts on the original Rab (Fig. 5b, top), while simple sharing of effectors, e.g., both Rab5 and Rab4 bind rabenosyn 5, while Rab4 effector GRASP1 likely couples to Rab11 via syntaxn 13 [84] is common and may also act as an integrator (Fig. 5b, bottom). There are also several examples of Rab-modulating activities embedded within tether complexes, for example the HOPS complex is a GEF for Rab7, and is itself activated by interaction with Rab5, while conversely Rab7 can inactivate Rab5 by activating a GAP protein, essentially a transducer and oscillator circuit [83, 85], while cascade mechanisms have been suggested to define subcompartmental boundaries [86]. The presence of GEF activity in TRAPP, a second tethering complex, suggests that GEF activity may be a general tether feature, providing an interesting mechanism for the coordination of function, and hence the evolution of new pathways. This, coupled with the close physical proximity of many Rab proteins on various membrane, for example Rab4, 5, and 11 on endosomes and Rab1 and Rab2 in the early exocytic systems [83, 87, 88] suggests that such coupling may be very common indeed. Further, the Rab system does not function in isolation and is interconnected to additional signaling pathways, for example Rho [89]. However, at present, our knowledge of the evolution of such aspects remains very incomplete. Sculpting of these systems may lead to alterations in the pathways, however, for example metazoan Rab4 clearly mediates recycling pathways [90] but the trypanosome orthologue participates more in lysosomal delivery than recycling [91, 92].
The Rab GAP family is dominated by TBC (tre-2/USP6, BUB2, cdc16)-domain proteins, which account for ~90% of known Rab-GAPs [93]. While a minority are not part of the TBC family, the predominance of the TBC domain allows an accurate estimate of RabGAP family numbers; the family is usually similar, or slightly smaller than the number of Rab proteins encoded in the genome (Gabernet-Castello, Dacks and Field, unpublished data). While the specificity of most TBC GAPs remains to be defined, studies in S. cerevisiae indicate a degree of promiscuity [94, 95], raising the issue of how activity is regulated, and if there is a mechanism for the coordination of TBC GAP activity against subsets of Rab proteins; if such exists it is not clear, and certainly beyond present in silico predictive capabilities. Further, some TBC domain proteins can function as GAPs for non-Rab GTPases.
At a simplistic level, the evolution of Rabs and TBC GAPs might be expected to be one of close co-evolution, as the tally for both factors appears to track across genomes and Rab families of many sizes, i.e., Rab families of 6 to ~70 members (Gabernet-Castello, Dacks and Field, unpublished data). In organisms with extremely large Rab families, Trichomonas vaginalis and Entamoeba histolytica, this correlation breaks down, and there are fewer TBC genes than Rab genes. Therefore, for the most part, orthologous pairs of Rabs and TBC GAPs could be expected to share the same functions, while the exceptional genomes with expanded Rab families may reflect promiscuity between TBCs and the large cohort of Rabs. As the highly expanded Rab family of T. vaginalis is structured into small clades, or clusters, this may suggest that a single TBC acts as GAP for all Rabs in such clusters [54]. However, experimental evidence for this is scant, and a systematic analysis of TBC evolution and family structure has only just been achieved (Gabernet-Castello, Dacks and Field, unpublished data). This analysis shows that innovation of TBC GAPs is complex, with a large cohort present in the LECA, but that clear lineage-specific innovations postdate the LECA. However, while this analysis is a step forward, and facilitates phylogenetically based investigations of TBC GAP function, there remains a major confounding issue as most TBC-containing proteins also possess additional domains, many of which have important functions. When comparing Rab GAP proteins between highly divergent taxa, despite being able to reconstruct evolutionary history for the TBC domains, there is clear evidence for extensive domain swapping, insertion, and deletion. This in turn has an obvious influence on TBC evolutionary trajectory and function for the corresponding trafficking pathways that the TBC GAP controls (Gabernet-Castello, Dacks and Field, unpublished data). Such complexity is challenging to chart and is not predictable at this time as the datasets available are too few and the underlying principles, if any, poorly defined. This is most definitely an area where further work is urgently needed, and represents a challenge to both computational and experimental biologists.
In terms of effectors, it is even less clear how specificity has arisen, and this is compounded by multiple binding modes, roles in disparate cellular systems, and the involvement of nonparalogous protein families. For example, Rab proteins may interact directly with cargo receptors (Rab9 with TIP47, Rab5 with adaptins and clathrin during coated pit formation) [96], act on vesicle coat removal, or provide a bridge to cytoskeletal elements, either indirectly (Rab27 with myosin Va via melanophillin) [97] or directly (Rab6 with KIF20A, a kinesin). Interactions with coiled-coil proteins mediating vesicle fusion and targeting are known for several Rabs (Rab11 and the FIP family proteins, Rab5 and EEA1) [78, 98, 99]. Multiple interactions with SNARE proteins, utilizing several mechanisms, have also been reported to operate either via the SM proteins (Rab5 via rabenosyn5 to Vps45 or Ypt1 direct with Sly1) or coiled-coil proteins, (Rab5 to SynE via EEA1) (see [37], for recent review). What many of these interactions do share is a coiled-coil architecture, which further confounds in silico prediction attempts, and together with the inherent errors in many cell biological studies of interaction networks, attempting to identify a common theme or unifying principle has failed so far (Fig. 6).
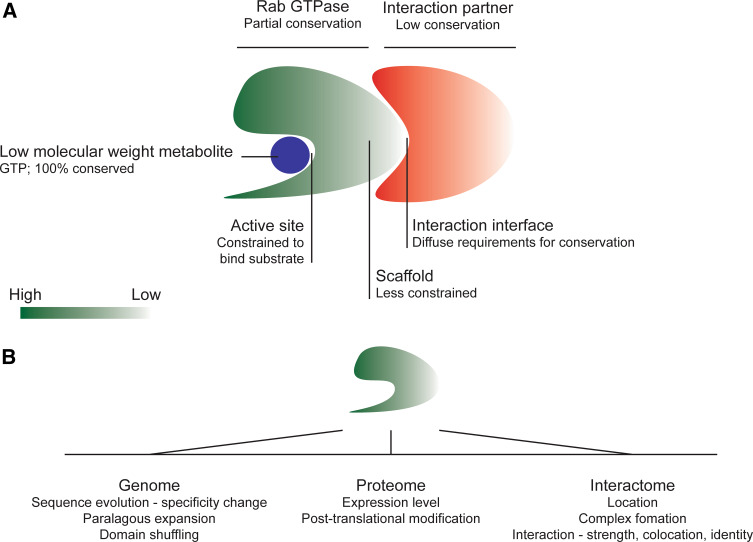
Fig. 6.
Evolutionary pressures and constraints. a Different aspects of a small GTPase and interactions with effectors or other proteins modulating function are differentially constrained. At the most basic level, Rab proteins are nucleosidases, and this constraint is reflected in strong conservation of the framework regions that mediate GTP binding and hydrolytic function. Importantly, as GTP is invariant, the selective pressure is very strong. Other regions of the molecule suffer less severe constraints, which is reflected in the variability in many surface loops of the Rab family, many of which offer binding sites for interaction partners. As these interaction sites may vary, coevolution between Rab and effector is important, but sequence conservation is less so, and regions not involved in such interaction may be subject to even weaker pressure, resulting in increased divergence at the sequence level and extreme difficulty in identification in silico. b Potential selective pressures operating on Rab proteins at the genomic, proteomic, and interactome levels. Additional feedback and constraints operating cooperatively between these various facets of functional evolution make the network of pressures highly complex and challenging to predict
A more recent attempt to examine effectors in a systematic manner (Rodriguez, Tavares-Cadete, and Pereira-Leal, pers. comm.) highlights this issue rather well. In this analysis, the study was restricted to two well-characterized opisthokonts, Homo sapiens and S. cerevisiae. Even across a restricted branch of the Eukaryota, their major findings were that Rab effectors are not conserved between species, and seem to rarely arise by paralogous expansion; while this also may reflect undersampling, it is unlikely that this can fully explain the result (Fig. 6). Hence this analysis sets the effectors apart from the TBC GAPs, the latter being a clear example of paralogous expansion, albeit with the remaining uncertainty on coevolution and specificity.
Regardless of the high degree of diversity within the effectors, there does exist a clearly conserved core of Rab-interacting proteins, including the prenylation machinery. Recent evidence suggests that a small number of factors are sufficient to mediate the most central fusogenic functions of Rab proteins [100]. This study also indicated that full functionality requires cooperative interactions between Rabs, Rab effectors and SNARE proteins, and even then the kinetics of membrane fusion are considerably lower for this cell-free system than in vivo. Further, a comparison between Rab11-interaction networks from Opisthokonta (principally Metazoa) and T. brucei, an excavate, does illustrate that some effectors are conserved (Gabernet-Castello and Field, unpublished data). Specifically, it was found that Rab11 interaction with Sec15 is present in both metazoan cells and T. brucei. Further, while FIPs are restricted to metazoa, a novel coiled-coil protein, TbRBP74, was identified as a Rab11 interaction partner in trypanosomes. Similarly to FIPs, TbRBP74 is a homodimer and binds exclusively to the GTP-bound form of Rab11, suggesting a similar mechanism and architecture for these two groups of proteins. If this represents some evolutionary architectural restriction is unclear, but the high number of coiled-coil proteins participating in Rab-mediated activities suggests a favored configuration, and certainly provides some promise that prediction of function may become possible. The exciting consequence of such predictions is that in silico reconstruction of transport pathways in experimentally nontractable organisms would become possible, providing a much expanded view of how endomembrane transport has been sculpted across the eukaryotes, as well as providing predictive power for understanding the cell biology.
Conclusions
Drawing together comparative genomic data with biochemical and cell biological evidence and examining it in an evolutionary framework has proven remarkably powerful. Here we have highlighted perhaps the best characterized example—the Rab and Rab-associated machinery. Hopefully, future considerations of a similar scope will be possible for additional aspects of the membrane-trafficking machinery. Using this approach, we have seen our evolutionary understanding of the membrane-trafficking system progress from essentially a side-note in explanations for other organelles [101], to increasingly detailed reconstructions of the machinery in the last common ancestor [12, 102], to drawing mechanistic inferences of how non-endosymbiotic organelles arose and evolve [27]. The latter allows (and indeed necessitates) incorporation of system-level data encompassing not only the list of which components are present but how they interact and respond to different evolutionary challenges.
In common with other cellular systems, selective pressures operate on multiple aspects of the intracellular trafficking system, for the creation of new pathways, modulated specificity and novel functions (Fig. 3). There have been considerable advances in this field recently, with the availability of new genomic data and improved analytical methods; with falling sequencing costs this is a very exciting time for evolutionary molecular biology. Coupling detailed knowledge of trafficking systems derived from classical model organisms, together with studies in representative taxa from several supergroups, is making possible an improved view of the range of evolutionary innovations after the LECA. Critically, the development of several non-opisthokont organisms as models provides an opportunity to analyze such innovations in a functional context. A vital next step must be the characterization and prediction of interaction networks; the combination of diverse mechanisms of molecular evolution, large families, incomplete datasets, and considerable noise within the data we have to hand makes this a formidable challenge, but one that is essential to providing the insight into the way that specificity and functionality within the endomembrane system has arisen.
Acknowledgments
We are very grateful to our funding agencies, principally the Wellcome Trust and an NSERC Discovery grant to JBD who have made this work possible. We also thank members of our laboratories and colleagues who have contributed unpublished data or insights.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Maynard Smith J, Szathmáry E. The major transitions in evolution. Oxford, UK: Oxford University Press; 1995. [Google Scholar]
- 2.Fuerst JA. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 3.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 4.Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 6.Barr FA, Short B. Golgins in the structure and dynamics of the Golgi apparatus. Curr Opin Cell Biol. 2003;15:405–413. doi: 10.1016/S0955-0674(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 8.Scita G, Di Fiore PP. The endocytic matrix. Nature 463:464–473 [DOI] [PubMed]
- 9.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 10.Jackson CL. Mechanisms of transport through the Golgi complex. J Cell Sci. 2009;122:443–452. doi: 10.1242/jcs.032581. [DOI] [PubMed] [Google Scholar]
- 11.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T. Eukaryotes with no mitochondria. Nature. 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 14.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 15.Roger AJ, Simpson AG. Evolution: revisiting the root of the eukaryote tree. Curr Biol. 2009;19:R165–R167. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 16.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 17.Burki F, Inagaki Y, Brate J, Archibald JM, Keeling PJ, Cavalier-Smith T, Sakaguchi M, Hashimoto T, Horak A, Kumar S, Klaveness D, Jakobsen KS, Pawlowski J, Shalchian-Tabrizi K. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field MC, Sergeenko T, Wang YN, Bohm S, Carrington M. Chaperone requirements for biosynthesis of the trypanosome variant surface glycoprotein. PLoS ONE 5:e8468 [DOI] [PMC free article] [PubMed]
- 23.Ackers JP, Dhir V, Field MC. A bioinformatic analysis of the RAB genes of Trypanosoma brucei . Mol Biochem Parasitol. 2005;141:89–97. doi: 10.1016/j.molbiopara.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Field MC, Natesan SK, Gabernet-Castello C, Koumandou VL. Intracellular trafficking in the trypanosomatids. Traffic. 2007;8:629–639. doi: 10.1111/j.1600-0854.2007.00558.x. [DOI] [PubMed] [Google Scholar]
- 25.Ebine K, Ueda T. Unique mechanism of plant endocytic/vacuolar transport pathways. J Plant Res. 2009;122:21–30. doi: 10.1007/s10265-008-0200-x. [DOI] [PubMed] [Google Scholar]
- 26.Rutherford S, Moore I. The Arabidopsis Rab GTPase family: another enigma variation. Curr Opin Plant Biol. 2002;5:518–528. doi: 10.1016/S1369-5266(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 27.Dacks JB, Poon PP, Field MC. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci USA. 2008;105:588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denny PW, Field MC, Smith DF. GPI-anchored proteins and glycoconjugates segregate into lipid rafts in Kinetoplastida. FEBS Lett. 2001;491:148–153. doi: 10.1016/S0014-5793(01)02172-X. [DOI] [PubMed] [Google Scholar]
- 29.Kierszniowska S, Seiwert B, Schulze WX. Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol Cell Proteomics. 2009;8:612–623. doi: 10.1074/mcp.M800346-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsui K, Hatakeyama K, Matsushita M, Kanazawa H. Saccharomyces cerevisiae Na +/H + antiporter Nha1p associates with lipid rafts and requires sphingolipid for stable localization to the plasma membrane. J Biochem. 2009;145:709–720. doi: 10.1093/jb/mvp032. [DOI] [PubMed] [Google Scholar]
- 31.Lakhan SE, Sabharanjak S, De A. Endocytosis of glycosylphosphatidylinositol-anchored proteins. J Biomed Sci. 2009;16:93. doi: 10.1186/1423-0127-16-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- 33.Nichols B. Caveosomes and endocytosis of lipid rafts. J Cell Sci. 2003;116:4707–4714. doi: 10.1242/jcs.00840. [DOI] [PubMed] [Google Scholar]
- 34.Allen CL, Goulding D, Field MC. Clathrin-mediated endocytosis is essential in Trypanosoma brucei . EMBO J. 2003;22:4991–5002. doi: 10.1093/emboj/cdg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 36.Dunn B, Stearns T, Botstein D. Specificity domains distinguish the Ras-related GTPases Ypt1 and Sec4. Nature. 1993;362:563–565. doi: 10.1038/362563a0. [DOI] [PubMed] [Google Scholar]
- 37.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 38.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 39.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeffer S. A model for Rab GTPase localization. Biochem Soc Trans. 2005;33:627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 41.Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 42.Buvelot Frei S, Rahl PB, Nussbaum M, Briggs BJ, Calero M, Janeczko S, Regan AD, Chen CZ, Barral Y, Whittaker GR, Collins RN. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol Cell Biol. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120:3905–3910. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 44.Mackiewicz P, Wyroba E. Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol Biol. 2009;9:101. doi: 10.1186/1471-2148-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elias M, Patron NJ, Keeling PJ. The RAB family GTPase Rab1A from Plasmodium falciparum defines a unique paralog shared by chromalveolates and rhizaria. J Eukaryot Microbiol. 2009;56:348–356. doi: 10.1111/j.1550-7408.2009.00408.x. [DOI] [PubMed] [Google Scholar]
- 46.Pereira-Leal JB. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic. 2008;9:27–38. doi: 10.1111/j.1600-0854.2007.00667.x. [DOI] [PubMed] [Google Scholar]
- 47.Saito-Nakano Y, Mitra BN, Nakada-Tsukui K, Sato D, Nozaki T. Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica . Cell Microbiol. 2007;9:1796–1808. doi: 10.1111/j.1462-5822.2007.00915.x. [DOI] [PubMed] [Google Scholar]
- 48.Quevillon E, Spielmann T, Brahimi K, Chattopadhyay D, Yeramian E, Langsley G. The Plasmodium falciparum family of Rab GTPases. Gene. 2003;306:13–25. doi: 10.1016/S0378-1119(03)00381-0. [DOI] [PubMed] [Google Scholar]
- 49.Langsley G, van Noort V, Carret C, Meissner M, de Villiers EP, Bishop R, Pain A. Comparative genomics of the Rab protein family in Apicomplexan parasites. Microbes Infect. 2008;10:462–470. doi: 10.1016/j.micinf.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 51.Nevin WD, Dacks JB. Repeated secondary loss of adaptin complex genes in the Apicomplexa. Parasitol Int. 2009;58:86–94. doi: 10.1016/j.parint.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis . Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lal K, Field MC, Carlton JM, Warwicker J, Hirt RP. Identification of a very large Rab GTPase family in the parasitic protozoan Trichomonas vaginalis. Mol Biochem Parasitol. 2005;143:226–235. doi: 10.1016/j.molbiopara.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642 [DOI] [PubMed]
- 56.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Kelly WG, Logsdon JM, Jr, Schurko AM, Harfe BD, Hill-Harfe KL, Kahn RA. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. Faseb J. 2004;18:1834–1850. doi: 10.1096/fj.04-2273com. [DOI] [PubMed] [Google Scholar]
- 58.Price HP, Stark M, Smith DF. Trypanosoma brucei ARF1 plays a central role in endocytosis and Golgi-lysosome trafficking. Mol Biol Cell. 2007;18:864–873. doi: 10.1091/mbc.E06-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang I, Robinson DG. Transport vesicle formation in plant cells. Curr Opin Plant Biol. 2009;12:660–669. doi: 10.1016/j.pbi.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 60.Yahara N, Ueda T, Sato K, Nakano A. Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol Biol Cell. 2001;12:221–238. doi: 10.1091/mbc.12.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders N, Jurgens G. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol Life Sci. 2008;65:3433–3445. doi: 10.1007/s00018-008-8227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 63.Seabra MC. Membrane association and targeting of prenylated Ras-like GTPases. Cell Signal. 1998;10:167–172. doi: 10.1016/S0898-6568(97)00120-4. [DOI] [PubMed] [Google Scholar]
- 64.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K. Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell. 2004;117:749–760. doi: 10.1016/j.cell.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 66.Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci USA. 2007;104:12294–12299. doi: 10.1073/pnas.0701817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Z, Wu YW, Das D, Delon C, Cramer J, Yu S, Thuns S, Lupilova N, Waldmann H, Brunsveld L, Goody RS, Alexandrov K, Blankenfeldt W. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 2008;27:2444–2456. doi: 10.1038/emboj.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasteiro R, Pereira-Leal JB. Multiple domain insertions and losses in the evolution of the Rab prenylation complex. BMC Evol Biol. 2007;7:140. doi: 10.1186/1471-2148-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol 20:247–252 [DOI] [PubMed]
- 70.Hala M, Elias M, Zarsky V. A specific feature of the angiosperm Rab escort protein (REP) and evolution of the REP/GDI superfamily. J Mol Biol. 2005;348:1299–1313. doi: 10.1016/j.jmb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Larijani B, Hume AN, Tarafder AK, Seabra MC. Multiple factors contribute to inefficient prenylation of Rab27a in Rab prenylation diseases. J Biol Chem. 2003;278:46798–46804. doi: 10.1074/jbc.M307799200. [DOI] [PubMed] [Google Scholar]
- 72.Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature. 1991;353:769–772. doi: 10.1038/353769a0. [DOI] [PubMed] [Google Scholar]
- 73.Ali BR, Wasmeier C, Lamoreux L, Strom M, Seabra MC. Multiple regions contribute to membrane targeting of Rab GTPases. J Cell Sci. 2004;117:6401–6412. doi: 10.1242/jcs.01542. [DOI] [PubMed] [Google Scholar]
- 74.Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawasaki M, Nakayama K, Wakatsuki S. Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr Opin Struct Biol. 2005;15:681–689. doi: 10.1016/j.sbi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 76.Somsel Rodman J, Wandinger-Ness A. Rab GTPases coordinate endocytosis. J Cell Sci. 2000;113(Pt 2):183–192. doi: 10.1242/jcs.113.2.183. [DOI] [PubMed] [Google Scholar]
- 77.Vitale G, Rybin V, Christoforidis S, Thornqvist P, McCaffrey M, Stenmark H, Zerial M. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 79.McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/S0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 80.Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thoma N, Rybin V, Zerial M, Scheidig AJ, Goody RS. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol. 2001;310:141–156. doi: 10.1006/jmbi.2001.4735. [DOI] [PubMed] [Google Scholar]
- 81.Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 2003;22:78–88. doi: 10.1093/emboj/cdg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shirane M, Nakayama KI. Protrudin induces neurite formation by directional membrane trafficking. Science. 2006;314:818–821. doi: 10.1126/science.1134027. [DOI] [PubMed] [Google Scholar]
- 83.Del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol. 2008;4:206. doi: 10.1038/msb.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoogenraad CC, Popa I, Futai K, Sanchez-Martinez E, Wulf PS, van Vlijmen T, Dortland BR, Oorschot V, Govers R, Monti M, Heck AJ, Sheng M, Klumperman J, Rehmann H, Jaarsma D, Kapitein LC, van der Sluijs P. Neuron specific Rab4 effector GRASP-1 coordinates membrane specialization and maturation of recycling endosomes. PLoS Biol. 2010;8:e1000283. doi: 10.1371/journal.pbio.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 86.Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci USA. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dhir V, Goulding D, Field MC. TbRAB1 and TbRAB2 mediate trafficking through the early secretory pathway of Trypanosoma brucei . Mol Biochem Parasitol. 2004;137:253–265. doi: 10.1016/j.molbiopara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 89.Sirokmany G, Szidonya L, Kaldi K, Gaborik Z, Ligeti E, Geiszt M. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab and Rho GTPases. J Biol Chem. 2006;281:6096–6105. doi: 10.1074/jbc.M510619200. [DOI] [PubMed] [Google Scholar]
- 90.Deneka M, Neeft M, Popa I, van Oort M, Sprong H, Oorschot V, Klumperman J, Schu P, van der Sluijs P. Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes. EMBO J. 2003;22:2645–2657. doi: 10.1093/emboj/cdg257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hall BS, Pal A, Goulding D, Acosta-Serrano A, Field MC. Trypanosoma brucei: TbRAB4 regulates membrane recycling and expression of surface proteins in procyclic forms. Exp Parasitol. 2005;111:160–171. doi: 10.1016/j.exppara.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 92.Hall BS, Pal A, Goulding D, Field MC. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J Biol Chem. 2004;279:45047–45056. doi: 10.1074/jbc.M407271200. [DOI] [PubMed] [Google Scholar]
- 93.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 94.Will E, Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J Biol Chem. 2001;276:12135–12139. doi: 10.1074/jbc.M011451200. [DOI] [PubMed] [Google Scholar]
- 95.Rak A, Fedorov R, Alexandrov K, Albert S, Goody RS, Gallwitz D, Scheidig AJ. Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. EMBO J. 2000;19:5105–5113. doi: 10.1093/emboj/19.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McLauchlan H, Newell J, Morrice N, Osborne A, West M, Smythe E. A novel role for Rab5-GDI in ligand sequestration into clathrin-coated pits. Curr Biol. 1998;8:34–45. doi: 10.1016/S0960-9822(98)70018-1. [DOI] [PubMed] [Google Scholar]
- 97.Wu X, Wang F, Rao K, Sellers JR, Hammer JA., 3rd Rab27a is an essential component of melanosome receptor for myosin Va. Mol Biol Cell. 2002;13:1735–1749. doi: 10.1091/mbc.01-12-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jing J, Prekeris R. Polarized endocytic transport: the roles of Rab11 and Rab11-FIPs in regulating cell polarity. Histol Histopathol. 2009;24:1171–1180. doi: 10.14670/hh-24.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eathiraj S, Mishra A, Prekeris R, Lambright DG. Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J Mol Biol. 2006;364:121–135. doi: 10.1016/j.jmb.2006.08.064. [DOI] [PubMed] [Google Scholar]
- 100.Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, Drechsel D, Kalaidzidis Y, Zerial M. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 101.Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 102.Field MC, Gabernet-Castello C, Dacks JB. Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv Exp Med Biol. 2007;607:84–96. doi: 10.1007/978-0-387-74021-8_7. [DOI] [PubMed] [Google Scholar]
- 103.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]