Abstract
Fungi, particularly the white rot basidiomycetes, have an extraordinary capability to degrade and/or mineralize (to CO2) the recalcitrant fused-ring high molecular weight (≥ 4 aromatic-rings) polycyclic aromatic hydrocarbons (HMW PAHs). Despite over 30 years of research demonstrating involvement of P450 monooxygenation reactions in fungal metabolism of HMW PAHs, specific P450 monooxygenases responsible for oxidation of these compounds are not yet known. Here we report the first comprehensive identification and functional characterization of P450 monooxygenases capable of oxidizing different ring-size PAHs in the model white rot fungus Phanerochaete chrysosporium using a successful genome-to-function strategy. In a genome-wide P450 microarray screen, we identified six PAH-responsive P450 genes (Pc-pah1 - Pc-pah6) inducible by PAHs of varying ring size, namely naphthalene, phenanthrene, pyrene, and benzo(a)pyrene (BaP). Using a co-expression strategy, cDNAs of the six Pc-Pah P450s were cloned and expressed in Pichia pastoris in conjunction with the homologous P450 oxidoreductase (Pc-POR). Each of the six recombinant P450 monooxygenases showed PAH-oxidizing activity albeit with varying substrate specificity towards PAHs (3–5 rings). All six P450s oxidized pyrene (4-ring) into two monohydroxylated products. Pc-Pah1 and Pc-Pah3 oxidized BaP (5-ring) to 3-hydroxyBaP whereas Pc-Pah4 and Pc-Pah6 oxidized phenanthrene (3-ring) to 3-, 4-, and 9-phenanthrol. These PAH-oxidizing P450s (493 – 547 aa) are structurally diverse and novel considering their low overall homology (12–23%) to mammalian counterparts. To our knowledge, this is the first report on specific fungal P450 monooxygenases with catalytic activity toward environmentally persistent and highly toxic HMW PAHs.
Keywords: Phanerochaete chrysosporium, Cytochrome P450 monooxygenase, P450 oxidoreductase, Polycyclic aromatic hydrocarbons, Biodegradation, Pichia pastoris
1. Introduction
Fused-ring polycyclic aromatic hydrocarbons (PAHs) constitute an important group of highly toxic and significant environmental chemicals that are generated from different anthropogenic and industrial processes and accidents such as crude oil spills [1]. The persistence and genotoxicity of PAHs increase with increasing aromatic rings. Consequently, HMW PAHs (≥4 benzene rings) are particularly a significant problem both from the point of view of environmental clean up as well as human health as these are recalcitrant to biodegradation [2] and are mutagenic and/or carcinogenic to living systems [3]. Among the microorganisms (bacteria, yeasts, and filamentous fungi) investigated for their ability to break down these hazardous chemicals [1,4,5], only a few species belonging to actinomycetes and fungi have the ability to oxidize HMW PAHs. In particular, white rot group of basidiomycete fungi have shown an extraordinary capability to completely degrade (to CO2) both low molecular weight (LMW) and high MW PAH compounds [5,6,7].
Phanerochaete chrysosporium (henceforth abbreviated as PC) is the most intensively studied model white rot fungus for mechanisms to degrade lignin and a wide range of xenobiotics including PAHs [6]. Originally, its aromatic ring-oxidizing activity was ascribed to the non-specific extracellular peroxidases [8,9] that are differentially expressed under nutrient-limited (ligninolytic) culture conditions [10]. Subsequent studies by us and others have led to increasing evidences on peroxidase-independent degradation of several aromatics including PAHs under nutrient-sufficient (non-ligninolytic) culture conditions and involvement of P450 monooxygenation reactions [11–17]. Role of P450 monooxygenation in the rate-limiting initial oxidation of HMW PAHs and certain LMW PAHs with high ionization potential (> 7.35 eV) such as phenanthrene has also been reported in other basidiomycete and non-basidiomycete fungi [4,18]. However, despite the past over 30 years of research on PAH biodegradative fungal species [1,4,5], the specific P450 monooxygenase enzymes responsible for this oxidation activity have not been reported. Collectively considering the above discussion, there is a critical need for identification of specific fungal P450 genes and enzymes responsible for the oxidation of HMW and/or both LMW and HMW PAH compounds, in order to understand the P450-mediated mechanisms and to facilitate development of improved biotransformation processes and applications.
Cytochrome P450 monooxygenases are a superfamily of heme-thiolate proteins that catalyze a broad range of reactions such as carbon hydroxylation, heteroatom oxygenation, dealkylation, epoxidation, reduction, dehalogenation [19]. The typical eukaryotic P450 monooxygenase system contains a P450 monooxygenase and a P450 oxidoreductase (POR), both of which are normally membrane-associated. This family of proteins has seen a tremendous growth in the genomic era [20, http://drnelson.uthsc.edu/CytochromeP450.html]. However, post-genomic functional characterization of orphan P450s in fungal and other genomes has been a challenging task due to their poor amenability to heterologous expression in simpler hosts. Whole genome sequencing of PC [21] has uncovered the presence of a large P450 diversity, comprised of about 150 P450 genes [22]. While functional genomic studies on these P450s are emerging [16,23,24], majority remain orphan with virtually unknown function. Hence, there is a great need to characterize the role of individual P450s in this model organism. Our recent studies using the first custom-designed genome-wide P450 microarray [25,26], have led to a working hypothesis that substrate-specific inducibility could be key to identifying the xenobiotic substrates for orphan P450 enzymes in this organism [16,23,25]. In this study, we therefore employed a two-stage genome-to-function strategy, including a genome-wide (microarray-based) transcriptional induction profiling to identify candidate P450 genes responsive to PAHs of varying ring-size followed by co-expression and catalytic characterization of the identified recombinantly expressed P450 proteins. These efforts led to identification of a set of six PAH-responsive P450 monooxygenase genes in the PC genome (designated Pc-Pah1 through Pc-Pah6). Subsequent co-expression along with the reductase partner and catalytic analysis revealed their diverse PAH substrate specificity particularly towards HMW (4–5 ring) PAHs. Part of the data reported in this manuscript was presented at the 104th annual general meeting of the American Society for Microbiology, New Orleans, LA [27].
2. Materials and Methods
Microbial strains, chemicals, media, and culture conditions are detailed under Supplementary information. P450 inhibitor studies and induction of P450s in PC were performed as described in our recent study [16]. Preparation of fungal microsomes, CO-difference spectrum analysis, and calculation of total P450 concentration were performed using established methods [28, 29]. Differential gene expression profiling was performed using a genome-wide custom-designed 70 mer oligos-based P450 microarray developed in our previous studies [25,26]. Full-length cDNAs for the identified PAH-inducible P450 genes (designated Pc-Pah1 through Pc-Pah6) were cloned using gene-specific RT-PCR as described previously [26]. cDNA sequences are available in the GenBank with accession numbers listed in Table 1. Heterologous co-expression of the Pc-Pah genes along with the homologous Pc-POR [30] in the yeast Pichia pastoris (PP) was performed using the pPICZB expression vector system (Invitrogen). The experimental strategy used for creating the binary constructs is depicted in Fig. S1 (Supplementary information). Expression analysis and PAH compound oxidation in yeast whole cell assays were performed as detailed in Supplementary information. P450 hydroxylated metabolites of the PAH compounds were analyzed by HPLC followed by liquid chromatography-electrospray mass spectrometry (LC-ESI/MS) (Thermo Finnigan) as described in Supplementary information.
Table 1.
Multiple PAH-responsive P450 genes identified using genome-wide P450 microarray analysis on P. chrysosporiuma.
| P450 ID | CYP name | NCBI Gene Accession number | Protein ID (JGI WGS Ver. 2) | Transcriptional induction (fold-change)b | |||
|---|---|---|---|---|---|---|---|
| Naphthalene | Phenanthrene | Pyrene | Benzo(a)pyrene | ||||
| Pc-Pah1 | CYP5136A2 | AY515588 | 5852 | 7.8 ± 4.64 | 3.24 ± 1.95 | ||
| Pc-Pah2 | CYP5145A3 | GU270838 | 6327 | 2.72 ± 0.04 | 3.28 ± 0.73 | 1.67 ± 0.65 | 1.71 ± 0.11 |
| Pc-Pah3 | CYP5144A7 | AY515589 | 133311 | 7.34 ± 0.01 | 2.87 ± 1.24 | 15.12 ± 7.19 | |
| Pc-Pah4 | CYP5136A3 | GU270839 | 5001 | 9.52 ± 2.38 | 1.32 ± 0.08 | ||
| Pc-Pah5 | CYP5142A3 | AY515590 | 28946 | 7.37 ± 0.26 | |||
| Pc-Pah6 | CYP5144A5 | AY515591 | 132481 | 4.43 ± 0.6 | |||
PAH treatments and transcriptional profiling were performed using nutrient-rich (malt extract) cultures as described under Materials and Methods section.
The fold change values are means ± standard errors obtained by evaluating eight independent spots for each gene and are statistically significant (P ≤ 0.05 and FDR of 0.1).
Abbreviations: CYP, Cytochrome P450; JGI, Joint Genome Institute of the US Department of Energy; WGS, Whole genome sequence
3. Results and discussion
3.1. Involvement of P450 enzyme system in biodegradation of HMW PAH compounds in PC
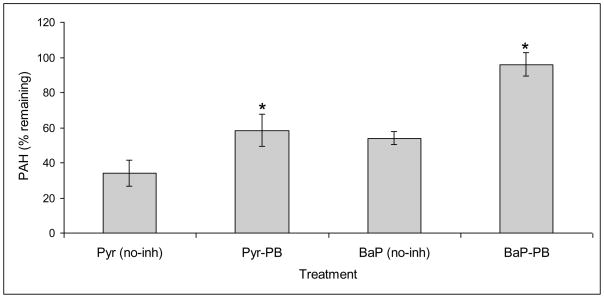
Addition of the P450 inhibitor piperonyl butoxide (PB) in the nutrient-rich malt extract (ME) cultures of PC led to a significant abrogation of the degradation activity towards pyrene and benzo(a)pyrene (BaP) (Fig. 1). This suggested a key role of P450 monooxygenase(s) in initial oxidation of these HMW PAH compounds in PC, analogous to that reported for the lower PAH phenanthrene [11]. Comparison between the chemically-killed (CK) control cultures and uninoculated control revealed that unextractable mycelial adsorption partially contributed toward the apparent total disappearance in case of pyrene (~16.6%) but not in BaP degradation. The PB attenuation effect on biodegradation was relatively higher for BaP (Fig. 1), implying the involvement of divergent sets of P450s for the two PAHs. Role of P450s in PAH oxidation was further evidenced by our initial P450 enzyme induction studies on the whole fungus grown under similar conditions. Results showed about 2-fold induction of total P450 enzyme content (based on typical CO-difference spectrum) in response to 10 ppm pyrene (Table S2). Considering this, we performed a genome-wide induction profiling for identification of specific PAH-inducible P450 genes as part of our genome-to-function strategy to characterize those catalyzing PAH oxidation.
Figure 1.
Effect of P450 inhibitor piperonyl butoxide (PB) on biodegradation of pyrene and benzo(a)pyrene by P. chrysosporium. Nutrient-rich malt extract broth cultures and negative controls were prepared as described under Materials and Methods section. Final concentrations of the test PAH and PB in the cultures were 20 ppm and 0.5 mM, respectively. Asterisk indicates statistically significant (P ≤0.05) effect of the inhibitor. The plotted values represent means ± standard deviations for three biological replicates. Abbreviations: Pyr, pyrene; BaP, benzo(a)pyrene; no-inh, no inhibitor.
3.2. Microarray-based identification of PAH-responsive P450 monooxygenases (Pc-Pah P450s)
Genome-wide P450 transcriptional profiling using the P450ome microarray first developed in our previous studies [25,26] showed significant induction (up to ~15-fold) of multiple P450 genes in response to the four individual representative PAHs of increasing ring size (2–5 rings) in nutrient-rich ME cultures (Table 1 and Fig. S2). A total of six candidate PAH-responsive P450 genes (designated Pc-Pah1 through Pc-Pah6) were selected, based on a cut off criterion of ≥ 3-fold induction by at least one of the inducer PAHs. Subsequent qRT-PCR analysis on selected genes and treatments verified the microarray results. For instance, qRT-PCR based fold-change values for Pc-Pah2 were 2.45 ± 0.03 (naphthalene), 4.21 ± 0.44 (phenanthrene), and 2.47 ± 0.11 (BaP) as against 2.72 ± 0.04, 3.28 ± 0.73, and 1.71 ± 0.11, respectively in microarray analysis. The house-keeping gene GAPDH remained unchanged. Of the six PAH-responsive genes, a set of four genes each was inducible by naphthalene ((Pc-Pah2, Pc-Pah3, Pc-Pah5 and Pc-Pah6) and phenanthrene (Pc-Pah1 through 4) whereas a set of two genes each was inducible by pyrene (Pc-Pah1 and Pc-pah4), and BaP (Pc-pah2 and Pc-pah3) (Table 1). The two pyrene-inducible genes (Pc-Pah1 and Pc-Pah4) were also inducible in the presence of the lower PAH phenanthrene. The two BaP-inducible genes (Pc-Pah2 and Pc-Pah3) were also inducible by the lower PAHs naphthalene and phenanthrene. Taken together, a Pc-Pah gene inducible by the higher PAH (4- or 5- ring) was also inducible by a lower PAH (2- and/or 3- ring)
Interestingly, five of the six PAH-responsive P450 genes identified here also showed upregulation under defined high nitrogen (HN) culture conditions in our earlier study [26], further confirming their optimal expressibility under non-ligninolytic conditions. We also observed inducibility of two of the genes (Pc-Pah4 and Pc-Pah5) in response to alkylphenols [16], indicating their broader substrate range against environmental xenobiotics.
3.3. Recombinant co-expression of Pc-Pah P450s and Pc-POR in Pichia pastoris
Bioinformatic analyses on the six Pc-Pah genes and their corresponding cDNAs, cloned and sequenced in this study for recombinant expression, revealed variable structural features such as 8 to 12 introns with variable distribution (Fig. S3) and genome-wide localization. Interestingly, two of the genes Pc-Pah3 and Pc-Pah6, are part of a tandem gene cluster of 12 P450 members in the PC genome. Pc-Pah5 is also on the same genome scaffold and is separated from the upstream 4 tandem P450s cluster by the intervening 3 non-P450 genes.
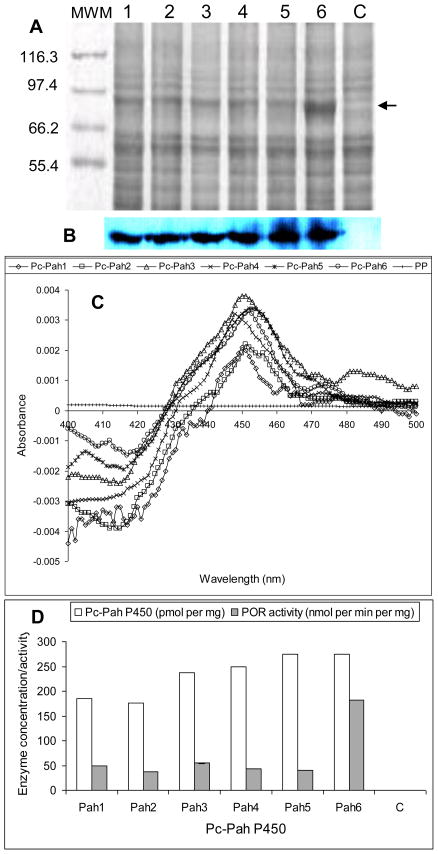
The co-expression strategy enabled a successful active expression of the individual Pc-Pah P450s proteins. This strategy assumes significance considering the expression difficulties encountered in our previous efforts on independent expression of Pc-Pah P450 without the homologous POR. SDS-PAGE analysis of the six microsomal preparations showed a clear band coinciding with Pc-POR when compared to the control (Fig. 2A). Western blot analysis using anti-His antibody showed full-length expression of all six Pc-Pah P450 proteins (Fig. 2B). Active nature of the expressed recombinant P450 proteins was evident from their characteristic CO-difference spectra (Fig. 2C). The observed maximum absorption peaks were at 448 nm (Pc-Pah4), 450 nm (Pc-Pah1 and Pc-Pah3), 451 nm (Pc-Pah2), and 452 nm (Pc-Pah5 and Pc-Pah6). P450 and POR expression levels varied for the six Pc-Pah clones (Fig. 2D), with Pc-Pah5 and 6 showing the highest P450 concentration. Pc-Pah6 had the highest co-expressing POR activity, an observation consistent with the corresponding band intensity in the SDS-PAGE gel (Fig. 2A).
Figure 2.
Co-expression and activity characterization of Pc-Pah P450 monooxygenases and the homologous P450 reductase partner (Pc-POR) in Pichia pastoris. The expressed enzymes were assessed in the yeast microsomes prepared as described under Materials and Methods section. (A) SDS-PAGE analysis of the microsomal protein preparations from recombinant P450-POR yeast clones Pc-Pah1 through Pc-Pah6 (lanes 1–6) and negative control yeast (lane c). Proteins were separated on 7% SDS-PAGE and detected by staining with SYPRO Ruby (BioRad). The protein band coinciding with Pc-POR (estimated 81.6 kDa) is shown by an arrow. (B) Western blot analysis of Pc-Pah proteins using anti-his antibody. (C) Characteristic CO-difference spectra for the individual Pc-Pah proteins (Pc-Pah1 through Pc-Pah6) and negative control. (D) P450 enzyme concentrations and POR activity levels of the individual Pc-Pah clones versus the negative control (C). P450 concentration values are based on the CO-difference spectra shown in panel C. POR activity values are the means for three biological replicates (the SD values varied between 0.07 and 0.23).
3.4. Catalytic analysis of the recombinant fungal P450 enzymes
3.4.1. Substrate specificity
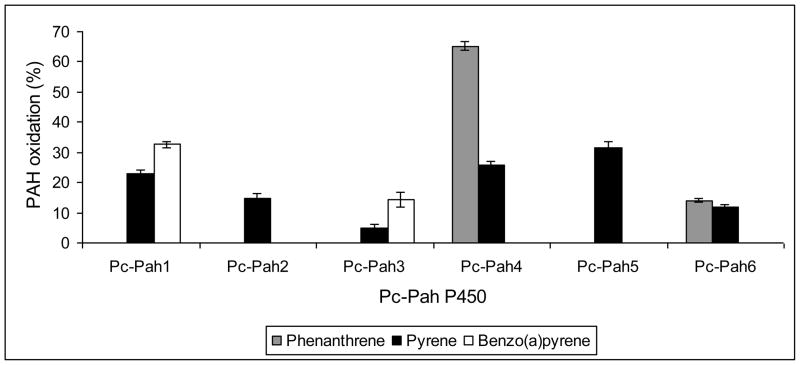
Whole-cell yeast biocatalysts are considered ideal for screening of hydrophobic compounds such as PAHs as the use of cells allows ready uptake as well as alleviates the need for isolation of microsomes and in vitro reconstitution of the P450 assay [31]. In whole cell assays, the six recombinant Pc-Pah enzymes showed catalytic activity toward PAHs, albeit with varying activity and substrate specificity (Fig. 3). No activity was observed in the empty vector control yeast. While all six Pc-Pah P450s showed pyrene oxidation, quantitative differences in the activity were observed among the individual enzymes; Pc-Pah5 and Pc-Pah4 showed relatively higher pyrene oxidation activity as compared to the other four Pc-Pah P450s. In comparison, a tighter substrate specificity was observed for phenanthrene as only Pc-Pah4 (~65%) and Pc-Pah6 (~14%) could oxidize this compound (Fig. 3). This is significant considering that phenanthrene is not a substrate for lignin peroxidases because of its high ionization potential (> 7.35 eV). Anthracene, another 3-ring PAH, was however, not detectably oxidized by any of the six Pc-Pah P450s in this study. Only Pc-Pah1 and Pc-Pah3 showed activity towards BaP, with the former showing relatively higher levels of oxidation (32.6% vs. 14.3%). The co-expressed POR levels (Fig. 2D) did not seem to quantitatively correlate either with the relative P450 expression levels or with the PAH oxidation activity (Fig. 3) of the individual Pc-Pah P450s. Common specificity of Pc-Pah4 and Pc-Pah6 towards phenanthrene, albeit to a varying extent, may be because of their evolution from a common progenitor gene as has been hypothesized for members of the same P450 genomic cluster in our previous genome-wide bioinformatic study on P450ome of this organism [22].
Figure 3.
Relative PAH-oxidizing activities of the six recombinant Pc-Pah P450 enzymes towards varying ring-size PAH compounds in whole cell assays. Each culture was spiked with a test PAH and the oxidation activity was assessed at 36 h of incubation. The values represent means ± standard deviations for three biological replicates and are statistically significant (P ≤0.05).
The 450 inhibitor PB almost completely abrogated the observed PAH-oxidizing activity of all six Pc-Pah enzymes in the recombinant yeast cultures (Table S3) confirming their role in the observed PAH catalysis. PB did not exert any growth inhibition, confirming the enzymatic basis of the observed activity inhibition. Comparison of the P450 inhibitor effect on the whole fungus versus the recombinant Pc-Pah P450 yeast clones showed complete inhibition of BaP-oxidizing P450 activity in both the biological systems. In contrast, only partial inhibition of activity was observed for pyrene in the whole fungus (unlike the yeast clones), indicating the likely presence of additional pyrene-oxidizing P450s (non-responsive to PB) in the PC genome. This is not unexpected considering the observed general amenability of pyrene to oxidation by all six P450s in this study.
3.4.2. Catalytic specificity
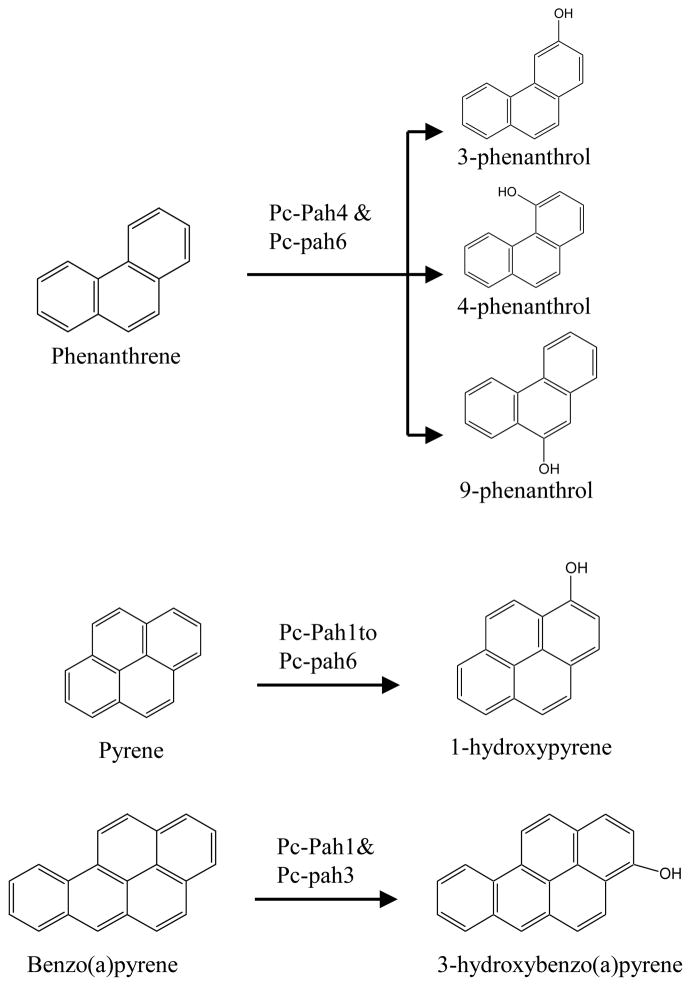
Initial HPLC screening on the PAH oxidation reaction extracts for the individual reactive P450s showed variable number of metabolite peaks for individual PAHs. No such peaks were observed in the empty vector control yeast. Subsequent LC-ESI/MS analysis (Fig. S4) confirmed the observed HPLC patterns of the metabolite peaks and allowed identification of the oxidation metabolites by accurate mass measurement on the whole reaction extracts using authentic standards (Table S4). Phenanthrene reaction extracts for the Pc-Pah4 and Pc-Pah6 clones showed three peaks (I, II, III) with same measured monoisotopic mass (m/z) but different retention time (Rt). The Rt of the three peaks exactly matched with those of the authentic standards (Table S4). Based on the Rt values and m/z comparisons, the peaks were thus identified as 3-phenanthrol (peak I), 4-phenanthrol (peak III), and 9-phenanthrol (peak II). Among the three metabolites, 9-phenanthrol was the most abundant followed by 4- and 3-phenanthrols, respectively. In contrast, pyrene reaction extracts for each of the 6 Pc-Pah P450s showed two peaks exhibiting the same m/z but different Rt. The second peak (II) showed Rt coinciding with those of the authentic standard 1-hydroxypyrene. In contrast with phenanthrene and pyrene, a single peak was observed in the BaP reaction extracts from Pc-Pah1 and Pc-Pah3. As the Rt and m/z values of the peak were the same as that of the authentic standard, this peak was identified as 3-hydroxybenzo(a)pyrene (Table S4).
Taken together, catalytic activity of the Pc-Pah P450s towards varying ring-size PAHs revealed their functional diversity albeit with some redundancy. While all Pc-Pah P450s catalyzed the oxidation of pyrene, specificity was observed in respect of phenanthrene and BaP (Fig. 4). It is interesting that the multifunctional Pc-Pah P450s (those catalyzing oxidation of more than one PAH) showed a pattern of ring-size specificity. For instance, the P450s showed activity either with phenanthrene and pyrene (3 and 4 ring PAH) or pyrene and BaP (4 and 5 ring PAH) but none of them oxidized both phenanthrene and BaP (3 and 5 ring PAH) (Fig. 4). While the substrate responsiveness (induction) profiling strategy proved critical in successful identification of relevant PAH-oxidizing P450s (Table 1), the induction patterns did not show absolute correlation with the respective substrate specificities (Fig. 3).
Figure 4.
PAH oxygenation reactions catalyzed by the individual recombinant Pc-Pah enzymes (Pc-Pah1 to Pc-Pah6) of P. chrysosporium.
Formation of oxidized products in the single P450 enzyme-based oxidation reactions confirmed that Pc-Pah P450s are involved in the initial oxidation step of the PAH biodegradation process. P450-mediated oxidation of the PAH ring in eukaryotes occurs via formation of an epoxide which in turn is converted into hydroxylated products by non-enzymatic isomerization and/or by the action of epoxide hydrolase [4]. The relative abundance of the three phenanthrols was consistent with the regiospecificity profile observed for the whole fungus [1]; 9-phenanthrol was found to be the most abundant whereas 3-phenanthrol the least. In this context, it is interesting that recent whole fungus-based studies using nutrient-limited (defined low nitrogen) cultures showed the formation of a single monohydroxylated product, 9-phenanthrol [17]. In previous PC studies [11], it was unclear whether a single P450 or multiple P450s are involved in the formation of multiple phenanthrols via formation of different epoxides (9, 10-oxide versus 3, 4-oxide). In this context, our results showed involvement of a single P450 in the formation of all three phenanthrols, suggesting that individual P450 (Pc-Pah4 or Pc-Pah6) can oxidize phenanthrene at both positions (9,10 or 3,4). Each of the six Pc-Pah P450s oxidized pyrene into two mono hydroxylated products, of which one was identified as 1-hydroxypyrene. BaP was oxidized to 3-hydroxybenzo(a)pyrene by Pc-Pah1 and Pc-Pah3. This is consistent with the whole fungus study on PC wherein 3-hydroxybenzo(a)pyrene was identified as the sole oxidation product [15].
Taken together, matching regiospecificity of the recombinant Pc-Pah P450s and the parent whole fungus (PC) towards oxidation of different PAHs and gene inducibility under in vivo conditions confirm the authenticity and physiological relevance of the identified P450s in PC. Considering that the white rot fungi possess a comprehensive enzymatic machinery to completely degrade (to CO2) PAHs, the P450 monooxygenases may link up with the PAH mineralization pathway components under appropriate conditions; such common pathway components (non-ligninolytic and ligninolytic) for cycling of PAH metabolites have been indicated in previous studies [18,32]. Hence, the presence of P450s in white rot fungi assumes greater significance than in other biological systems that have a standalone P450 enzyme system for PAH metabolism such as in non-basidiomycete fungi [4] and animals [19]. Interestingly, there was a partial overlap in catalytic regiospecificity [33,34] between the fungal Pc-Pah P450 enzymes and mammalian PAH-oxidizing P450s (human/rat/mouse CYP1A1, 1A2, and 1B1), possibly because of low overall protein level homology (12–23%) (Table S5). This phenomenon holds promise for future basic structure-activity relationship studies to generate improved versatile biocatalysts for detoxification and biotransformation applications.
4. Conclusions
We report a successful genome-to-function approach to functionally characterize orphan P450s in the P. chrysosporium genome, a strategy that could be extendable to other genomes (prokaryotes and other eukaryotes such as fungi, insects, plants, and animals) with orphan P450s. To our knowledge, this report constitutes the first genome-wide comprehensive identification and functional characterization of fungal P450 monooxygenases catalyzing oxidation of varying ring-size PAH compounds namely phenanthrene, pyrene and BaP. This study opens up avenues for investigating similar PAH-oxidizing P450 enzymes in other fungal systems and for exploiting these structurally and catalytically diverse P450 enzymes for future basic and applied research on P450-mediated mechanisms and processes in bioremediation and biotechnological applications.
Supplementary Material
Acknowledgments
The work was supported by the NIH’s National Institute of Environmental Health Science (NIEHS) grants R01ES10210 and R01ES015543 to JSY.
Abbreviations
- PAH
polycyclic aromatic hydrocarbon
- CYP
cytochrome P450
- POR
cytochrome P450 oxidoreductase
- PC
Phanerochaete chrysosporium
- PP
Pichia pastoris
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 2.Shuttleworth KL, Cerniglia CE. Environmental aspects of PAH biodegradation. Appl Biochem Biotechnol. 1995;54:291–302. doi: 10.1007/BF02787927. [DOI] [PubMed] [Google Scholar]
- 3.Xue W, Warshawsky D. Molecular mechanisms of action of selected organic carcinogens. In: Warshawsky D, Landolph JR, editors. Molecular carcinogenesis and molecular biology of human cancer. Taylor and Francis Group, CRC Press; Boca Raton, FL, USA: 2006. pp. 45–77. [Google Scholar]
- 4.Cerniglia CE. Fungal metabolism of polycyclic aromatic hydrocarbons: past, present and future applications in bioremediation. J Indus Microbiol Biotechnol. 1997;19:324–33. doi: 10.1038/sj.jim.2900459. [DOI] [PubMed] [Google Scholar]
- 5.Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, Tian YS, Yao QH. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol Rev. 2008;32:927–955. doi: 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 6.Pszczynski A, Crawford RL. Potential for bioremediation of xenobiotic compounds by the white rot fungus Phanerochaete chrysosporium. Biotechnol Prog. 1995;11:368–379. [Google Scholar]
- 7.Sack U, Heinze TM, Deck J, Cerniglia CE, Cazau MC, Fritsche W. Novel metabolites in phenanthrene and pyrene transformation by Aspergillus niger. Appl Environ Microbiol. 1997;63:2906–2909. doi: 10.1128/aem.63.7.2906-2909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bumpus JA, Tien M, Wright D, Aust D. Oxidation of persistent environmental pollutants by a white rot fungus. Science. 1985;228:1434–1436. doi: 10.1126/science.3925550. [DOI] [PubMed] [Google Scholar]
- 9.Hammel KE, Kalyanaraman B, Kirk TK. Oxidation of polycyclic aromatic hydrocarbons and dibenzo[p]dioxins by Phanerochaete chrysosporium ligninase. J Biol Chem. 1986;261:16948–16952. [PubMed] [Google Scholar]
- 10.Kersten P, Cullen D. Extracellular oxidative systems of the lignin-degrading Basidiomycete Phanerochaete chrysosporium. Fungal Genet Biol. 2007;44:77–87. doi: 10.1016/j.fgb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland JB, Selby AL, Freeman JP, Evans FE, Cerniglia CE. Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl Environ Microbiol. 1991;57:3310–3316. doi: 10.1128/aem.57.11.3310-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav JS, Reddy CA. Non-involvement of lignin peroxidases and manganese peroxidases in 2,4,5-trichlorophenoxyacetic acid degradation by Phanerochaete chrysosporium. Biotechnol Lett. 1992;14:1089–1092. [Google Scholar]
- 13.Yadav JS, Reddy CA. Degradation of benzene, toluene, ethylbenzene, and xylenes (BTEX) by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:756–762. doi: 10.1128/aem.59.3.756-762.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kullman SW, Matsumura F. Metabolic pathways utilized by Phanerochaete chrysosporium for degradation of the cyclodiene pesticide endosulfan. Appl Environ Microbiol. 1996;62:593–600. doi: 10.1128/aem.62.2.593-600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL. Evidence for cytochrome P450 and P450-mediated benzo(a)pyrene hydroxylation in the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett. 1996;135:51–55. doi: 10.1111/j.1574-6968.1996.tb07965.x. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian V, Yadav JS. Role of P450 monooxygenases in the degradation of the endocrine-disrupting chemical nonylphenol by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 2009;75:5570–5580. doi: 10.1128/AEM.02942-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ning D, Wang H, Ding C, Lu H. Novel evidence of cytochrome P450-catalyzed oxidation of phenanthrene in Phanerochaete chrysosporium under ligninolytic conditions. Biodegradation. 2010 doi: 10.1007/s10532-010-9349-9. (in press) [DOI] [PubMed] [Google Scholar]
- 18.Bezalel L, Hadar Y, Cerniglia CE. Enzymatic mechanisms involved in phenanthrene degradation by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1997;63:2495–2501. doi: 10.1128/aem.63.7.2495-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;24:128–45. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Park J, Lee S, Choi J, Ahn K, Park B, Park J, Kang S, Lee YH. Fungal cytochrome P450 database. BMC Genomics. 2008;9:402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez D, Larrondo LF, Putnam N, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 22.Doddapaneni H, Chakraborty R, Yadav JS. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white-rot fungus Phanerochaete chrysosporium: Evidence for gene duplications and extensive gene clustering. BMC Genomics. 2005;6:1–24. doi: 10.1186/1471-2164-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doddapaneni H, Yadav JS. Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white rot fungus Phanerochaete chrysosporium. Appl Microbiol Biotechnol. 2004;65:559–565. doi: 10.1007/s00253-004-1645-z. [DOI] [PubMed] [Google Scholar]
- 24.Kasai N, Ikushiro SI, Hirosue S, et al. Enzymatic properties of cytochrome P450 catalyzing 3−-hydroxylation of naringenin from the white-rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun. 2009;387:103–108. doi: 10.1016/j.bbrc.2009.06.134. [DOI] [PubMed] [Google Scholar]
- 25.Yadav JS, Doddapaneni H. Genome-wide expression profiling and xenobiotic inducibility of P450 monooxygenase genes in the white rot fungus Phanerochaete chrysosporium. In: Aznenbacher P, Hudecek J, editors. Proceedings of the 13th International Conference on Cytochromes P450 Biochemistry, Biophysics and Drug Metabolism, Monduzzi Editore. Bologna, Italy: 2003. pp. 333–340. [Google Scholar]
- 26.Doddapaneni H, Yadav JS. Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathways in the white-rot fungus Phanerochaete chrysosporium. Mol Genet Genomics. 2005;274:454–466. doi: 10.1007/s00438-005-0051-2. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian V, Doddapaneni H, Yadav JS. Microarray-based identification, cloning, and heterologous expression of PAH-inducible P450 monooxygenase genes of the white rot fungus Phanerochaete chrysosporium. 104th Annual ASM General Meeting; May 24–27; New Orleans, LA. 2004. Abs # K-063. [Google Scholar]
- 28.Subramanian V, Doddapaneni H, Syed K, Yadav JS. P450 Redox Enzymes in the white rot fungusPhanerochaete chrysosporium: gene transcription, heterologous expression, and activity analysis on the purified proteins. Curr Microbiol. 2010 doi: 10.1007/s00284-010-9612-7. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guengerich P. Analysis and characterization of enzymes. In: Hayes AW, editor. Principles and Methods of Toxicology. 2. Raven Press Ltd; New York, NY: 1989. pp. 777–814. [Google Scholar]
- 30.Yadav JS, Loper JC. Cytochrome P450 oxidoreductase gene and its differentially terminated cDNAs from the white rot fungus Phanerochaete chrysosporium. Curr Genetics. 2000;37:65–73. doi: 10.1007/s002940050010. [DOI] [PubMed] [Google Scholar]
- 31.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 32.Tatarko M, Bumpus JA. Biodegradation of phenanthrene by Phanerochaete chrysosporium: on the role of lignin peroxidase. Lett Appl Microbiol. 1993;17:20–24. [Google Scholar]
- 33.Chaturapit S, Holder GM. Studies on the hepatic microsomal metabolism of (14C) Phenanthrene. Biochem Pharmacol. 1978;27:1865–1871. doi: 10.1016/0006-2952(78)90034-5. [DOI] [PubMed] [Google Scholar]
- 34.Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet. 2006;21:257–76. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.