Abstract
Penta-1,2,3,4,6-O-galloyl-beta-D-glucose (PGG) has been shown by us and others to inhibit the in vivo growth of human prostate cancer (PCa) xenografts in athymic nude mice and mouse lung cancer allograft in syngenic mice without evident adverse effect on their body weight. We observed a rapid inhibition of DNA synthesis in S-phase cells in PGG-exposed cancer cells and in PGG-treated isolated nuclei. The purpose of the present study was to test the hypothesis that PGG inhibits DNA replicative synthesis through a direct inhibition of one or more DNA polymerases (pols). Using purified pols, we show that PGG exhibited a selective inhibition against the activities of B-family replicative pols (α, δ and ε) and Y-family (η, ι and κ) of bypass synthesis pols, and the inhibitory effect of PGG on pol α was the strongest with IC50 value of 13 nM. PGG also inhibited pol β, but the potency was an order of magnitude less than against pol α. PGG inhibition of pol α and κ activity was non-competitive with respect to the DNA template-primer and the dNTP substrate; whereas it inhibited pol β competitively. Docking simulation on pol β, which is the only mammalian pol with solved crystal structure, suggests several favorable interactions with the catalytic pocket/binding site for the incoming dNTP. These results support PGG as a novel inhibitor of select families of mammalian pols by distinct mechanisms, and suggest that the potent pol inhibition may contribute to its anti-cancer efficacy.
Keywords: penta-1,2,3,4,6-O-galloyl-beta-D-glucose (PGG); DNA polymerase (pol); enzyme inhibitor; DNA replication; anticancer effect
1. Introduction
DNA replication, recombination and repair in eukaryotes are key systems to support cell proliferation and differentiation and maintain the integrity of the genome [1]. DNA polymerases (pols) play crucial roles in each of these processes. Pols catalyze the addition of deoxyribonucleotides (dNTP) to the 3′-hydroxyl terminus of primed double-stranded DNA molecules [2].
The mammalian genome encodes at least 15 pols to conduct nuclear and cellular DNA synthesis [3, 4]. Eukaryotic cells contain three replicative pols (α, δ and ε), mitochondrial pol γ, and at least eleven non-replicative pols [β, ζ, η, θ, ι, κ, λ, μ, ν, terminal deoxynucleotidyl transferase (TdT) and REV1] [3–5]. Pols have a highly conserved structure, which means that their overall catalytic subunits, on the whole, vary little among species. Conserved structures usually indicate important, irreplaceable functions of the cell, the maintenance of which provides evolutionary advantages. Based on sequence homology, eukaryotic pols can be divided into four main different families, A, B, X, and Y [6]. Family A includes mitochondrial pol γ, and pols θ and ν, and family B includes three replicative pols (α, δ, and ε) and pol ζ. Family X comprises pols β, λ, μ, and TdT, and family Y includes bypass synthesis pols η, ι, κ, and REV1. Because not all functions of eukaryotic pols have been fully elucidated, selective inhibitors of pols are useful reagents for distinguishing pols and clarifying their biological functions. Due to increased proliferation in malignant cancer cells with respect to normal cells, selective inhibitors of pol could be a class of potentially useful anti-cancer agents. Indeed some inhibitors suppress human cancer cell proliferation in vitro and exert anti-cancer efficacy in vivo [7]. The Mizushina laboratory has concentrated intense efforts on investigating eukaryotic pols and their naturally occurring inhibitors [8].
The Lü laboratory and collaborators have shown that the hydrolysable gallotannin compound 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG), present abundantly in some Oriental medicinal herbs, suppresses the in vivo growth of human DU145 prostate cancer (PCa) xenografts in nude mice at a tested daily intra-peritoneal injection dose of 20 mg/kg body weight [9] and mouse Lewis lung cancer allografts in syngenic mice in a dose-dependent manner in the range of 4 and 20 mg/kg [10], respectively. Very recently, Kuo et al have shown a positive inhibitory efficacy of PGG (i.p. injection, every other day, 25 mg/kg) against the growth of the aggressive human PCa PC-3 cells inoculated into the tibia in nude mice, simulating metastatic growth [11]. These studies support the promising efficacy of PGG as a cancer chemotherapeutic or chemopreventive agent to selectively inhibit malignancy without overt host toxicity.
Chemically and functionally, PGG appears to be distinct from its constituent gallic acid or tea polyphenols [12]. For anti-cancer activity, potential mechanisms include anti-angiogenesis [10], anti-proliferative actions through G1 arrest, induction of apoptosis, anti-inflammation and anti-oxidation (for a comprehensive review, see [12]). Putative molecular targets include p53, Stat3, Cox-2, VEGF-receptor 2 (Flk/KDR), AP-1, SP-1, Nrf-2, MMP-9 and fatty acid synthase [12]. In particular relevance to prostate cancer, PGG was shown as an inhibitor of rat liver microsomal 5 α-reductase (EC 1.3.99.5), which catalyzes the conversion of androgen testosterone (T) to a more active androgen dihydrotestosterone (DHT) with an IC50 at 7.8 μM [13]. Kinetic studies showed that PGG was a competitive inhibitor for NADPH while a non-competitive inhibitor for testosterone. The mode of inhibition suggests that PGG may inhibit 5α-reductase by competing with NADPH binding site.
The cell cycle effects of PGG in prostate cancer (PCa) cells have recently been evaluated [9, 14]. The data show that treatment with PGG induced not only G1 arrest but also S-arrest. Irrespective of the p53 functional status of the PCa cell lines, PGG exerted a rapid (within 2 h) and potent inhibition (IC50 ~ 6 μM) of BrdU incorporation into S phase cells. In isolated nuclei, PGG inhibited DNA replicative synthesis with superior efficacy to a known pol α inhibitor, aphidocolin. In addition to the S-arrest action, a close association of down regulation of cyclin D1 was found with G1 arrest induced by PGG exposure in the pro-apoptotic range. Over-expressing this G1 cyclin abolished G1 arrest, but hastened the S-arrest induction by PGG. Together, the data indicate that PGG induces PCa S-arrest probably through DNA replicative blockage and induces G1-arrest via cyclin D1 down regulation to contribute to anti-cancer activity. The G1 and S arrest actions and the inhibition of DNA replication by PGG are also observed in breast cancer cell lines (to be published elsewhere). The data raise the hypotheses that PGG may be a novel inhibitor of pols, especially replicative pols and applicable to inhibiting malignancies of other organ sites.
In this study, we tested whether and how PGG directly inhibits pols and other DNA metabolic enzymes. The data show that PGG selectively inhibits replicative pols (α, δ and ε) with IC50 ranging from 13 to 66 nM. PGG also inhibits bypass synthesis pols (e.g., η, ι, κ) with IC50 in 30–45 nM and base excision repair pol β with IC50 108–160 nM. The inhibitory mechanism of pol α and κ by PGG is non-competitive with respect to dNTP and DNA template-primer, different from that for pol β, which is competitive with respect to dNTP and DNA template-primer. In silico docking of PGG to pol β, the only mammalian pol with solved crystal structures, suggests tight binding of PGG, consistent with a competitive inhibition of this pol.
2. Materials and Methods
2.1 Chemicals and reagents
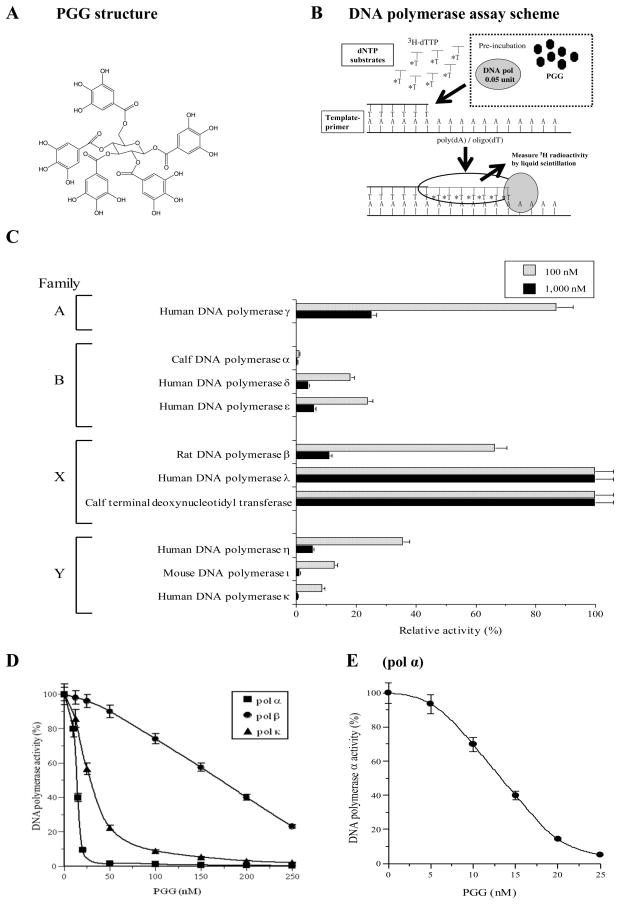
PGG (Fig. 1A) was prepared by methanolysis of tannic acid (Fisher Chemical, Pittsburgh, USA) per a published method [15]. The purity was ~99%. Nucleotides such as [3H]-deoxythymidine 5′-triphosphate (dTTP) (43 Ci/mmol), and chemically synthesized DNA template, such as poly(dA) were purchased from GE Healthcare Bio-Sciences (Little Chalfont, UK). DNA primer, such as oligo(dT)18, was custom-made by Sigma-Aldrich K.K. (Hokkaido, Japan).
Fig. 1.
(A) Chemical structure of PGG. (B) Scheme for DNA polymerase assays. (C) Differential inhibitory effects of PGG on the activities of various mammalian pols. 100 nM (gray bars) and 1,000 nM (black bars) of PGG were incubated with each enzyme (0.05 unit). Enzymatic activity was measured as described previously [17, 18]. Enzyme activity in the absence of the compounds was taken as 100%. Data are shown as the means ± SEM of four independent experiments. (D) Dose-response inhibition curves of PGG against mammalian pol α, κ and β. PGG was incubated with calf pol α (square) as a representative B family pols, mouse pol κ (triangle) as a representative Y family pols and rat pol β (circle) as a representative X family of pols (0.05 unit of each). (E) More refined dose–response curve for calf pol α to define IC50 for PGG. Pol activity in the absence of the compounds was taken as 100 %. For D and E, data are shown as the mean ± SEM of three independent experiments.
2.2 Enzymes
Mammalian pols (calf pol α, rat pol β, human pols γ, δ, ε, η, κ and λ, and mouse pol ι) and plant pols (cauliflower pol α and rice pol λ), which were purified and have high activity, were prepared as described in our previous report [16]. Calf thymus TdT and bovine pancreas deoxyribonucrease I (DNase I) were obtained from Stratagene Cloning Systems (La Jolla, CA, USA). Human immunodeficiency virus type-1 (HIV-1) reverse transcriptase (recombinant) and the Klenow fragment of pol I from E. coli were purchased from Worthington Biochemical Corp. (Freehold, NJ, USA). T4 pol, Taq pol, T7 RNA polymerase and T4 polynucleotide kinase were purchased from Takara Bio (Tokyo, Japan). Purified human placenta DNA topoisomerases I and II were purchased from TopoGen, Inc. (Columbus, OH, USA).
2.3 Pol assays
The reaction mixtures for pol α, pol β, plant pols and prokaryotic pols were described previously [17, 18]. Those for pol γ, and pols δ and ε were as described by Umeda et al [19] and Ogawa et al [20], respectively. The reaction mixtures for pols η, ι and κ were the same as for pol α, and the reaction mixture for pol λ was the same as for pol β. For pols (See assay scheme in Fig. 1B), poly(dA)/oligo(dT)18 (A/T = 2/1) and dTTP were used as the DNA template-primer and nucleotide (i.e., dNTP) substrate, respectively. For HIV-1 reverse transcriptase, poly(rA)/oligo(dT)18 (A/T = 2/1) and dTTP were used as the template-primer and nucleotide substrate, respectively. For TdT, oligo(dT)18 (3′-OH) and dTTP were used as the DNA primer and nucleotide substrate, respectively.
PGG was dissolved in distilled dimethyl sulfoxide (DMSO) at various concentrations and sonicated for 30 sec. Aliquots of 4 μl sonicated samples were mixed with 16 μl of each enzyme (final amount 0.05 unit) in 50 mM Tris-HCl (pH7.5) containing 1 mM dithiothreitol, 50% glycerol and 0.1 mM EDTA, and kept at 0 °C for 10 min. These inhibitor-enzyme mixtures (8 μl) were added to 16 μl of each of the enzyme standard reaction mixtures, and incubation was carried out at 37 °C for 60 min, except for Taq pol, which was incubated at 74 °C for 60 min. Activity without the inhibitor was considered 100%, and the remaining activity at each concentration of the inhibitor was determined relative to this value (Fig. 1B). One unit of pol activity was defined as the amount of enzyme that catalyzed the incorporation of 1 nmol dNTP (i.e., dTTP) into synthetic DNA template-primers in 60 min at 37 °C under the normal reaction conditions for each enzyme [17, 18].
2.4 Other DNA metabolic enzymes assays
The activities of calf primase of pol α, human telomerase, T7 RNA polymerase, mouse inosine 5′-monophosphate (IMP) dehydrogenase, human DNA topoisomerases I and II, T4 polynucleotide kinase and bovine DNase I were measured in standard assays according to the manufacturer’s specifications, as described by Tamiya-Koizumi et al [21], Oda et al [22], Nakayama et al [23], Mizushina et al [24], Ishimaru et al [25], Soltis et al [26] and Lu et al [27], respectively.
2.5 Thermal transition of DNA
The thermal transition of double-stranded to single-stranded DNA with or without PGG was profiled with a spectrophotometer (U3210, Hitachi, Tokyo) equipped with a thermoelectric cell holder. Calf thymus double-stranded DNA (dsDNA) (6 μg/ml) was dissolved in 0.1 M sodium phosphate buffer (pH 7.0) containing 1% DMSO. The solution temperature was equilibrated at 78 °C for 10 min, and then increased by 1 °C at 2 min intervals for each measurement point. Any change in the absorbance of the compound itself at each temperature point was automatically subtracted from that of DNA plus the compound in the spectrophotometer.
3. RESULTS
3.1 Differential inhibitory effects of PGG on various families of mammalian pols and a lack of inhibition on other DNA metabolic enzymes
First, we tested whether PGG inhibited the activities of ten mammalian DNA pols, such as families A (pol γ), B (pols α, δ and ε), X (pols β and λ and TdT), and Y (pols η, ι and κ). As shown in Fig. 1C, this compound at 100 and 1,000 nM was found to significantly inhibit the activities of the B- and Y-families of pols, and calf pol α was the most strongly inhibited among these pols. PGG inhibited dose-dependently the activity of replicative pol α and bypass synthesis pol κ (Fig. 1D), and 50% inhibition was observed at concentrations (IC50) of 13 nM (Fig. 1D and E) and 30 nM (Fig. 1D), respectively. PGG also inhibited base excise repair pol β with IC50 of ~160 nM (Fig. 1D). The Mizushina laboratory has been screening mammalian pol inhibitors for 15 years, and PGG was by far the strongest pol inhibitor ever tested [7, 8].
The inhibitory activity of PGG on families A and X of mammalian pols were lower, especially the activities of pol λ and TdT of X-family pols were not affected (Fig. 1C). On the other hand, higher plant pols (cauliflower pol α and rice pol λ), and prokaryotic pols (the Klenow fragment of E. coli pol I, T4 pol and Taq pol) were not influenced by PGG (supplemental Fig. S1A).
PGG at the highest tested concentration (i.e., 1 μM) did not suppress activities of other DNA metabolic enzymes such as calf primase of pol α, HIV-1 reverse transcriptase, human telomerase, T7 RNA polymerase, mouse IMP dehydrogenase (type II), human topoisomerases I and II, T4 polynucleotide kinase, and bovine DNase I (supplemental Fig. S1B).
These results suggest that PGG is a potent inhibitor of selected mammalian pols, especially those involved in replicative and bypass synthesis. PGG also inhibited base excision repair pol β but an order of magnitude less potent than against pol α.
3.2 Lack of effects/interactions of PGG with nucleic acids and general proteins
To determine whether PGG caused an alteration of DNA double strand structure, the interaction of PGG with calf thymus DNA was investigated based on the thermal transition of dsDNA in the presence or absence of PGG. The melting temperature (Tm) of dsDNA with an excess amount of PGG (15 μM) was measured using a spectrophotometer equipped with a thermoelectric cell holder. In the concentration range used, no effect of PGG was seen on Tm, the temperature at which a thermal transition occurs; whereas 15 μM of ethidium bromide (EtBr), a typical intercalating compound used as a positive control, produced a clear shift of thermal transition (supplemental Fig. S2). These results indicated that PGG did not intercalate to dsDNA such as existing in DNA template-primer region of the pol assay. PGG might therefore directly bind to the enzymes and inhibit their activity.
To determine the effects of a non-ionic detergent on the binding of PGG to different class of mammalian pols, we selected pols α, κ and β, which are representative of B, Y and X pol family, respectively, for testing (supplemental Table S1). Nonidet P-40 (NP-40) was added to the reaction mixture at a concentration of 0.05%. In the absence of PGG, the activity of pols was not affected by addition of NP-40, and we designated the activities in these cases as 100%. The inhibitory effect of PGG (at 10 and 100 nM for pols α, κ, at 100 nM and 200 nM for pol β) was not reversed by the addition of 0.05% NP-40 to the reaction mixture. These results suggested that PGG could bind to and interact with the hydrophilic region of the enzyme protein. We also tested whether an excess amount of a substrate DNA analog, poly(rC) (50 μM), or a protein, bovine serum albumin (BSA) (200 μg/ml), could prevent the inhibitory effects of PGG. If PGG bound to the enzymes by non-specific adhesion, the addition of the nucleic acid and/or protein would be expected to reduce its inhibitory activity. The fact that neither poly(rC) nor BSA influenced the inhibitory effects on PGG suggest that PGG could selectively bind to specific site(s) on the pol enzymes and not to the nucleic acid.
3.3 Mode of inhibition of mammalian pols α and κ by PGG
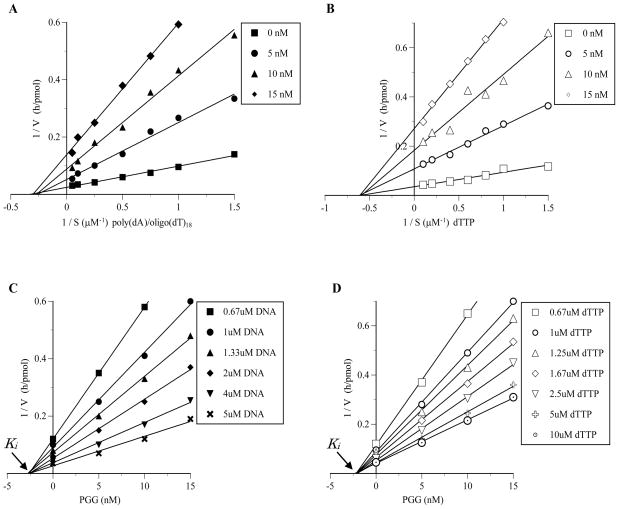
Next, to elucidate the mechanism by which PGG inhibited calf pol α, we examined the extent of inhibition as a function of the DNA template-primer or 2′-deoxynucleoside 5′-triphosphate (dNTP) substrate concentration (Fig. 2). In kinetic analysis, poly(dA)/oligo(dT)18 and 2′-deoxythymidine 5′-triphosphate (dTTP) were used as the synthetic DNA template-primer and dNTP substrate, respectively. The inhibitory mode against DNA template-primer site was analyzed using the reaction condition of various concentrations (0.67 – 10 μM) of poly(dA)/oligo(dT)18 and high concentration (50 μM) of dTTP. The inhibitory mode against dNTP substrate site was analyzed using the reaction condition of various concentrations (0.4 – 10 μM) of dTTP and high concentration (50 μM) of poly(dA)/oligo(dT)18. Lineweaver-Burk double reciprocal plots of the obtained data showed that the PGG-induced inhibition of pol α activity was non-competitive with respect to both the DNA template-primer and the dNTP substrate. For the DNA template-primer, the apparent Michaelis constant (Km) was unchanged at 3.6 μM, whereas the apparent maximum velocity (Vmax) was observed to decrease to 32.2%, 18.9% and 13.6% in the presence of 5, 10 and 15 nM of PGG, respectively (Fig. 2A). The apparent Km for the dNTP substrate was unchanged at 1.65 μM, and the apparent Vmax for the dNTP substrate decreased from 29.2 to 3.77 pmol/h in the presence of 15 nM of PGG (Fig. 2B). Inhibition constant (Ki) values, obtained from Dixon plots, were found to be 2.5 nM and 2.2 nM for the DNA template-primer and dNTP substrate, respectively (Fig. 2C and D). Because the Ki value for the DNA template-primer was almost the same as that for the dNTP substrate, the affinity of PGG for the pol enzyme/DNA template-primer binary complex was similar as for the pol enzyme/nucleotide substrate complex.
Fig. 2.
Kinetic analysis of the inhibition of calf thymus pol α by PGG. (A and B) Lineweaver–Burk double-reciprocal plots obtained by varying DNA template-primer (i.e., poly(dA)/oligo(dT)18) concentrations (A), and dNTP substrate (i.e., dTTP) concentrations (B). (C and D) The inhibition constant (Ki) were determined as 2.5 and 2.2 nM from a Dixon plot made on the basis of the same data for A and B, respectively. The amount of pol α in the assay mixture was 0.05 unit.
It is important to note that when activated DNA (i.e., calf thymus dsDNA with gaps digested by bovine DNase I) was used as the DNA template-primer and with 4 dNTPs as the substrates, the mode of inhibition of pol α by PGG was the same as that with the above synthetic DNA template-primer (Data not shown).
The kinetics of PGG inhibition of pol κ is the same as for pol α (data not shown), except the Ki with respect to dNTP or DNA template-primer was higher than for pol α, being 15.1 and 9.3 nM, respectively (Table 1).
Table 1.
Summary of enzymes reported to be inhibitable by PGG
| Enzyme | IC50 | Ki | Mode of Inhibition | Reference |
|---|---|---|---|---|
| Pol α | 13 nM | 2.2 nM 2.5 nM |
Non-competitive for dNTP Non-competitive for DNA template-primer |
Current work |
| Pol κ | 30 nM | 15.1 nM 9.3 nM |
Non-competitive for dNTP Non-competitive for DNA template-primer |
Current work |
| Pol β | 108–160 nM | 23.8 nM 20.1 nM |
Competitive for dNTP Competitive for DNA template-primer |
Current work |
| H+, K+- ATPase | 166 nM | NR* | Non-competitive for K+ | [12]** |
| HCV NS3 protease | 1,600 nM | NR | NR | [12] |
| Fatty acid synthase | 2,980 nM | NR | NR | [12] |
| Xanthine oxidase | 3,200 nM | NR | NR | [12] |
| Beta-secretase | 3,760 nM | 5,130 nM | Non-competitive for Rh-EVNLDAEFK-Quencher | [39] |
| Aminopeptidase N | 6,500 nM | NR | NR | [12] |
| 5-alpha-Reductase | 7,800 nM | 480 nM 2,680 nM |
Competitive for NADPH Non-competitive for testosterone |
[12],[13] |
| Neutral endopeptidase | 12,500 nM | NR | NR | [12] |
| HIV integrase | 13,700 nM | NR | NR | [12] |
| Angiotensin converting enzyme | 73,100 nM | NR | NR | [12] |
| HIV reverse transcriptase | >100,000 nM | NR | NR | [12] |
Not reported
Original papers were cited in this comprehensive review
3.4 Mode of inhibition of mammalian pol β by PGG
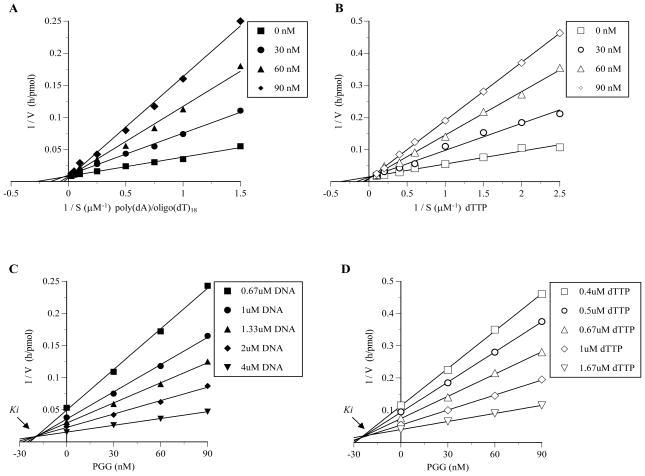
Double reciprocal plots of the obtained data showed that the PGG-induced inhibition of pol β activity was consistent with the premise that PGG competitively inhibits this enzyme with respect to both the DNA template-primer and the dNTP substrate (Fig. 3). For the DNA template-primer, the apparent Michaelis constant (Km) changed from 6.74 μM to 9.5 μM, 14.3 μM and 28.6 μM in the presence of 30, 60 and 90 nM of PGG, respectively, whereas the apparent maximum velocity (Vmax) remained unchanged at 111 pmol/h (Fig. 3A). The Km for the dNTP substrate was changed from 3.05 μM to 6.10 μM, 12.2 μM and 24.4 μM, in the presence of 30, 60 and 90 nM of PGG, respectively, whereas apparent maximum velocity (Vmax) remained unchanged at 62.5 pmol/h (Fig. 3B). Inhibition constant (Ki) values, obtained from Dixon plots, were found to be 20.1 nM PGG and 23.8 nM PGG for the DNA template-primer and dNTP substrate, respectively (Fig. 3C and D).
Fig. 3.
Kinetic analysis of the inhibition of rat pol β by PGG. (A and B) Lineweaver–Burk double-reciprocal plots obtained by varying DNA template-primer (i.e., poly(dA)/oligo(dT)18) concentrations (A), and dNTP substrate (i.e., dTTP) concentrations (B). (C and D) The inhibition constant (Ki) were determined as 20.1 and 23.8 nM from a Dixon plot made on the basis of the same data for A and B, respectively. The amount of pol β in the assay mixture was 0.05 unit.
3.5 Defining PGG interaction region on mammalian family X pols
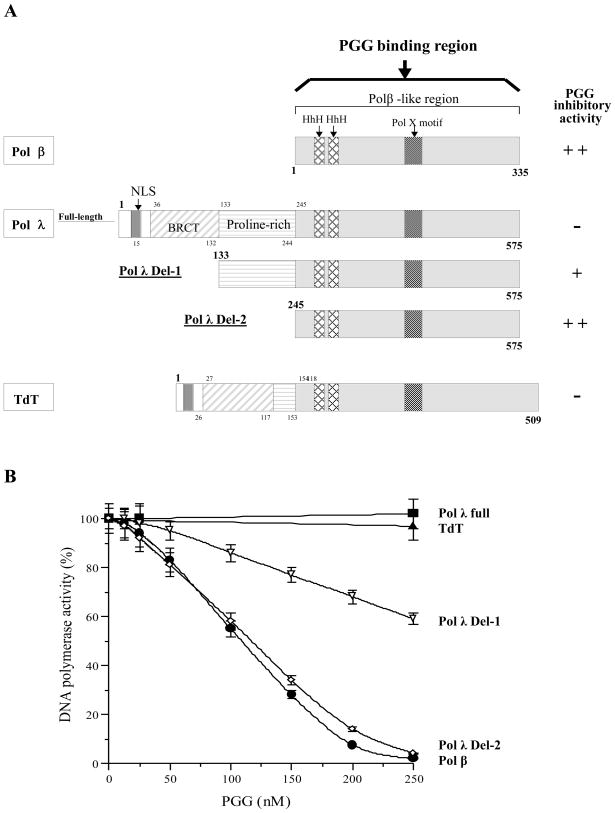
Because the activity of rat pol β was inhibited by PGG, but that of other mammalian family X pols such as human pol λ and calf TdT were not sensitive to PGG (see Fig. 1C), we next focused on comparing their structural features to define possible interaction regions of PGG.
As schematically illustrated in Fig. 4A, human pol λ (63.4 kDa) consists of a nuclear localization signal (NLS) (residues 1–35), a BRCA1 C-terminus (BRCT) domain (residues 36–132), a proline-rich region (residues 133–244) and a pol β-like region containing two helix-hairpin-helixes (HhHs) and a pol X motif (residues 245–575). The C-terminal part of pol λ (residues 245–575) is composed of a catalytic core which is similar to pol β [28]. Pol λ possesses multiple enzymatic activities, including pol, terminal transferase, 5′-deoxyribose-5-phosphate (dRP) lyase, and polynucleotide synthetase, all located in the C-terminal part containing the pol β-like region [29, 30]. A truncated pol λ, in which the BRCT motif and/or proline-rich region was deleted from the N-terminal part (i.e., the C-terminal part of the pol β-like region), retained pol activity. Pol λ shares this with the highly homologous TdT, which has template-independent (i.e., terminal transferase) DNA synthetic activity. The biochemical properties of TdT have been extensively characterized [31, 32], and this enzyme is known to be able to add several nucleotides to the 3′-ends of single-stranded DNA (ssDNA) or double-stranded DNA templates [33].
Fig. 4.
(A) Schematic representation of pol β, pol λ and TdT of the X family of pols. The NLS (nuclear localization signal), BRCT (BRCA1 C-terminus) domain, proline-rich region, HhH (helix-hairpin-helix) and pol X motif are indicated. The pol β-like region includes two HhHs and a pol X motif. The inhibitory activity of PGG against these enzymes is indicated, “++” is an IC50 value of < 200 nM, and “+” is an IC50 value of 200 to 400 nM, based on data from C. (B) Inhibition dose-response curves of PGG against mammalian X pol family and deletion mutants. PGG was incubated with rat pol β (closed circle, ●), full length of human pol λ (closed square, ■), del-1 of human pol λ (open reverse-triangle, △), del-2 of human pol λ (open diamond, ◇), and calf TdT (closed triangle, ▲) (0.05 unit of each). Pol activity in the absence of PGG was taken as 100%. Data are shown as the means ± SEM of three independent experiments.
We prepared full-length pol λ (residues 1–575) and N-terminal-deleted mutants, del-1 pol λ (133–575) and del-2 pol λ (245–575)[34]. Fragments of human pol λ were dose-dependently inhibited by PGG (Fig. 4B), and in order of their inhibitory sensitivity, the pol λ mutants ranked as follows: del-2 > del-1 > full-length pol λ (Fig. 4B). The inhibitory effects on del-2 pol λ mutant and on rat pol β were strongest in the pol X family, with IC50 values of 117 and 108 nM, respectively. The effect seemed to be relatively selective for pol β and del-2 pol λ mutant, both lack NLS, the BRCT domain and proline-rich region. It is remarkable that in screening mammalian pol inhibitors for 15 years by the Mizushina lab, PGG was the strongest pol β inhibitor tested [7, 8].
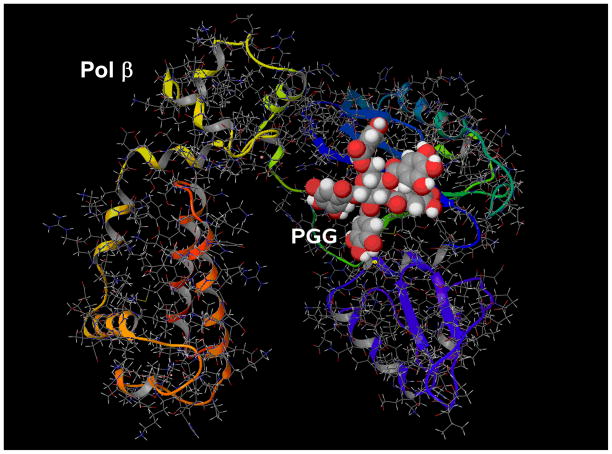
3.6 Super-Computer docking simulation suggests possible binding of PGG to active site of pol β
We used a IBM Bluegene Supercomputer to dock PGG to the active site of human pol β X-ray crystal structure (PDB code:1ZQI) [35] (Fig. 5). Mammalian pol β is the smallest in the pol families and therefore crystal structures have been solved [36, 37]. The docking results show that PGG could form several favorable interactions with the polymerase catalytic pocket/binding site for the incoming dNTP. The free energy of binding is predicted to be −10.26 kcal/mol and the docking runs gave only one possible solution (pose). In addition the compound seems to bind in a way that sterically obstructs two amino acids (Asp192 and Asp196) that are part of the catalytic core of the protein, potentially inhibiting the enzyme’s activity. The docking simulation provides important molecular insights into how PGG inhibits pol β in a competitive manner with respect to dNTP substrate and DNA template-primer. The structure for mammalian pol α or others have not been completely solved due to large size/multiple units.
Fig. 5.
Docking of PGG into human DNA pol β using a IBM Bluegene Supercomputer at the Hormel Institute.
4. DISCUSSION
The enzymology data described above provide experimental support for a possible direct and potent inhibition by PGG of the human and mammalian replicative pols, especially pol α, and DNA bypass synthesis pols, e.g. pol κ. We ruled out non-specific protein binding by PGG as a possible cause of the pol inhibition by inclusion of BSA in the assay (Supplemental Table S1). PGG is known to bind proteins such as albumin [15], as do all tannins by definition. The mammalian pol specificity is also evident in that pols from plant, fish and prokaryote are not sensitive to PGG (Supplemental Fig. S1A) and neither are many other human or mammalian DNA metabolizing enzymes tested (Supplemental Fig. S1B). However, we want to caution that the pol screening assay (Fig. 1B) used simple DNA template-primer for ease of detection and provided a good measure for pol enzyme processivity, but may not realistically assess substrate specificity for many of the non-replicative pols.
As it is understood today, pol α will be crucial to extend the primed nascent DNA strand but the other two replicative pols δ and ε with higher processivity will elongate the growing Okazaki fragments as well as the continuous strand [38]. The fact that PGG inhibited pol α and δ and ε, all with IC50 values lower than 100 nM, could implicate them as the direct targets for PGG to cause DNA replicative inhibition and S-arrest of PGG-exposed cancer cells. With respect to target identification and off-target activities of PGG, Table 1 summarized some enzymes reported to be inhibited by PGG in comparison with the pols. Of all the reported enzymes, only H+, K+-ATPase is inhibited by PGG with a potency within the same order of magnitude as pol β. The nano-molar potency of PGG against replicative pols, especially pol α in comparison with other reported enzymes, such as the testosterone activating 5-α-reductase with IC50 value of 7,800 nM, or fatty acid synthase with IC50 of 2,980 nM [13], suggests the good likelihood for PGG to work through the pol inhibition mechanism in vivo in cancer cells.
The inhibition kinetic studies suggest that PGG does not compete with DNA template-primer and dNTP binding sites of the catalytic domain of mammalian pols α and κ (Fig. 2). Rather PGG may bind to another site(s), and simultaneously disturbs dNTP substrate incorporation into the DNA template-primer of pols α and κ. In contrast, PGG inhibited pol β by competitive mechanism(s) (Fig. 3). Computational docking of PGG to the catalytic domain of pol β (Fig. 5) is consistent with PGG competitively inhibits this enzyme with respective to the DNA template-primer and dNTP its binding sites. The comparison of deletion mutants (del-1, del 2) of pol λ with its full length counterpart and other X-family members including pol β and TdT for their capacity to be inhibited by PGG (Fig. 4A and B) offered some insights of the protein region for interaction with PGG. Since pol β represents the minimal structure to be inhibited by the competitive mechanism, the N-terminal regions of pol λ and TdT may fold in a manner to mask the site(s) of interaction between PGG and the pol-β like structures. These speculations will require additional structural information of the more complex pols and experimental data to verify. Co-crystallization of PGG with pols and their mutagenesis of specific amino acids might be necessary to provide definitive proof.
In summary, we have presented enzymology data that support PGG as a novel, potent and selective inhibitor of human and mammalian pols involved in DNA replicative and bypass repair synthesis, as well as for base excision repair pol β, albeit with less potency. The findings may provide a relevant in vivo biochemical mechanism to account for the reported S-phase arrest action and anti-cancer and anti-angiogenesis efficacy of PGG. In addition to its obvious medicinal applications, PGG may be a novel research reagent for understanding pol structure and functions.
Supplementary Material
Acknowledgments
YM is grateful for the donations of calf pol α by Dr. M. Takemura of Tokyo University of Science (Tokyo, Japan), rat pol β, human pols δ and ε by Dr. K. Sakaguchi of Tokyo University of Science (Chiba, Japan), human pol γ by Dr. M. Suzuki of Nagoya University School of Medicine (Nagoya, Japan), human pols η and ι by Dr. F. Hanaoka and Dr. C. Masutani of Osaka University (Osaka, Japan), human pol κ by Dr. H. Ohmori of Kyoto University (Kyoto, Japan), and human pol λ by Dr. O. Koiwai of Tokyo University of Science (Chiba, Japan). The support of Prof. Zigang Dong of Hormel Institute for computational analysis is grateful acknowledged, as is of Drs. Ahmad Shaik and Chengguo Xing at Department of Medicinal Chemistry, University of Minnesota for preparing the PGG used.
Funding supports
*This work was supported in part by the “Academic Frontier” Project for Private Universities: matching fund subsidy from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT), 2006 – 2010, (Y. M.). Y. M. acknowledges a Grant-in-Aid for Young Scientists (A) (No. 19680031) from MEXT, and The Salt Science Research Foundation, No. 09S3 (Japan). Support for this work also came from the US National Institutes of Health grant CA136953 (JL).
Nomenclature and Abbreviations
- pol
DNA polymerase (EC 2.7.7.7)
- dTTP
2′-deoxythymidine 5′-triphosphate
- dNTP
2′-deoxynucleoside 5′-triphosphate
- TdT
terminal deoxynucleotidyl transferase
- HIV-1
human immunodeficiency virus type-1
- IMP
inosine 5′-monophosphate
- DNase I
deoxyribonuclease I
- DMSO
dimethyl sulfoxide
- dsDNA
double-stranded DNA
- Tm
melting temperature
- EtBr
ethidium bromide
- NP-40
Nonidet P-40
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DePamphilis M. DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 2.Kornberg K, Baker TA. DNA replication. New York: Freeman W. D. and Co; 1992. [Google Scholar]
- 3.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annual review of biochemistry. 2002;71:133–63. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek K, Kunkel TA. Functions of DNA polymerases. Advances in protein chemistry. 2004;69:137–65. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 5.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. The Journal of biological chemistry. 2006;281:23445–55. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5681–3. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi K, Sugawara F, Mizushina Y. Inhibitors of eukaryotic DNA polymerases. Seikagaku. 2002;74:244–51. [PubMed] [Google Scholar]
- 8.Mizushina Y. Specific inhibitors of mammalian DNA polymerase species. Bioscience, biotechnology, and biochemistry. 2009;73:1239–51. doi: 10.1271/bbb.90121. [DOI] [PubMed] [Google Scholar]
- 9.Hu H, Lee HJ, Jiang C, Zhang J, Wang L, Zhao Y, et al. Penta-1,2,3,4,6-O-galloyl-beta-D-glucose induces p53 and inhibits STAT3 in prostate cancer cells in vitro and suppresses prostate xenograft tumor growth in vivo. Molecular cancer therapeutics. 2008;7:2681–91. doi: 10.1158/1535-7163.MCT-08-0456. [DOI] [PubMed] [Google Scholar]
- 10.Huh JE, Lee EO, Kim MS, Kang KS, Kim CH, Cha BC, et al. Penta-O-galloyl-beta-D-glucose suppresses tumor growth via inhibition of angiogenesis and stimulation of apoptosis: roles of cyclooxygenase-2 and mitogen-activated protein kinase pathways. Carcinogenesis. 2005;26:1436–45. doi: 10.1093/carcin/bgi097. [DOI] [PubMed] [Google Scholar]
- 11.Kuo PT, Lin TP, Liu LC, Huang CH, Lin JK, Kao JY, et al. Penta-O-galloyl-beta-D-glucose suppresses prostate cancer bone metastasis by transcriptionally repressing EGF-induced MMP-9 expression. Journal of agricultural and food chemistry. 2009;57:3331–9. doi: 10.1021/jf803725h. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Li L, Kim SH, Hagerman AE, Lu J. Anti-cancer, anti-diabetic and other pharmacologic and biological activities of penta-galloyl-glucose. Pharmaceutical research. 2009;26:2066–80. doi: 10.1007/s11095-009-9932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HH, Ho CT, Lin JK. Theaflavin-3,3′-digallate and penta-O-galloyl-beta-D-glucose inhibit rat liver microsomal 5alpha-reductase activity and the expression of androgen receptor in LNCaP prostate cancer cells. Carcinogenesis. 2004;25:1109–18. doi: 10.1093/carcin/bgh106. [DOI] [PubMed] [Google Scholar]
- 14.Hu H, Zhang J, Lee HJ, Kim SH, Lu J. Penta-O-galloyl-beta-D-glucose induces S- and G(1)-cell cycle arrests in prostate cancer cells targeting DNA replication and cyclin D1. Carcinogenesis. 2009;30:818–23. doi: 10.1093/carcin/bgp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Hagerman AE. Characterization of soluble non-covalent complexes between bovine serum albumin and beta-1,2,3,4,6-penta-O-galloyl-D-glucopyranose by MALDI-TOF MS. Journal of agricultural and food chemistry. 2004;52:4008–11. doi: 10.1021/jf035536t. [DOI] [PubMed] [Google Scholar]
- 16.Mizushina Y, Motoshima H, Yamaguchi Y, Takeuchi T, Hirano K, Sugawara F, et al. 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Marine drugs. 2009;7:624–39. doi: 10.3390/md7040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushina Y, Tanaka N, Yagi H, Kurosawa T, Onoue M, Seto H, et al. Fatty acids selectively inhibit eukaryotic DNA polymerase activities in vitro. Biochimica et biophysica acta. 1996;1308:256–62. doi: 10.1016/0167-4781(96)00121-2. [DOI] [PubMed] [Google Scholar]
- 18.Mizushina Y, Yoshida S, Matsukage A, Sakaguchi K. The inhibitory action of fatty acids on DNA polymerase beta. Biochimica et biophysica acta. 1997;1336:509–21. doi: 10.1016/s0304-4165(97)00067-6. [DOI] [PubMed] [Google Scholar]
- 19.Umeda S, Muta T, Ohsato T, Takamatsu C, Hamasaki N, Kang D. The D-loop structure of human mtDNA is destabilized directly by 1-methyl-4-phenylpyridinium ion (MPP+), a parkinsonism-causing toxin. European journal of biochemistry/FEBS. 2000;267:200–6. doi: 10.1046/j.1432-1327.2000.00990.x. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa A, Murate T, Suzuki M, Nimura Y, Yoshida S. Lithocholic acid, a putative tumor promoter, inhibits mammalian DNA polymerase beta. Jpn J Cancer Res. 1998;89:1154–9. doi: 10.1111/j.1349-7006.1998.tb00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamiya-Koizumi K, Murate T, Suzuki M, Simbulan CM, Nakagawa M, Takemura M, et al. Inhibition of DNA primase by sphingosine and its analogues parallels with their growth suppression of cultured human leukemic cells. Biochemistry and molecular biology international. 1997;41:1179–89. doi: 10.1080/15216549700202271. [DOI] [PubMed] [Google Scholar]
- 22.Oda M, Ueno T, Kasai N, Takahashi H, Yoshida H, Sugawara F, et al. Inhibition of telomerase by linear-chain fatty acids: a structural analysis. The Biochemical journal. 2002;367:329–34. doi: 10.1042/BJ20021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama C, Saneyoshi M. Inhibitory effects of 9-beta-D-xylofuranosyladenine 5′-triphosphate on DNA-dependent RNA polymerase I and II from cherry salmon (Oncorhynchus masou) Journal of biochemistry. 1985;97:1385–9. doi: 10.1093/oxfordjournals.jbchem.a135192. [DOI] [PubMed] [Google Scholar]
- 24.Mizushina Y, Dairaku I, Yanaka N, Takeuchi T, Ishimaru C, Sugawara F, et al. Inhibitory action of polyunsaturated fatty acids on IMP dehydrogenase. Biochimie. 2007;89:581–90. doi: 10.1016/j.biochi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Ishimaru C, Yonezawa Y, Kuriyama I, Nishida M, Yoshida H, Mizushina Y. Inhibitory effects of cholesterol derivatives on DNA polymerase and topoisomerase activities, and human cancer cell growth. Lipids. 2008;43:373–82. doi: 10.1007/s11745-007-3149-y. [DOI] [PubMed] [Google Scholar]
- 26.Soltis DA, Uhlenbeck OC. Isolation and characterization of two mutant forms of T4 polynucleotide kinase. The Journal of biological chemistry. 1982;257:11332–9. [PubMed] [Google Scholar]
- 27.Lu BC, Sakaguchi K. An endo-exonuclease from meiotic tissues of the basidiomycete Coprinus cinereus. Its purification and characterization. The Journal of biological chemistry. 1991;266:21060–6. [PubMed] [Google Scholar]
- 28.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. Faseb J. 1997;11:68–76. [PubMed] [Google Scholar]
- 29.Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hubscher U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. Journal of molecular biology. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 30.Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu and terminal deoxyribonucleotidyl transferase. Journal of molecular biology. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 31.Pandey V, Modak MJ. Biochemistry of terminal deoxynucleotidyltransferase (TdT): characterization and mechanism of inhibition of TdT by P1, P5-bis(5′-adenosyl) pentaphosphate. Biochemistry. 1987;26:2033–8. doi: 10.1021/bi00381a036. [DOI] [PubMed] [Google Scholar]
- 32.Pandey VN, Modak MJ. Biochemistry of terminal deoxynucleotidyltransferase. Identification and unity of ribo- and deoxyribonucleoside triphosphate binding site in terminal deoxynucleotidyltransferase. The Journal of biological chemistry. 1989;264:867–71. [PubMed] [Google Scholar]
- 33.Roychoudhury R, Jay E, Wu R. Terminal labeling and addition of homopolymer tracts to duplex DNA fragments by terminal deoxynucleotidyl transferase. Nucleic acids research. 1976;3:863–77. doi: 10.1093/nar/3.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–51. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 35.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–61. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 36.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science (New York, NY) 1994;264:1891–903. [PubMed] [Google Scholar]
- 37.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science (New York, NY) 1994;264:1930–5. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 38.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. The Journal of biological chemistry. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HJ, Seong YH, Bae KH, Kwon SH, Kwak HM, Nho SK, et al. Beta-secretase (BACE1) inhibitors from Sanguisorbae Radix. Archives of pharmacal research. 2005;28:799–803. doi: 10.1007/BF02977345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.