Abstract
Adult skeletal muscles retain an adaptive capacity to switch between slow- and fast-twitch properties that are largely dependent on motoneuron activity. Our studies on the transcriptional regulation of the Troponin I slow (TnIs) and fast (TnIf) genes uncovered a dual mechanism of transcriptional enhancement and repression by a single activity pattern, that promotes the phenotypic differences among myofibers while preserving their adaptive capacity. Using the Tnf Fast Intronic Regulatory Element (FIRE), we initially demonstrated that fast-patterned activity (infrequent, high frequency depolarization) is necessary to up-regulate FIRE-dependent transcription and that its effect differs dramatically from muscle denervation. Hence, the "fast muscle program" is not a default state mimicked simply by denervation or muscle inactivity. Next, we found that slow-patterned activity (tonic, slow frequency stimulation) selectively represses FIRE-dependent transcription while enhancing transcription from the TnIs Slow Upstream Regulatory Element (SURE). Unexpectedly, repression of the TnIf FIRE by slow-patterned activity is mediated by an NFAT element that directly binds NFATc1, a transcription factor that translocates to the nucleus selectively by slow-pattern depolarization and has been implicated in the up-regulation of the slow muscle program. Transfection of siRNAs targeting NFATc1 or mutation of the TnIFIRE NFAT site result in the upregulation of FIRE-dependent transcription in slow muscle, but have no effect in fast muscle. These findings demonstrate a novel function of NFAT as a repressor of transcription of fast contractile genes in slow muscles and, more importantly, they illustrate how specific activity patterns can enhance the phenotypic differences among fibre-types by differentially regulating transcription in a use-dependent manner while retaining the adaptive properties of adult muscles.
Keywords: Skeletal-muscle, Fibre-types, Activity NFATc1, Gene-regulation
Introduction
The contractile properties of skeletal muscle are highly adaptive to environmental conditions. They are modified during aging, physical training, immobilization, in the absence of gravity (as occurs in space flight), under pathological conditions such as diabetes, cancer, neuromuscular disease (see rev. (Sartorelli and Fulco 2004)) or experimentally by stimulation of either the motor neuron axons or the muscle directly. These changes are largely regulated by the selective transcriptional repression or activation of genes encoding contractile proteins in response to distinct nerve derived electrical patterns, making transcription an important regulatory switch (see rev. (Buonanno and Rosenthal 1996; Gundersen 1998; Schiaffino et al. 2007; Spangenburg and Booth 2003)).
Development, nerve-activity and denervation
Developmental studies suggest that the initial specialization into distinct fibre types is influenced by innate properties of differentiating myoblasts (i.e., imprinting) and/or distinct cell lineages that give rise to slow- and fast-twitch myofibres. Muscle diversity is apparent prior to innervation and during perinatal development (DiMario and Stockdale 1997 and 1993; Milner et al. 1998; Rafuse et al. 1996; Thompson et al. 1990; Vogel and Landmesser 1987). Cell autonomous differentiation into distinct fibre-types, in absence of external influences from innervation, has been reported during in vivo and in vitro development both in rodent and avian muscles (Butler-Browne et al. 1982; Calvo et al. 1996; Calvo et al. 2001; Condon et al. 1990; DiMario et al. 1993; Hallauer et al. 1993; Miller and Stockdale 1986).
During the first weeks after birth, as rodents are beginning to walk and coordinate movements, muscles largely receive unpatterned activity. It is not until the second week that differential patterns of activity become evident (Personius and Balice-Gordon 2001; Vrbova et al. 1985), and by P15 they resemble those of adult muscle fibres (Hennig and Lomo 1985; Personius and Belice-Gordon 2001). In the spinal cord, gap-junctional coupling is present among motor neurons at birth, but is no longer present by P7 (Chang et al. 1999). The timing of the loss of motor neuron gap-junctional coupling closely matches the loss of temporally correlated motoneuron activity (Vrbova et al. 1985), and the eventual emergence of different muscle types (see Buonanno and Rosenthal, 1996).
In 1960 Buller and Eccles conducted an elegant experiment where denervated fast-twitch muscles were cross-re-innervated by a "slow nerve", and the muscle adopted slow-twitch properties; on the other hand when denervated slow muscles were re-innervated by "fast nerves" the muscles adopted fast phenotypes (Buller et al. 1960). Based on these observations, the authors hypothesized that a substance released from the ectopic grafted nerves crossed the neuromuscular junctions and traversed the length of myofibers,to transform their contractile properties to match either the slow or fast firing properties of the motoneuron. To evaluate this hypothesis and determine directly if the change in muscle contractile properties (twitch, tetanic and stimulation response) in response to re-innervation were due to electrical activity or chemical factors from the nerve, Lømo and collaborators implanted electrodes around denervated muscles that would deliver firing patterns mimicking the endogenous nerve activity (Lomo et al. 1974). Denervated slow muscles that were stimulated directly with a fast activity pattern via implanted electrodes changed their muscle phenotypes in the fast direction, while denervated fast muscles that were stimulated with a slow activity pattern changed their muscle phenotypes in the slow direction. Later it was shown that the motoneuron-derived signals were largely provided by the depolarization patterns, since cross-innervation, stimulation of the nerve or direct stimulation of denervated muscle all gave similar results (Eken and Gundersen 1988; Pette and Vrbova 1992; Windisch et al. 1998). The instructive signals provided by the depolarization and their effectiveness is dependent on the pattern and amount of action-potentials evoked in the muscle (Eken and Gundersen 1988; Gundersen and Eken 1992; Pette and Vrbova 1992; Salmons and Sreter 1976).
Denervation generally leads to altered contractile properties in both fast and slow muscles, and the re-expression of many embryonic/fetal genes (Buonanno and Rosenthal 1996). The fast EDL becomes slower with time while the slow soleus becomes faster (Gundersen 1985; Gutmann et al. 1972). A gradual loss of fibre-type-specific myofibrillar profiles associated with the up-regulation of isoforms not normally detected or only found during development is observed. In denervated rat soleus muscle there is an increase in type IIa and IIx fibres at the expense of type I fibres. The loss of slow type I fibres in favor of type IIa and IIx, and the fact that fast muscles are depolarized more infrequently by motoneurons, has lead to the suggestion that a fast "default program" prevails in the absence of innervation (Butler-Browne et al. 1982; Esser et al. 1993; Jerkovic et al. 1997; Windisch et al. 1998). However, as will be discussed below, studies on the transcriptional regulation of the TnI genes support the existence of a fast muscle regulatory program that differs from denervation. Instead, these studies support the idea that activity serves to enhance the phenotypic differences between muscles, and that in its absence, the degree of phenotypic differences is diminished.
We chose the genes encoding TnIs and TnIf as models to investigate the mechanism underlying nerve activity dependent regulation of fibre-type-specificity because the two isoforms are restricted either to slow-twitch muscles (i.e, expressing MHC type-I) or fast twitch muscles (i.e., expressing MHC type-IIb, IIx, IIa), respectively. Expression of TnIs mRNA, and to a lesser extent the TnIf mRNA, decrease after denervation in the rat soleus and EDL muscles, respectively. This observation is consistent with experiments in which unloading of the soleus muscle results in a slow to fast transition, and is accompanied by a reduction in slow troponins and increase of their fast counter parts (Yu et al. 2007). Electrical stimulation of denervated soleus muscles via implanted electrodes selectively results in an increase of TnIs mRNA levels when “slow” electrical patterns are used, or of TnIf transcript levels when “fast” electrical patterns are used (Calvo et al. 1996). Taken together, these findings suggest that nerve-derived activity can both down- and up-regulate gene expression selectively.
Cis-regulatory elements in Troponin I
The transcriptional regulation of the rat TnIs and quail TnIf genes were initially studied in transfected muscle cells in culture, where the cell-type and developmental regulation of TnI during myoblast differentiation could be analyzed. Regulatory sequences within the upstream region of TnIs gene (Banerjee-Basu and Buonanno 1993) and the intronic region of TnIf gene (Hastings and Emerson 1982; Yutzey et al. 1989) were identified using deletion studies. However, given the limited differentiation of myotubes in culture that fail to manifest adult muscle properties, experiments to identify the DNA elements that confer muscle-type specificity were performed in transgenic mice. These studies led to the identification of the TnIs Slow Upstream Regulatory Element (SURE) and the TnIf Fast Intronic Regulatory Element (FIRE) that are sufficient and necessary to confer slow and fast fibre-type-specific transcription, respectively (Nakayama et al. 1996). In addition, these elements also confer tissue- and developmental-specific transcription. The adult muscle expression patterns driven by SURE and FIRE emerge in three developmental steps, namely: 1) activation in primary fibres, 2) activation, but to markedly higher levels, in secondary fibres, and 3) differential augmentation during postnatal maturation of the fast fibre population, (Calvo et al. 2001; Hallauer and Hastings 2002). These results indicate the operation of multiple developmental mechanisms of TnI fibre-type–related gene regulation, including both gene activation and gene repression mechanisms.
Electroporation and in vivo analysis of adult muscles
In transgenic animals where genes have been injected into fertilized eggs, the genes may undergo regulation throughout development. Genes transfected by electroporation into adult muscles, on the other hand, are not subject to developmental regulation, and manifest fibre-type-specific properties of mature muscles. For this reason, this type of studies serve as a useful model to study activity-dependent regulation of muscle genes.
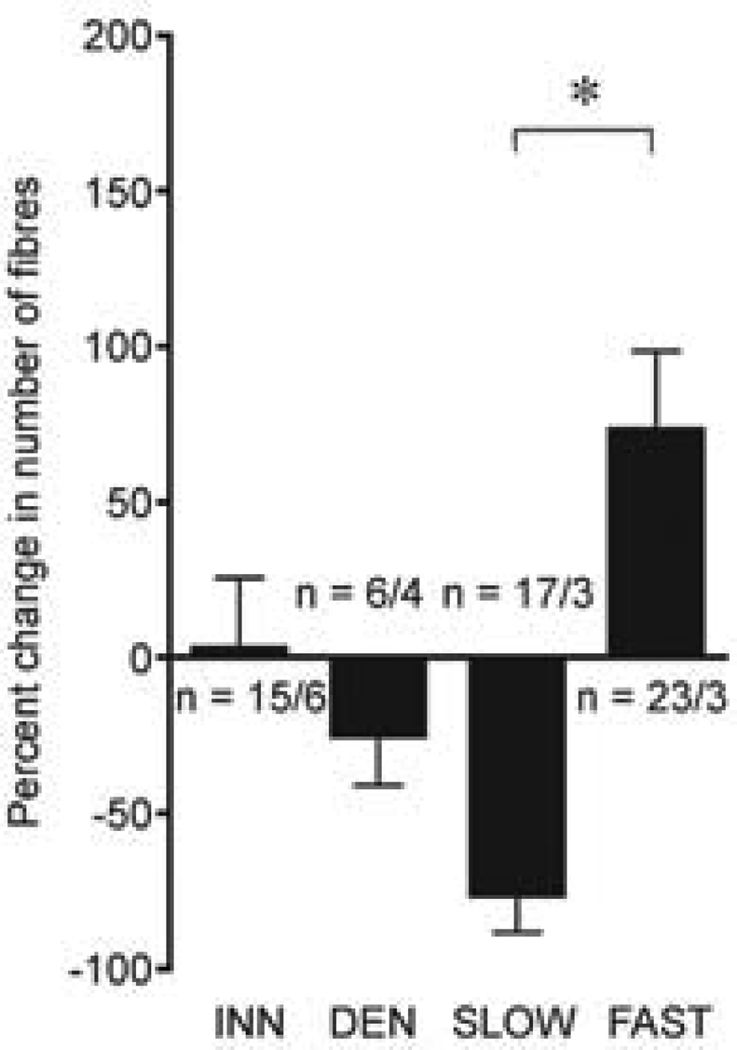
To this end, we used a combination of in vivo transfection, live muscle imaging and fluorescence quantification to investigate the role of electrical activity pattern in the transcriptional control of TnIf regulatory sequences by directly stimulating denervated muscles with patterns that mimic fast and slow motor units (Rana et al. 2005). Rat soleus muscles were electroporated with green fluorescent protein (GFP) reporter constructs harbouring 2.1 kb of TnIf regulatory sequence. One week later, electrodes were implanted and muscles stimulated for 12 days. The change in GFP fluorescence of individual muscle fibres before and after the stimulation was used as a measure for transcriptional responses to different patterns of action- potentials. Our results indicated that the response of TnI promoter sequences to electrical stimulation was consistent with the regulation of the endogenous genes (Calvo et al. 1996). The TnIf enhancer was activated by a matching fast pattern. Removal of nerve-evoked activity by denervation, or stimulation with a mismatching pattern, reduced the transcriptional activity (fig. 1) (Rana et al. 2005). These results differ from the previously proposed “default program” that prevails in the absence of innervation (Butler-Browne et al. 1982; Esser et al. 1993; Jerkovic et al. 1997), and provides evidence for the existence of a separate 'fast muscle program'.
Fig. 1. TnIf2100 promoter is activated selectively by fast-patterned electrical stimulation.
Myofibres transfected by electroporation with the TnIf2100 EGFP reporter construct were studied in innervated and denervated solei, as well as denervated soleus muscles stimulated with either fast or slow activity patterns. Bar graphs are showing the average change (%) in the number of fluorescent fibres. The number of fibres/muscles (n/n) included in the analysis is shown for each treatment. * P < 0.05. This panel is from Rana et al. 2005.
Repression of TnIf gene by NFATc1
The FIRE shares several cis-acting elements with the slow TnI SURE enhancer. Some are present in the general muscle-specific region of both enhancers, and others map to the fibre-type-specific region. The SURE enhancer has one putative NFAT site, which partially overlaps an E-box-like site, while the FIRE has three NFAT-like sites spread across the FIRE region. Studies have shown that GTF3 (also known as MusTRD1 or GTF2IRD1) binds to the fibre-type-specific region of SURE and mediates slow gene repression (Calvo et al. 2001), whereas FIRE does not share any GTF3 binding sequence with SURE.
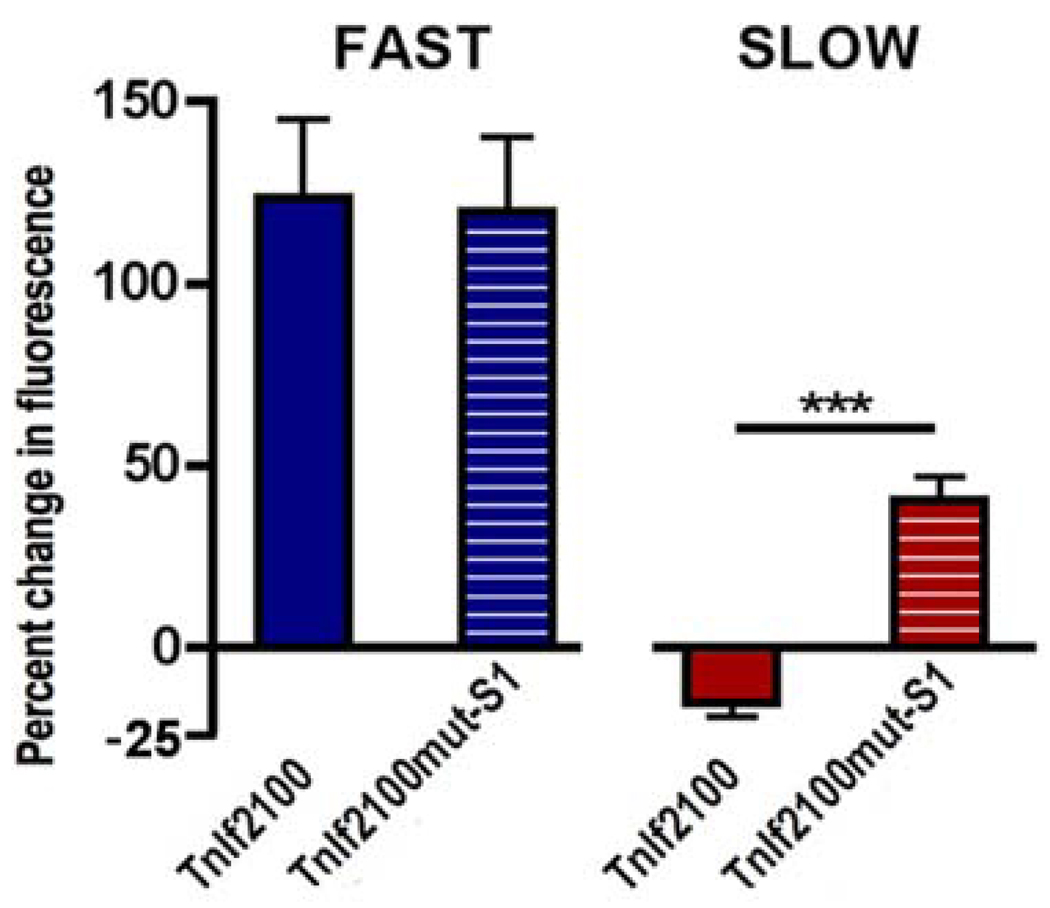
By utilizing EMSAs, we identified one site in the fibre-type-specific region of FIRE that showed binding capacity to NFATc1 (Rana et al. 2008). Through in vivo transfection, live muscle imaging and fluorescence quantification, as previously described (Rana et al. 2005), we tested the FIRE promoter with and without point mutations in the NFATc1 binding site. Interestingly, this site indicated that it is important for repression of FIRE activity in the slow soleus muscle. At the same time the mutation of this site had no effect on FIRE activity in the fast EDL (fig. 2). Consistent with this result, transfection of siRNAs targeting NFATc1 prevented the down-regulation of TnI FIRE by slow activity; no effect was observed in the EDL. In conclusion, either mutation of the NFATc1 site in FIRE or the knock-down of NFATc1 result in the inability of slow activity to repress TnIf transcription in slow muscles, but have no measurable effect in fast muscles (Rana et al. 2008). The selective reduction of TnIf transcription in slow muscles constitutes an activity-dependent mechanism that broadens the differences of TnI isotype expression in slow muscles.
Fig. 2. Mutation of the NFAT site selectively prevents TnIf FIRE repression by slow-patterned activity.
Soleus myofibres transfected by electroporation with TnIf2100 and TnIf2100mut-S1 EGFP reporter constructs and stimulated with either fast or slow activity patterns. To measure enhancer function, myofibres were imaged 1 week after transfection, at the time of denervation and onset of stimulation. A second image was taken after 12 days of stimulation. Bars represent relative changes (%) in EGFP in single soleus fibres stimulated for 12 days with either fast (blue) or slow (red) patterns. Values represent the mean SEM (40 – 72 myofibres from six muscles per condition; ***, P 0.001; one-way ANOVA with Bonferroni post test). This panel is from Rana et al. 2008.
A model of gene regulation in skeletal muscle
The findings discussed herewith provide a new way of thinking about mechanisms that lead to differential gene expression in fast and slow muscles. Traditionally, people have considered how patterned activity increases or induces gene expression. However, our data suggest that the mechanisms are more complex. We have shown that activity can serve to both increase and decrease expression of genes as to functionally match the depolarization patterns, but that it can also serve to down-regulate those genes that are not functionally matched to motoneuron activity. We have proposed that it is the combination of both stimulatory and repressive effects of activity that results in such exquisite separation of functional properties between slow- and fast-twitch fibres, in such way that the capacity of muscles to retain their receptiveness to environmental changes within a dynamic range is not sacrificed.
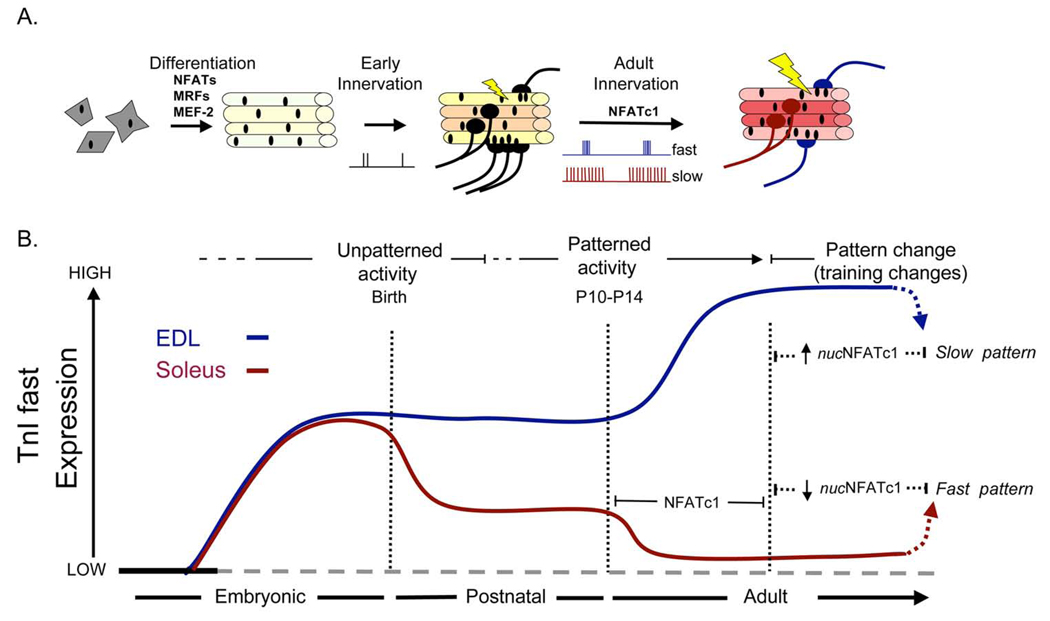
We propose the following model for regulation of fast and slow TnI genes (fig. 3). TnIf is not only up-regulated in fast fibres but also actively down-regulated in slow fibres. This promotes the idea that the TnIf is transcribed at intermediate levels in any given fibre-type, but is down- or up-regulated according to cell-lineage (Calvo et al. 2001), and by patterned electrical activity after birth (Personius and Belice-Gordon 2001). During the embryonic stage TnIf is being expressed non-selectively, but closer to birth the TnIf expression is repressed in future slow muscles. After birth when motoneuron innervating muscles start firing distinct electrical patterns, TnIf is up-regulated in the fast muscles and down-regulated in the slow. With removal of the nerve the transcription levels return to post innervation levels at birth. By reversing the external influence by imposed electrical patterns, the gene activity can be reversed. The intermediate levels of gene expression allows for greater dynamic range for changes in either direction and retains the plastic properties of adult muscles.
Fig. 3. Model representing the developmental and activity-dependent regulation of TnIf in slow- and fast-twitch myofibres.
(A) Depiction of the progressive stages of skeletal muscle differentiation, motoneuron innervation and retraction, and the role of motoneuron patterned activity. While motoneuron and muscle diversification are evident during embryonic/fetal and perinatal development, further specialization of adult slow- and fast-twitch myofibres after P7 coincides with their stimulation by slow and fast patterns of motoneuron activity. The different colour intensities of myofibres are to emphasize that slow and fast motoneuron activity enhances the differences between slow- and fast-twitch muscles, but yet adult muscles retain an adaptive capacity. (B) Illustration of the transcriptional regulation of TnIf in developing and maturing fast (blue) and slow (red) myofibres. This panel is from Rana et al. 2008.
References
- Banerjee-Basu S, Buonanno A. cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol Cell Biol. 1993;13(11):7019–7028. doi: 10.1128/mcb.13.11.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller AJ, Eccles JC, Eccles RM. Interactions between motoneurones and muscles in respect of the characteristic speeds of their responses. J Physiol. 1960;150:417–439. doi: 10.1113/jphysiol.1960.sp006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A, Rosenthal N. Molecular control of muscle diversity and plasticity. Dev Genet. 1996;19(2):95–107. doi: 10.1002/(SICI)1520-6408(1996)19:2<95::AID-DVG1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Butler-Browne GS, Bugaisky LB, Cuenoud S, Schwartz K, Whalen RG. Denervation of newborn rat muscle does not block the appearance of adult fast myosin heavy chain. Nature. 1982;299(5886):830–833. doi: 10.1038/299830a0. [DOI] [PubMed] [Google Scholar]
- Calvo S, Stauffer J, Nakayama M, Buonanno A. Transcriptional control of muscle plasticity: differential regulation of troponin I genes by electrical activity. Dev Genet. 1996;19(2):169–181. doi: 10.1002/(SICI)1520-6408(1996)19:2<169::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Calvo S, Vullhorst D, Venepally P, Cheng J, Karavanova I, Buonanno A. Molecular dissection of DNA sequences and factors involved in slow muscle-specific transcription. Mol Cell Biol. 2001;21(24):8490–8503. doi: 10.1128/MCB.21.24.8490-8503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Gonzales M, Pinter MJ, Balice-Gordon RJ. Gap junctional coupling and petterns of connexin expression among neonatal rat lumbar spinal motor neurons. J. Neurosci. 1999;19:10813–10828. doi: 10.1523/JNEUROSCI.19-24-10813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon K, Silberstein L, Blau HM, Thompson WJ. Differentiation of fiber types in aneural musculature of the prenatal rat hindlimb. Dev Biol. 1990;138(2):275–295. doi: 10.1016/0012-1606(90)90197-q. [DOI] [PubMed] [Google Scholar]
- DiMario JX, Fernyak SE, Stockdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature. 1993;362(6416):165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- DiMario JX, Stockdale FE. Both myoblast lineage and innervation determine fiber type and are required for expression of the slow myosin heavy chain 2 gene. Dev Biol. 1997;188(1):167–180. doi: 10.1006/dbio.1997.8619. [DOI] [PubMed] [Google Scholar]
- Eken T, Gundersen K. Electrical stimulation resembling normal motor-unit activity: effects on denervated fast and slow rat muscles. J Physiol (Lond) 1988;402:651–659. doi: 10.1113/jphysiol.1988.sp017227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser K, Gunning P, Hardeman E. Nerve-dependent and -independent patterns of mRNA expression in regenerating skeletal muscle. Dev Biol. 1993;159(1):173–183. doi: 10.1006/dbio.1993.1231. [DOI] [PubMed] [Google Scholar]
- Gundersen K. Early effects of denervation on isometric and isotonic contractile properties of rat skeletal muscles. Acta. Phsiol. Scand. 1985;124(4):549–555. doi: 10.1111/j.1748-1716.1985.tb00047.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K. Determination of muscle contractile properties: the importance of the nerve. Acta Physiol Scand. 1998;162(3):333–341. doi: 10.1046/j.1365-201X.1998.0336e.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K, Eken T. The importance of frequency and amount of electrical stimulation for contractile properties of denervated rat muscles. Acta. Phsiol. Scand. 1992;145(1):49–57. doi: 10.1111/j.1748-1716.1992.tb09335.x. [DOI] [PubMed] [Google Scholar]
- Gutmann E, Melichna J, Syrovy I. Contraction properties and ATPase activity in fast and slow muscle of the rat during denervation. Exp. Neurol. 1972;36:488–497. doi: 10.1016/0014-4886(72)90008-8. [DOI] [PubMed] [Google Scholar]
- Hallauer PL, Bradshaw HL, Hastings KE. Complex fiber-type-specific expression of fast skeletal muscle troponin I gene constructs in transgenic mice. Development. 1993;119(3):691–701. doi: 10.1242/dev.119.3.691. [DOI] [PubMed] [Google Scholar]
- Hallauer PL, Hastings KE. TnIfast IRE Enhancer: Multistep Developmental Regulation During Skeletal Muscle Fiber Type Differentiation. Dev. Dyn. 2002;224:422–431. doi: 10.1002/dvdy.10122. [DOI] [PubMed] [Google Scholar]
- Hastings KEM, Emerson CP. cDNA clone analysis of six co-regulated mRNAs encoding skeletal muscle contractile proteins. Proc. Natl. Acad. Sci. USA. 1982;79:1553–1557. doi: 10.1073/pnas.79.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig R, Lomo T. Firing patterns of motor units in normal rats. Nature. 1985;314(6007):164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- Jerkovic R, Vitadello M, Kelly R, Buckingham M, Schiaffino S. Fibre type-specific and nerve-dependent regulation of myosin light chain 1 slow promoter in regenerating muscle. J Muscle Res Cell Motil. 1997;18(3):369–373. doi: 10.1023/a:1018630311208. [DOI] [PubMed] [Google Scholar]
- Lomo T, Westgaard RH, Dahl HA. Contractile properties of muscle: control by pattern of muscle activity in the rat. Proc R Soc Lond B Biol Sci. 1974;187(1086):99–103. doi: 10.1098/rspb.1974.0064. [DOI] [PubMed] [Google Scholar]
- Miller JB, Stockdale FE. Developmental origins of skeletal muscle fibers: clonal analysis of myogenic cell lineages based on expression of fast and slow myosin heavy chains. Proc Natl Acad Sci U S A. 1986;83(11):3860–3864. doi: 10.1073/pnas.83.11.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner LD, Rafuse VF, Landmesser LT. Selective fasciculation and divergent pathfinding decisions of embryonic chick motor axons projecting to fast and slow muscle regions. J Neurosci. 1998;18(9):3297–3313. doi: 10.1523/JNEUROSCI.18-09-03297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber- type-specific regulatory elements. Mol Cell Biol. 1996;16(5):2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personius KE, Balice-Gordon RJ. Loss of correlated motor neuron activity during synaptic competition at developing neuromuscular synapses. Neuron. 2001;31(3):395–408. doi: 10.1016/s0896-6273(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Personius KE, Belice-Gordon RJ. Loss of Correlated Motor Neuron Activity during Synaptic Competition at Developing Neuromuscular Synapses. Neuron. 2001;31:395–408. doi: 10.1016/s0896-6273(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Pette D, Vrbova G. Adaptation of mammalian skeletal muscle fibers to chronic electrical stimulation. Rev Physiol Biochem Pharmacol. 1992;120:115–202. doi: 10.1007/BFb0036123. [DOI] [PubMed] [Google Scholar]
- Rafuse VF, Milner LD, Landmesser LT. Selective innervation of fast and slow muscle regions during early chick neuromuscular development. J Neurosci. 1996;16(21):6864–6877. doi: 10.1523/JNEUROSCI.16-21-06864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana ZA, Gundersen K, Buoannao A. Activity-dependent repression of muscle genes by NFAT. Proc Natl Acad Sci. 2008;105(15):5921–5926. doi: 10.1073/pnas.0801330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana ZA, Gundersen K, Buonanno A, Vullhorst D. Imaging transcription in vivo: distinct regulatory effects of fast and slow activity patterns on promoter elements from vertebrate troponin I isoform genes. J Physiol. 2005;562(Pt 3):815–828. doi: 10.1113/jphysiol.2004.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmons S, Sreter FA. Significance of impulse activity in the transformation of skeletal muscle type. Nature. 1976;263(5572):30–34. doi: 10.1038/263030a0. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Fulco M. Molecular and cellular determinants of skeletal muscle atrophy and hypertrophy. Sci STKE. 2004;244 doi: 10.1126/stke.2442004re11. re11. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Sandri M, Murgia M. Activity-Dependent Signaling Pathways Controlling Muscle Diversity and Plasticity. Physiol. 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- Spangenburg EE, Booth FW. Molecular regulation of individual skeletal muscle fibre types. Acta Physiol Scand. 2003;178(4):413–424. doi: 10.1046/j.1365-201X.2003.01158.x. [DOI] [PubMed] [Google Scholar]
- Thompson WJ, Condon K, Astrow SH. The origin and selective innervation of early muscle fiber types in the rat. J Neurobiol. 1990;21(1):212–222. doi: 10.1002/neu.480210114. [DOI] [PubMed] [Google Scholar]
- Vogel M, Landmesser L. Distribution of fiber types in embryonic chick limb muscles innervated by foreign motoneurons. Dev. Biol. 1987;119(2):481–495. doi: 10.1016/0012-1606(87)90051-0. [DOI] [PubMed] [Google Scholar]
- Vrbova G, Navarrete R, Lowrie M. Matching of muscle properties and motoneurone firing patterns during early stages of development. J Exp Biol. 1985;115:113–123. doi: 10.1242/jeb.115.1.113. [DOI] [PubMed] [Google Scholar]
- Windisch A, Gundersen K, Szabolcs MJ, Gruber H, Lomo T. Fast to slow transformation of denervated and electrically stimulated rat muscle. J Physiol. 1998;510(Pt 2):623–632. doi: 10.1111/j.1469-7793.1998.623bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZB, Gao F, Feng HZ, Jin JP. Differential regulation of myofilament protein isoforms underlying the contractility changes in skeletal muscle unloading. Am J Physiol Cell Physiol. 2007;292(3):C1192–C1203. doi: 10.1152/ajpcell.00462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Kline RL, Konieczny SF. An internal regulatory element controls troponin I gene expression. Mol Cell Biol. 1989;9(4):1397–1405. doi: 10.1128/mcb.9.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]