Abstract
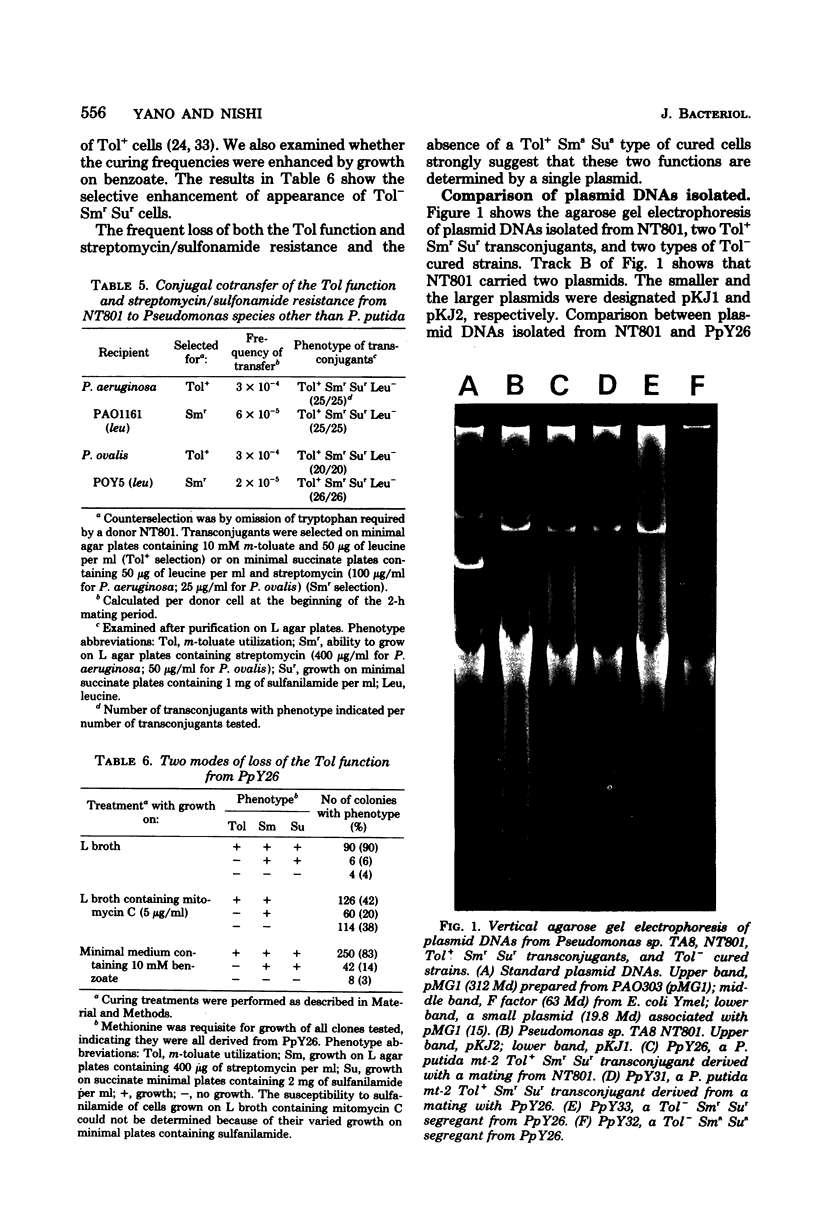
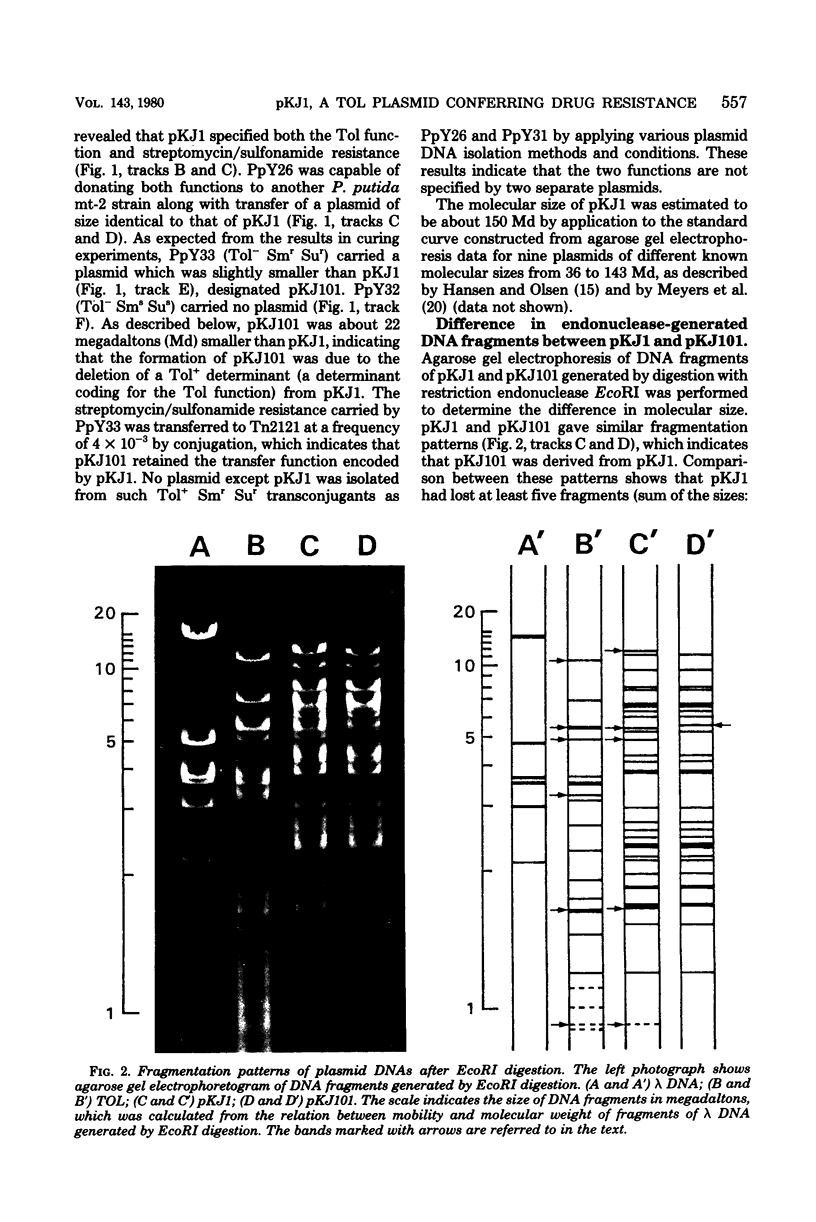
Pseudomonas sp. TA8 isolated by m-toluate enrichment from an aqueous sample metabolized toluene and m- and p-xylenes via the meta cleavage pathway, and manifested specific resistance to streptomycin and sulfonamides. A variety of experiments revealed that the pKJ1 plasmid of about 150 megadaltons carried by TA8 specified both the toluene and xylene degradative function (the Tol function) and streptomycin/sulfonamide resistance. The deletion of a segment of pKJ1 (about 22 megadaltons) resulted in the loss of the Tol function. pKJ1 was not assigned to Pseudomonas incompatibility group P-1, P-2, P-3, or P-9.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BAUCHOP T., ELSDEN S. R. The growth of micro-organisms in relation to their energy supply. J Gen Microbiol. 1960 Dec;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Morris D. W., Broda P. The relationship of degradative and resistance plasmids of Pseudomonas belonging to the same incompatibility group. Nature. 1979 Jul 26;280(5720):338–339. doi: 10.1038/280338a0. [DOI] [PubMed] [Google Scholar]
- Benedik M., Fennewald M., Shapiro J. Transposition of a beta-lactamase locus from RP1 into Pseudomonas putida degradative plasmids. J Bacteriol. 1977 Feb;129(2):809–814. doi: 10.1128/jb.129.2.809-814.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S., Shapiro J. TOL is a broad-host-range plasmid. J Bacteriol. 1978 Jul;135(1):278–280. doi: 10.1128/jb.135.1.278-280.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Dissociation of a degradative plasmid aggregate in Pseudomonas. J Bacteriol. 1974 Jun;118(3):815–820. doi: 10.1128/jb.118.3.815-820.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Friello D. A., Bopp L. H. Transposition of plasmid DNA segments specifying hydrocarbon degradation and their expression in various microorganisms. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3109–3112. doi: 10.1073/pnas.75.7.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Mylroie J. R., Friello D. A., Vacca J. G. Transformation of Pseudomonas putida and Escherichia coli with plasmid-linked drug-resistance factor DNA. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3647–3651. doi: 10.1073/pnas.72.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Plasmids in Pseudomonas. Annu Rev Genet. 1976;10:7–30. doi: 10.1146/annurev.ge.10.120176.000255. [DOI] [PubMed] [Google Scholar]
- Dunn N. W., Holloway B. W. Pleiotrophy of p-fluorophenylalanine-resistant and antibiotic hypersensitive mutants of Pseudomonas aeruginosa. Genet Res. 1971 Oct;18(2):185–197. doi: 10.1017/s0016672300012593. [DOI] [PubMed] [Google Scholar]
- Friello D. A., Mylroie J. R., Gibson D. T., Rogers J. E., Chakrabarty A. M. XYL, a nonconjugative xylene-degradative plasmid in Pseudomonas Pxy. J Bacteriol. 1976 Sep;127(3):1217–1224. doi: 10.1128/jb.127.3.1217-1224.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S. Gel electrophoretic separation of the complementary strands of bacteriophage DNA. Virology. 1972 Jul;49(1):342–344. doi: 10.1016/s0042-6822(72)80042-4. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Rogers J. E., Jacob A. E., Hedges R. W. Transposition of Pseudomonas toluene-degrading genes and expression in Escherichia coli. Nature. 1978 Jul 13;274(5667):179–180. doi: 10.1038/274179a0. [DOI] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Hayashi E., Yokota T., Ebina Y., Nakazawa A. Isolation of TOL and RP4 recombinants by integrative suppression. J Bacteriol. 1978 Apr;134(1):270–277. doi: 10.1128/jb.134.1.270-277.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Yokota T. Benzoate metabolism in Pseudomonas putida(arvilla) mt-2: demonstration of two benzoate pathways. J Bacteriol. 1973 Jul;115(1):262–267. doi: 10.1128/jb.115.1.262-267.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Yokota T. Isolation of a mutant TOL plasmid with increased activity and transmissibility from Pseudomonas putida (arvilla) mt-2. J Bacteriol. 1977 Jan;129(1):39–46. doi: 10.1128/jb.129.1.39-46.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Ornston M. K., Chou G. Isolation of spontaneous mutant strains of Pseudomonas putida. Biochem Biophys Res Commun. 1969 Jul 7;36(1):179–184. doi: 10.1016/0006-291x(69)90666-4. [DOI] [PubMed] [Google Scholar]
- Rheinwald J. G., Chakrabarty A. M., Gunsalus I. C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci U S A. 1973 Mar;70(3):885–889. doi: 10.1073/pnas.70.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Thacker R., Rørvig O., Kahlon P., Gunsalus I. C. NIC, a conjugative nicotine-nicotinate degradative plasmid in Pseudomonas convexa. J Bacteriol. 1978 Jul;135(1):289–290. doi: 10.1128/jb.135.1.289-290.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Hughes S. G., Broda P. Plasmid identification using specific endonucleases. Mol Gen Genet. 1974;133(2):141–149. doi: 10.1007/BF00264835. [DOI] [PubMed] [Google Scholar]
- White G. P., Dunn Evidence for transductional shortening of the plasmid obtained by recombination between the TOL catabolic plasmid and the R91 R plasmid. Genet Res. 1978 Feb;31(1):93–96. doi: 10.1017/s0016672300017833. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]