Identifying the structures that contribute to monoclonal antibody (mAb) binding sites (epitopes) within native G protein-coupled receptors (GPCRs) can be useful for developing topological models of the accessible receptor surface, for selecting the most relevant mAbs for therapeutic, diagnostic, and research applications and for distinguishing the intellectual property positions of otherwise similar mAbs. While conventional site-directed mutagenesis studies can identify individual amino acid residues that are critical to mAb binding, defining comprehensive epitopes is difficult and time-consuming for these structurally complex proteins. For example, in studies over the past decade, 13 residues (cumulatively) in the GPCR CCR5 have been reported to contribute to the interactions of five well-studied mAbs.1–5 However, crystallographically defined epitopes contain an average of 20 contact residues each,6 so these 13 residues likely represent only a portion of all the amino acids that constitute these five epitopes. Because of the importance of CCR5 in HIV infection and inflammation,7 more mAbs have been raised against the native form of this receptor than most other GPCRs, providing a useful set of tools to map its immunodominant structural regions. Here, we have used a high-throughput structure-function analysis strategy, which we refer to as “shotgun mutagenesis”, to comprehensively map the critical residues, and in some cases the critical atoms, for these five epitopes of CCR5.
To map mAb epitopes, we used an arrayed library of mutations covering nearly all the amino acids in the protein to identify amino acid changes that resulted in loss of mAb reactivity. This approach enabled each epitope to be rapidly mapped within a period of weeks. To create the mutant library, a parental CCR5 plasmid was first created, containing the full length (1059 bp) cDNA for wild type CCR5, flanked by a N-terminal HA epitope tag and a C-terminal V5 epitope tag. Cellular expression of the wild type tagged construct was confirmed by Western blot, immunofluorescence, and flow cytometry. Random mutations were next introduced into the parental CCR5 cDNA using a PCR-based method (Diversify PCR Random Mutagenesis kit, Clontech). Sequenced clones, most exhibiting one to two substitutions, were then selected from these random mutants to create a library with substitutions spanning the entire protein. The final library comprised 734 mutant CCR5 plasmids with substitutions in 346 of the 352 residues of CCR5 (>98% coverage). The average mutation rate per clone was 1.86 amino acids, and each amino acid position was substituted multiple times (an average of 3.95) across the entire library.
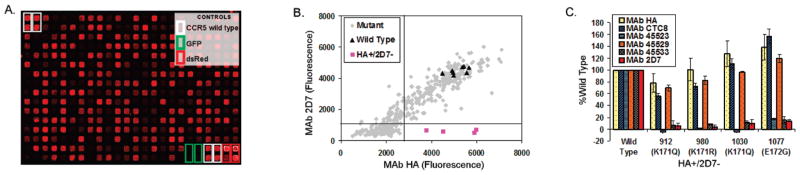
We used this selective library of CCR5 mutants to map the epitopes of the anti-CCR5 mAbs CTC8, 45523, 45529, 45533 (R&D Systems), and 2D7 (Becton Dickinson). All five mAbs were originally derived, in three independent immunizations, by injecting mice with cells transiently overexpressing human CCR5.4,8 These mAbs are therefore representative of the murine immune response to a human GPCR in its native conformation. All except CTC8 have been found to be conformation-dependent.4 Individual sequence-verified clones from the mutant plasmid library, plus controls, were arrayed in 384-well microplates and expressed in HEK-293T cells using a reverse-transfection protocol.9 After 24 h, cells were fixed and immunofluorescence was used to quantify the binding of each anti-CCR5 mAb, as well as mAbs against the HA and V5 epitope tags (Figure 1). 96% of the clones were fully translated, and 85% of the clones trafficked to the cell surface (Supplementary Figure 1). To identify the GPCR residues critical to each anti-CCR5 interaction, clones were identified that expressed on the surface at near-wild type levels (>50% of wild type HA epitope reactivity, thus eliminating mutants with gross defects in global folding) but that reacted with a given mAb at near background levels (<17% of wild type) (Table 1). To eliminate surface-expressed clones with defects in global structure, each clone was also tested for signaling activity in response to the chemokine ligand RANTES and for coreceptor function with the HIV-1 strain JRFL. Both of these receptor functions are known to require conformationally complex regions of CCR5, including extracellular loop 2 (ECL2).10–12 Clones that did not react with any conformation-dependent mAb, did not signal in response to RANTES, and did not function as a coreceptor were presumed to contain mutations that globally disrupt CCR5 structures and were eliminated from further consideration.
Figure 1.
Shotgun mutagenesis mapping of mAb epitopes. (A) Cells expressing a library of CCR5 mutants arrayed in 384-well microplates were fixed with paraformaldehyde, and immunoreactivity of each mAb (2D7 shown) was detected with a fluorescent secondary antibody and visualized on a NovaRay imager (AlphaInnotech). (B) Fluorescence intensity in each well was quantified using ArrayEase software. mAb binding was plotted as a function of surface expression, which was measured by immunoreactivity of an N-terminal HA epitope tag. Thresholds of 50% HA and 17% mAb (2D7) reactivity were then applied. Wild type clones are shown as triangles. Clones in the bottom right quadrant (squares) contain mutations that eliminate mAb epitope reactivity but that are still expressed on the surface at near wild-type levels. (C) The reactivities of the clones containing critical mutations for 2D7 are shown relative to their reactivity with other mAbs.
Table 1.
Immunofluorescence Mapping Dataa
| Residue | Clone | CTC8 | 45523 | 45529 | 45533 | 2D7 | RANTES | HIVJRFL |
|---|---|---|---|---|---|---|---|---|
| Y10 | 1429 | 16.7 (9.0) | 106.5 (17.3) | 107.8 (35.1) | 82.7 (23.5) | 100.6 (0.0) | 121 | 67 |
| D11 | 1490 | −2.1 (0.3) | 100.6 (4.6) | 89.8 (12.0) | 112.5 (10.4) | 87.2 (11.7) | 99 | 101 |
| I12 | 680 | 3.0 (3.7) | 92.7 (6.8) | 106 (11.0) | 66.8 (6.2) | 77.7 (0.5) | 120 | 93 |
| N13 | 727 | −4.4 (6.8) | 37.0 (2.7) | 36.5 (9.1) | 46.6 (0.9) | 52.3 (1.4) | 111 | 67 |
| Y15 | 621 | −9.9 (2.4) | 89.3 (2.9) | 87.9 (6.6) | 94.8 (6.8) | 81.0 (3.8) | 132 | 87 |
| I164 | 1438 | 80.7 (14.7) | −3.6 (3.2) | −9.8 (5.9) | 5.5 (3.7) | 35.1 (8.5) | 55 | 109 |
| F166 | 1312 | 78.7 (0.4) | 14.3 (10.0) | 0.7 (7.0) | 14.4 (6.5) | 60.4 (9.9) | 136 | 112 |
| K171 | 1030 | 92.9 (19.1) | −4.1 (1.5) | 80.7 (9.2) | 10.4 (2.8) | 6.8 (3.3) | 74 | 71 |
| E172 | 1077 | 82.3 (6.3) | −0.3 (1.4) | 62.8 (3.7) | 8.0 (3.0) | −5.3 (0.5) | n/a | 70 |
| F182 | 368 | 81.3 (13.8) | 64.5 (11.1) | 15.4 (12.2) | 74.4 (2.2) | 67.9 (6.0) | 21 | 91 |
| Y184 | 235 | 72.6 (4.7) | 66.2 (8.3) | −4.5 (4.2) | 78.8 (10.4) | 56.5 (1.4) | 93 | 111 |
| S185 | 976 | 79.9 (13.4) | 58.3 (10.1) | 12.9 (11.4) | 49.5 (5.2) | 58.3 (7.0) | 69 | 71 |
| Q186 | 967 | 177.9 (24.0) | 90.8 (15.9) | 3.2 (11.0) | 70.9 (0.7) | 168.7 (52.3) | 103 | 91 |
| W190 | 1042 | 64.8 (22.7) | 0.4 (8.0) | −26.9 (14.8) | 16.8 (10.7) | 119.7 (21.1) | 63 | 67 |
Results from two to three independent immunofluorescence experiments are shown (mean ± range in parentheses). To facilitate interpretation, results shown are background-subtracted, normalized to wild type reactivity with the same MAb, and normalized to surface expression of each mutant clone. Clones with reactivity of <17% with a given MAb are highlighted. The 17% threshold is approximately three standard deviations above background signals. Critical amino acids in clones containing more than one mutation were differentiated by comparing the reactivity of other clones with mutation of the same residues. Each residue was mutated to another random amino acid substitution, but only in one case did mutation to different residues disagree (for mAbs 45523, 45529, and 45533, I164T functioned at ~50% of wild type levels while I164N functioned at <17% of wild type, so I164 was considered critical for these mAbs). RANTES signaling of the E172 mutation was not measured (n/a) due to another mutation located in the G protein coupling region of the clone studied. Clone 1042, containing a mutation at W190, was expressed at <50% of wild type but was identified as critical upon analysis of all mutants in ECL2 regardless of surface expression.
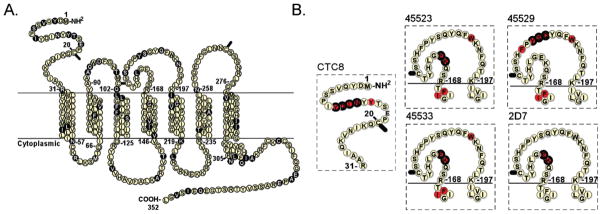
Each mAb epitope was found to comprise 2 to 7 critical amino acids (Figure 2). Shotgun mutagenesis identified all of the 13 critical residues identified by others over the past decade, as well as 11 novel critical residues. Similar to characterized epitopes on soluble proteins,6 aromatic and charged residues comprised a significant fraction of the CCR5 epitopes (17 of the 24 residues, 71%). mAb epitopes have been reported to comprise an average of 20 residues,6 but only a small percentage of these residues contribute significantly to the energetics of the interaction (e.g., ~5.4% contribute ≥2 kcal/mol).13 This suggests that the residues identified by shotgun mutagenesis likely include all or nearly all of the critical residues for these five mAb epitopes (the “hot spots” that are the energetically significant contact points). Other residues likely make contact with each mAb but might not be identified in this screen for several reasons, including weak interactions that do not substantially influence mAb binding, mutations that result in poor surface expression, or mAb interactions with amino acid C-alpha atoms.
Figure 2.
Epitopes of CCR5 mAbs. (A) Residues that differ between human and murine CCR5 (83% overall identity) are shown as shaded circles. (B) The epitopes of five mAbs identified using shotgun mutagenesis are shown, shaded in red. Residues previously reported as critical to these epitopes are circled in bold (residues that could not be distinguished individually as critical in published studies, e.g., chimeras, multiple substitutions, and mutations that only partially lowered activity, were not included).
Immunogenic regions are typically heterologous in sequence from the host species due to self-tolerance,6,14 and none of the mAbs characterized here react with murine CCR5 (data not shown). It was therefore surprising that most of the epitopes identified here exhibited high sequence conservation between the human and murine receptor. Of the 24 critical residues identified, 20 of the residues (83%), including all of the critical residues for 45523, 45533, and 2D7, were identical between human and murine CCR5. This suggests that identical amino acid sequences can still present immunogenically unique structures, possibly due to their context within a conformationally complex receptor or post-translational processing. Thus, sequence homology to murine GPCRs does not necessarily preclude deriving useful mAbs in mice. Subtle changes in structure may also explain how autoantibodies against human GPCRs are generated in certain autoimmune disorders.15
All critical residues identified by shotgun mutagenesis were localized to the predicted extracellular regions of the receptor. Interestingly, neither disruption nor enhancement of mAb epitopes by mutations in distant regions of the GPCR (e.g., allosteric effects caused by mutations buried in the transmembrane or intracellular regions) was observed. Collectively, the critical residues comprising the CCR5 epitopes were clustered in three regions: the N-terminus (Nt), the first half of ECL2 (ECL2a), and the second half of ECL2 (ECL2b). No critical residues were identified in ECL1 or ECL3, although some residues in these regions contributed weakly or had effects on global structure. Collectively, these data suggest that the Nt, ECL2a, and ECL2b likely comprise the sole immunodominant regions of the native CCR5 protein. Since epitope contact points are almost always surface-exposed,6 our results also imply that the Nt, ECL2a, and ECL2b are likely the most surface-accessible regions of CCR5 and that the specific CCR5 residues identified here have surface-accessible side chains.
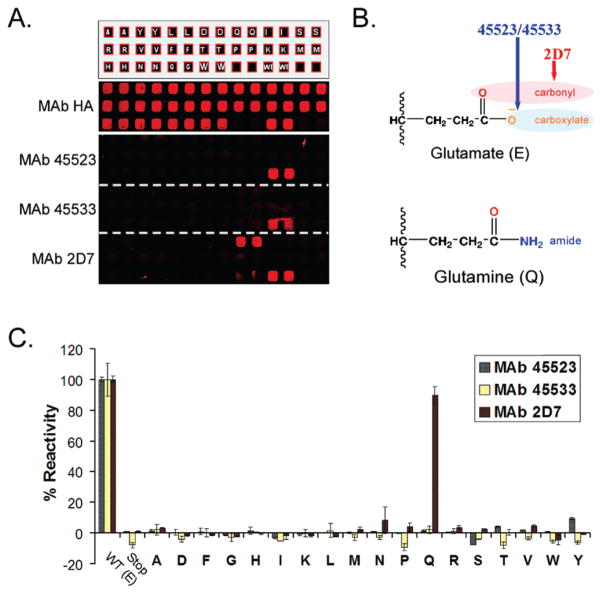
Our mapping results indicated that a number of amino acids are critical to the binding epitopes of more than one mAb. To test whether these mAbs interact with common residues in the same way, we mutated one such critical residue, E172, to 18 other amino acids and then tested the ability of each mutant to bind 45523, 45533, and 2D7. Because many amino acids differ by only single atoms in their side chains, we reasoned that this type of comprehensive substitution at a single position might reveal atomic-level structures that could differentiate each interaction. Indeed, we found that Q was an acceptable substitute for E in supporting the binding of mAb 2D7, while none of the mutations at this position permitted binding of mAbs 45523 or 45533 (Figure 3). These results suggest that similar antibodies can interact with common residues, such as E172, differently. Specifically, our data are consistent with (1) mAb 2D7 interacting with the carbonyl of E172 (common to both E and Q side chains) via interactions that are less charge dependent such as hydrogen bonds or dipolar interactions and (2) mAbs 45523 and 45533 interacting directly with the negatively charged carboxylate of E172 (unique to the E side-chain) via an ionic interaction. Interestingly, mAbs 45523 and 45533 (IgG2b and IgG1 isotypes, respectively) exhibited identical epitope maps that could not be distinguished at any level, suggesting that CCR5 presents a limited repertoire of immunodominant regions that may be represented by the mAbs analyzed here.
Figure 3.
Detailed mapping of epitopes. (A) Residue E172 of CCR5 was mutated to 18 other amino acids, and duplicate wells expressing each arrayed mutant were tested for binding of the mAbs 45523, 45533, and 2D7. None of the substitutions at position 172 supported binding of 45523 or 45533, while E172Q (alone) supported binding of 2D7. (B) These results suggest that 45523 and 45533 are interacting with the carboxylate of the E172 side chain (unique to E), while 2D7 is likely interacting with the carbonyl component of the side chain (exhibited by both E and Q). (C) Mean and range (error bars) of duplicate wells are quantified, and each experiment was repeated at least twice. All amino acid substitutions were detected at near-wild type levels for surface expression (HA), full-length translation (V5), and coreceptor function (HIV JRFL) (data not shown).
Our results suggest that a shotgun mutagenesis mapping approach can be used to accurately and comprehensively identify critical residues required for GPCR interactions with mAbs and other molecules. In our studies of CCR5, three structures—the Nt, ECL2a, and ECL2b—appear to be the sole immunodominant regions of the GPCR CCR5. Their immunodominance does not appear related to divergence from murine CCR5 but rather to their predicted surface-accessibility and side-chain physicochemistry (charged, aromatic). Epitopes could be distinguished by individual residues, and in some cases atoms, that were critical for mAb binding. GPCRs are a structurally conserved superfamily, so the epitope maps generated here for CCR5 may serve as a model for predicting and mapping the immunodominant regions of other GPCRs.
Supplementary Material
Acknowledgments
We thank Christopher Laing and Soma Banik-Banerjee for assistance in writing the manuscript and Bridget Puffer and David Sieg for molecular biology expertise and helpful discussions. We thank AlphaInnotech for supporting the application of a NovaRay imager in these studies. Funding for this work was provided by the National Institutes of Health (GM076779).
Footnotes
Supporting Information Available: Plot illustrating expression of CCR5 mutation library. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhang J, Rao E, Dioszegi M, Kondru R, DeRosier A, Chan E, Schwoerer S, Cammack N, Brandt M, Sankuratri S, Ji C. Antimicrob Agents Chemother. 2007;51:1386–1397. doi: 10.1128/AAC.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Lee B, Vakili J, Doranz BJ, Govaerts C, Migeotte I, Sharron M, Dupriez V, Vassart G, Doms RW, Parmentier M. J Biol Chem. 1999;274:18902–18908. doi: 10.1074/jbc.274.27.18902. [DOI] [PubMed] [Google Scholar]
- 4.Lee B, Sharron M, Blanpain C, Doranz BJ, Vakili J, Setoh P, Berg E, Liu G, Guy HR, Durell SR, Parmentier M, Chang CN, Price K, Tsang M, Doms RW. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 5.Olson WC, Rabut GE, Nagashima KA, Tran DN, Anselma DJ, Monard SP, Segal JP, Thompson DA, Kajumo F, Guo Y, Moore JP, Maddon PJ, Dragic T. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinstein ND, Mayrose I, Halperin D, Yekutieli D, Gershoni JM, Pupko T. Mol Immunol. 2008;45:3477–3489. doi: 10.1016/j.molimm.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Berger EA, Murphy PM, Farber JM. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 8.Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, Choe H, Sodroski J, Newman W, Koup RA, Mackay CR. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziauddin J, Sabatini DM. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 10.Rucker J, Samson M, Doranz BJ, Libert F, Berson JF, Yi Y, Smyth RJ, Collman RG, Broder CC, Vassart G, Doms RW, Parmentier M. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 11.Doranz BJ, Lu ZH, Rucker J, Zhang TY, Sharron M, Cen YH, Wang ZX, Guo HH, Du JG, Accavitti MA, Doms RW, Peiper SC. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanpain C, Doranz BJ, Vakili J, Rucker J, Govaerts C, Baik SS, Lorthioir O, Migeotte I, Libert F, Baleux F, Vassart G, Doms RW, Parmentier M. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 13.Bogan AA, Thorn KS. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 14.Berzofsky JA. Science. 1985;229:932–940. doi: 10.1126/science.2410982. [DOI] [PubMed] [Google Scholar]
- 15.Mobini R, Magnusson Y, Wallukat G, Viguier M, Hjalmarson A, Hoebeke J. J Autoimmun. 1999;13:179–186. doi: 10.1006/jaut.1999.0310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.