Understanding the electrostatic environment within the idiosyncratic interior of folded proteins and its connection to biomolecular function remains a key challenge in biochemistry and biophysics. Vibrational probes incorporated into proteins on specific residues or ligands are exquisitely sensitive reporters of the local environment and how it is altered by pH changes, mutations, structural perturbations, or variations in bound ligands1-6. While IR frequency shifts associated with various environments can be considered at a qualitative level (e.g., is the probe buried or on a solvent exposed surface?7,8), our goal has been to extract quantitative information on electric fields in proteins1,2. Although several studies have suggested a connection between observed vibrational band shifts and local electrostatic fields due to the organized environment around the probe3,6, in the absence of an independent experimental test it remains uncertain whether these shifts are due principally to electrostatic effects or are dominated by contributions from specific chemical interactions9, such as hydrogen bonds. We report herein a method for identifying and quantifying departures from an electrostatic mechanism for nitrile vibrational shifts that utilizes the relationship between IR frequency shifts and 13C NMR chemical shifts and demonstrate its utility in a protein.
The vibrational Stark effect (VSE) provides the connection between observed IR frequency shifts, (in cm-1), and the difference in the local electrostatic field, (in MV/cm), experienced by a probe at two different sites in a protein or as a result of a pH change, mutation, or ligand binding1,2. The sensitivity of an oscillator to electrostatic field is the Stark tuning rate, [in cm-1/(MV/cm)], which is obtained by measuring the effect of an applied electric field on the IR spectrum11-13. The nitrile stretch offers an optimal combination of oscillator strength, frequency, and Stark tuning rate for measurements in biological systems11-14. In this case, observed frequency shifts, , can be used to obtain information on variations in the projection of the protein electrostatic field on the probe through:
| (1) |
where h is Planck's constant and c is the speed of light. A powerful feature of nitrile probes is knowledge of both the magnitude and direction of , as it is typically parallel to the C-N bond12 and the absolute direction of this bond in a protein can be obtained by X-ray crystallography.
Nitrile probes can potentially form hydrogen bonds or other specific chemical interactions with nearby groups that can affect the IR frequency via a mechanism not described by Eqn. 115-18, thereby complicating interpretation of observed shifts19. In what follows, we describe an experimental method to identify cases in which protein-bound nitriles participate in such an interaction, using the relationship between IR frequency shifts and 13C NMR chemical shifts for the nitrile. We provide evidence that for shifts that do not involve a change of the nitrile's state from free to hydrogen-bonded or vice versa, Eqn. 1 can be used to determine the accompanying . In the presence of a hydrogen bond of the type detected by this study, we show that this method can be used to decompose the observed IR frequency shift into an electrostatic and a non-electrostatic contribution.
We focus on ethyl thiocyanate (EtSCN) as a model for nitriles introduced site-specifically into proteins by direct modification of unique cysteine residues21 (Cys-SCN). These probes can be incorporated at different sites in a protein to report on site-to-site differences or to examine a specific perturbation from multiple distances and orientations. Because the chemical modification of activated cysteines uses KCN as a reagent, it is trivial to incorporate 13C (or 15N or both) to perform independent NMR22 and IR measurements on the same probe in the same environment.
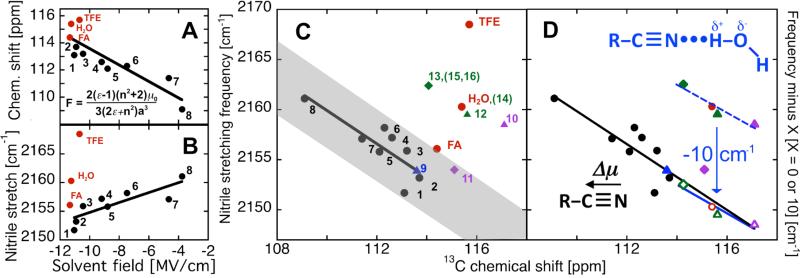
Figures 1A and B show the observed 13C NMR chemical shifts and IR frequency, respectively, for EtSCN in a wide range of solvents, divided between aprotic (black circles) and protic (red circles). For the aprotic solvents, a correlation is observed between both chemical shift and IR frequency and a classical measure of the solvent electric field calculated using the Onsager reaction field equation23,24 (inset in Fig. 1A). This simple equation, based on spherical solutes in an isotropic dielectric continuum, captures the general dependence of the magnitude of the electrostatic fields that solvents exert on dipolar solutes as a function of solvent dielectric. Focusing on the power of the model to capture general electrostatic trends, we note that the slope of correlation for the IR frequency, 0.9 cm-1/(MV/cm), is similar in magnitude to the linear Stark tuning rate, 0.72 cm-1/(MV/cm), measured for EtSCN by vibrational Stark spectroscopy in a frozen glass of 2-methyltetrahydrofuran26. The approximations of the Onsager model are worse for some combinations of solvent and solute, depending particularly on the shape of both molecules23, and this presumably accounts for some of the scatter in the correlation between the Onsager prediction and either IR frequency or NMR chemical shift. However, substantial, systematic deviations from the IR correlation are observed only for the hydrogen bond-donating solvents water and trifluoroethanol (TFE)20, with a smaller deviation observed for formamide (Fig. 1B, red circles). These departures of the IR frequency from the electric-field trend appear to result from the specific, blue-shifting effect of hydrogen bond formation to the nitrile, an effect that has been extensively discussed at both a qualitative7,8 and quantitative level15,16,18,27,28. In contrast to IR, no deviations for protic solvents are observed for the NMR chemical shift correlation (Fig. 1A, red circles).
Figure 1.
Chemical shifts vs. nitrile stretching frequencies in simple solvents and in the enzyme KSI. (A) 13C-chemical shift values and (B) IR stretching frequency of the nitrile of 1% v/v EtSCN in different solvents (0.5% in cyclohexane) plotted against the solvent electric field calculated by the Onsager equation23 (inset Fig. 1A). Black circles for aprotic solvents (in order of decreasing dielectric, from left to right on x-axis, dielectric, ε, in parenthesis): 1. dimethylsulfoxide (46.7), 2. dimethylformamide (36.7), 3. acetone (20.7), 4. CD2Cl2(9.1), 5. tetrahydrofuran (7.6), 6. CDCl3 (4.8), 7. toluene (2.4), and 8. cyclohexane (2.0). Black lines indicate the best fits for the 13C NMR data: 108 ppm – 0.6 ppm/(MV/cm), R2 = 0.76; and for the IR data: 2163.5 cm-1 + 0.9 cm-1/(MV/cm), R2 = 0.69. Omitted from the fit of the IR data are the hydrogen bonding solvents formamide, water and trifluoroethanol (red circles); R2 increases by only 0.04 if protic solvents are omitted from fit of chemical shifts, whereas the correlation of with solvent field for aprotic solvents is completely obscured if hydrogen-bonding solvents are included (R2 = 0.02). (C) 13C-NMR and IR from (A) and (B) plotted against each other in black and red circles, with line of best fit shown (-1.7 cm-1/ppm, R2 = 0.68) excluding H-bonding solvents (red circles). Observations for Cys-SCN-labeled KSI are numbered 9-16 (triangle for apo, diamond for liganded): apo M105C-CN (point 9), apo L61C-CN (point 10), L61C-CN•Equilenin (point 11), apo M116C-CN (point 12), M116C-CN•equilenin (point 13), M116C-CN•19-nortestosterone (point 14), M116C-CN•4-fluoro-3-methyl-phenol (point 15) and M116C-CN•2-naphthol (point16). Where points overlap (14 and H2O; 13, 15 and 16), only one example is plotted for clarity. (D) Same data as in (C) with and without IR correction for hydrogen bond contribution. All putative water-nitrile hydrogen bonded cases (all M116C-CN observations (12-16), apo L61C-CN (10), and EtSCN in water) shown before (closed symbols), and after (open symbols), subtraction by a 10 cm-1 correction for the hydrogen bond (blue line; slope = -1.4 cm-1/ppm, R2 = 0.93). The combined aprotic data and shifted data yield the trend shown by the dashed line (-1.7cm-1/ppm, R2 = 0.88). Protein spectra collected at 300 K in 40 mM phosphate buffer, pH 7.1. Frequencies are reported as the peak maximum; excluding TFE, EtSCN peaks are generally symmetric (see S.I.).
An individually measured IR frequency shift or change in 13C chemical shift for a nitrile probe in two different environments cannot by itself distinguish whether the observed shift is electrostatic in origin or complicated by specific effects. For example, in the IR, hydrogen bonding leads to similar frequencies for water and cyclohexane (Fig. 1B) while for NMR, aromatic ring-currents could affect a given chemical shift. However, if both are measured, then a distinction is possible. This strategy becomes apparent from a plot of 13C NMR chemical shifts versus IR frequency for EtSCN in different solvents (Fig. 1C). The common linear dependence on the solvent reaction field leads to the trend highlighted with the solid black line, with a significant departure from this trend observed for water and TFE (red circles)30. The magnitude of the deviation of formamide from the correlation lines in both Fig. 1 B and C is insufficient to unambiguously assign a hydrogen bond to the nitrile, so we attribute a hydrogen bond only for deviations greater than this example and outside the conservative zone suggested by the grey region in Fig. 1C29.
We have further tested and exploited this strategy to detect and quantify hydrogen bond contributions to observed frequency shifts for thiocyanate probes introduced around the active site of ketosteroid isomerase (KSI) D40N31 from P. putida. Three different KSI variants were studied, modified to incorporate a nitrile at residue 61 (L61C-CN), 105 (M105C-CN) or 116 (M116C-CN). To assess possible probe interactions within each modified protein, we determined X-ray structures for M105C-CN and M116C-CN and generated a molecular dynamics-based structural model for L61C-CN (see S.I.). These structural studies, (to be described in detail elsewhere33) revealed that the nitrile of M105C-CN is buried deep within the protein interior, well below the solvent-accessible surface of the active site and not within hydrogen bonding distance of any polar groups. The nitriles of L61CCN and M116C-CN, however, form part of the solvent-accessible active site surface, rendering them susceptible to hydrogen bond formation with water. Thus, we expected the M105C-CN nitrile to be free of hydrogen-bonding effects and to fall on the IR/NMR correlation line, whereas we expected both the M116C-CN and L61C-CN nitriles to defy the IR/NMR correlation due to hydrogen bond formation with water. As shown in Fig. 1C, these expectations were met: the IR/13C-NMR data for the –SCN probe at position 105 in the apoprotein (point 9) fall in the region characteristic of aprotic solvents, while the data for positions 61 (point 10) and 116 (point 12) fall in the region associated with protic solvents, indicating hydrogen bond formation.
Binding the steroid equilenin, a reaction intermediate analog, displaces bulk water from the KSI active site34. This change is predicted to dehydrate the L61C-CN side-chain (see S.I.) and therefore to shift the IR/NMR data for this residue into the region associated with aprotic solvents. As shown in Figure 1C, this prediction was met (point 11). The nitrile of equilenin-bound M116C-CN, however, was still observed (point 13) in the upper-right region of Fig. 1C, consistent with maintenance of the hydrogen bond in the presence of equilenin. The 1.7 Å resolution X-ray structure of M116C-CN with bound equilenin reveals sufficient space to accommodate a nearby water, although we did not observe electron density for an ordered water proximal to the nitrile33 (see S.I.). However, an ordered water molecule has been observed adjacent to residue 116 in a higher 1.3 Å resolution X-ray structure of the D40N/D103N mutant with bound equilenin (PDB: 3FZW)35. Further support for the presence of this water is provided by MD simulations of KSI with a bound steroid that also reported a water in this pocket36. From these observations, we conclude that the nitrile of equilenin-bound M116C-CN is hydrogen-bonded to a nearby water molecule that is present but is not detected in the 1.7 Å X-ray structure. When the ligands 19-nortestosterone, 4-fluoro-3-methyl-phenol, and 2-naphthol are bound (Fig, 1C, points 14-16, respectively), characteristic IR/13C-NMR signatures indicative of this hydrogen bond are similarly observed. Importantly, both IR and NMR observables for M116C-CN are shifted upon ligand binding in the direction expected for an increase in the projection of the electrostatic field on the nitrile, based on the field dependence determined in solvents (Fig 1A-B).
Considering only the set of IR/13C-NMR shifts assigned above to water-nitrile hydrogen bonds (all M116C-CN observations, apo L61C-CN, and EtSCN in water), another trend is observed with a slope that is similar to that obtained in aprotic solvents but is displaced by 10 cm-1 (Fig. 1D); a dataset comprised of the hydrogen-bonded set, displaced by 10 cm-1, combined with the aprotic solvent data has an identical slope to the set of aprotic solvents alone (Fig 1D). The 10 cm-1 displacement is similar to the 10 cm-1 difference for between subpopulations assigned to the hydrogen-bonded and free states of acetonitrile in ethanol15 and the 7 cm-1 frequency shift ascribed to the water-nitrile hydrogen bond in a survey of solvent effects on acetonitrile27. This constant offset and consistent slope suggest that the hydrogen bonding contribution and electrostatic effects from the local environment are additive, i.e., that in these examples, the hydrogen bond to water contributes a fixed amount to , with the shift due to the surrounding protein matrix, governed by Eqn. 1, superimposed. Thus, we use the observed for M116C-CN upon equilenin binding, the Stark tuning rate determined for EtSCN, and Eqn. 1 to estimate a 4 MV/cm increase in the projection of the local electric nitrile field on the upon equilenin binding. For L61-CN, after making the 10 cm-1 correction for the non-electrostatic portion of the shift due to the hydrogen bond, upon equilenin binding corresponds to an electric field increase of 8 MV/cm along the nitrile37. Nevertheless, the 10 cm-1 correction suggested by Fig. 1D cannot be generalized to all hydrogen bonds: the non-electrostatic portion of the shift likely depends on the chemical identity of the donor (cf. TFE and water in Fig 1B), as well as the geometry of the lowest energy configuration (as suggested by simulations4,17,27).
In conclusion, we have developed a method to distinguish whether observed IR peak shifts of nitrile probes arise principally from changes in local electrostatic field or contain contributions from both electrostatic and chemical effects, in this case hydrogen bond formation to the nitrile probe itself. A detailed application of this approach to reveal novel features of the KSI active site will be reported shortly35.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by grants to SGB (NIH GM27738) and DH (NSF MCB-0641393). PAS was supported in part by HHMI and G. Lieberman Predoctoral Fellowships.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Experimental methods, and Figs.S1-S4. This material is available free of charge at: (http://pubs.acs.org/page/jacsat/submission/authors.html).
REFERENCES
- 1.Park ES, Andrews SS, Hu RB, Boxer SG. J. Phys. Chem. B. 1999;103:9813–9817. [Google Scholar]
- 2.Suydam I, Snow C, Pande V, Boxer S. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 3.Thielges M, Case D, Romesberg F. J. Am. Chem. Soc. 2008;130:6597–6603. doi: 10.1021/ja0779607. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ye S, Huber T, Vogel R, Sakmar TP. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waegele MM, Gai F. J. Phys. Chem. Lett. 2010;1:781–786. doi: 10.1021/jz900429z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigala PA, Fafarman AT, Bogard PE, Boxer SG, Herschlag D. J. Am. Chem. Soc. 2007;129:12104–12105. doi: 10.1021/ja075605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Quillin ML, Phillips GN, Olson JS. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 7.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 8.McMahon HA, Alfieri KN, Clark KAA, Londergan CH. J. Phys. Chem. Lett. 2010;1:850–855. doi: 10.1021/jz1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A “specific chemical interaction” here implies stable complex formation that results in significant electronic wavefunction overlap between the probe and a particular chemical moiety in its environment. If this results in a rearrangement of the probe's molecular orbitals, it can have an effect on the frequency that defies description by Eqn 1. It is the aim of this work to provide an operational definition for when this contribution is significant10.
- 10.Decatur SM, Boxer SG. Biochem. Biophys. Res. Commun. 1995;212:159–164. doi: 10.1006/bbrc.1995.1950. [DOI] [PubMed] [Google Scholar]
- 11.Chattopadhyay A, Boxer SG. J. Am. Chem. Soc. 1995;117:1449–1450. [Google Scholar]
- 12.Andrews SS, Boxer SG. J. Phys. Chem. A. 2000;104:11853–11863. [Google Scholar]
- 13.Andrews SS, Boxer SG. J. Phys. Chem. A. 2002;106:469–477. [Google Scholar]
- 14.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 15.Eaton G, Pena-Nunez ES, Symons MCR. J. Chem. Soc. Faraday Trans. 1. 1988;84:2181–2193. [Google Scholar]
- 16.Purcell KF, Drago RS. J. Am. Chem. Soc. 1966;88:919–924. [Google Scholar]
- 17.Choi, Oh, Lee, Lee C, Cho M. J. Chem. Phys. 2008;128:134506. doi: 10.1063/1.2844787. [DOI] [PubMed] [Google Scholar]
- 18.Lindquist BA, Corcelli SA. J. Phys. Chem. B. 2008;112:6301–6303. doi: 10.1021/jp802039e. [DOI] [PubMed] [Google Scholar]
- 19.Other prospective IR probes with free electron lone-pairs (e.g., azide or carbonyls) not investigated herein are also prone to hydrogen bonding20 and possibly other specific chemical effects, whereas more inert species like the C-D moiety may be less susceptible to such effects.
- 20.Abraham MH, Grellier PL, Prior DV, Duce PP, Morris JJ, Taylor PJ. J. Chem. Soc. Perkin Trans. 2. 1989:699–711. [Google Scholar]
- 21.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. J. Am. Chem. Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty GM, Motherway R, Mayhew SG, Malthouse JPG. Biochemistry. 1992;31:7922–7930. doi: 10.1021/bi00149a025. [DOI] [PubMed] [Google Scholar]
- 23.Onsager L. J. Am. Chem. Soc. 1936;58:1486–1493. [Google Scholar]
- 24.Parameters used for the Onsager equation for EtSCN: μ0 = 4.0 Debye25, n=1.462, a3 =[(mol. weight )/density] = 0.14 nm3.
- 25.Bjørseth A, Marstokk KM. J. Mol. Struct. 1972;11:15–23. [Google Scholar]
- 26.Fafarman AT, Boxer SG. In preparation [Google Scholar]
- 27.Reimers JR, Hall LE. J. Am. Chem. Soc. 1999;121:3730–3744. [Google Scholar]
- 28.Aschaffenburg DJ, Moog RS. J. Phys. Chem. B. 2009;113:12736–12743. doi: 10.1021/jp905802a. [DOI] [PubMed] [Google Scholar]
- 29.Although the hydrogen bond is the only example described herein, other specific chemical interactions purported to defy prediction by Eqn. 1 (e.g. distal interaction sites on the probe scaffold4 or alternative hydrogen bonding geometry17,27) or significant, non-electrostatic perturbations to the NMR chem. shift could also be identified if they strayed from the grey region in Fig. 1C.
- 30.For carbonyls, where a correlation between stretching frequency and 13C chemical shift has also been reported Nyquist RA, Putzig CL, Hasha DL. Appl. Spectrosc. 1989;43:1049–1053.; Park KD, Guo K, Adebodun F, Chiu ML, Sligar SG, Oldfield E. Biochemistry. 1991;30:2333–2347. doi: 10.1021/bi00223a007. a large IR red-shift (and accompanying downfield chemical shift) could be due either to a hydrogen bond or a very large protein field, whereas for the nitrile, comparison of the two observables resolves this ambiguity.
- 31.The D40N mutation increases ligand binding affinity ~1000 fold32.
- 32.Petrounia IP, Blotny G, Pollack RM. Biochemistry. 2000;39:110–116. doi: 10.1021/bi9917838. [DOI] [PubMed] [Google Scholar]
- 33.Fafarman AT, Sigala PA, Schwans JP, Fenn TD, Herschlag D, Boxer SG. In Preparation. doi: 10.1073/pnas.1111566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SW, Cha SS, Cho HS, Kim JS, Ha HC, Cho MJ, Joo S, Kim KK, Choi KY, Oh BH. Biochemistry. 1997;36:14030–14036. doi: 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- 35.Sigala PA, Caaveiro JMM, Ringe D, Petsko GA, Herschlag D. Biochemistry. 2009;48:6932–6939. doi: 10.1021/bi900713j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feierberg I, Aqvist J. Biochemistry. 2002;41:15728–15735. doi: 10.1021/bi026873i. [DOI] [PubMed] [Google Scholar]
- 37.M116C-CN: (2161.8 - 2159.6 cm-1)/[0.7cm-1/(MV/cm)] = 4 MV/cm; L61C-CN: (2154.0 - [2158.6 -10] cm-1)/[ 0.7cm-1/(MV/cm)] = 8 MV/cm.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.