Abstract
Enzymatic depolymerization of lignocellulose by microbes in the bovine rumen and the human colon is critical to gut health and function within the host. Prevotella bryantii B14 is a rumen bacterium that efficiently degrades soluble xylan. To identify the genes harnessed by this bacterium to degrade xylan, the transcriptomes of P. bryantii cultured on either wheat arabinoxylan or a mixture of its monosaccharide components were compared by DNA microarray and RNA sequencing approaches. The most highly induced genes formed a cluster that contained putative outer membrane proteins analogous to the starch utilization system identified in the prominent human gut symbiont Bacteroides thetaiotaomicron. The arrangement of genes in the cluster was highly conserved in other xylanolytic Bacteroidetes, suggesting that the mechanism employed by xylan utilizers in this phylum is conserved. A number of genes encoding proteins with unassigned function were also induced on wheat arabinoxylan. Among these proteins, a hypothetical protein with low similarity to glycoside hydrolases was shown to possess endoxylanase activity and subsequently assigned to glycoside hydrolase family 5. The enzyme was designated PbXyn5A. Two of the most similar proteins to PbXyn5A were hypothetical proteins from human colonic Bacteroides spp., and when expressed each protein exhibited endoxylanase activity. By using site-directed mutagenesis, we identified two amino acid residues that likely serve as the catalytic acid/base and nucleophile as in other GH5 proteins. This study therefore provides insights into capture of energy by xylanolytic Bacteroidetes and the application of their enzymes as a resource in the biofuel industry.
Keywords: Bacterial Metabolism, Carbohydrate Metabolism, Microarray, Oligosaccharide, Polysaccharide, Bacteroidetes, Glycoside Hydrolase, Gut Microbiota, Xylan, Xylanase
Introduction
The hydrolysis and fermentation of plant cell wall polysaccharides are important metabolic processes that occur within the gut ecosystem of ruminants as well as humans. Xylan is the most abundant plant polysaccharide after cellulose, and microbes within the bovine rumen have evolved to efficiently degrade this hemicellulosic substrate. The depolymerization of xylan requires the coordinated action of a number of enzymes, including endoxylanases, β-xylosidases, α-l-arabinofuranosidases, α-glucuronidases, acetylxylan esterases, and ferulic acid esterases (1). Prevotella spp. are the most numerically dominant xylanolytic bacteria in the rumen (2, 3); therefore, the mechanism by which they degrade and utilize xylan has been an important topic of investigation (4–9). Prevotella bryantii B14 is frequently isolated from the rumen microbiome and can grow with xylan as the sole carbohydrate source (6). However, to date only six genes with roles in xylan hydrolysis have been cloned and characterized from P. bryantii B14, including two glycoside hydrolase (GH)2 family 10 endoxylanases (7, 8, 10), three GH family 3 β-xylosidases (4), and one GH family 43 β-xylosidase enzyme (7, 8). Given the capacity of this organism to grow efficiently with xylan substrates and considering the complexity in chemical linkages within natural xylans, it is likely that P. bryantii B14 uses additional as yet unidentified enzymes to degrade xylan.
The genome of P. bryantii B14 has recently been partially sequenced (11), and this bacterium harbors at least 109 genes predicted to encode either glycoside hydrolases or carbohydrate esterases. In this study, a transcriptional approach was employed to identify the genes harnessed by P. bryantii B14 to degrade xylan. Because it has been reported that endoxylanase activity of P. bryantii B14 is induced by medium to large sized xylo-oligosaccharides and not by monosaccharides (12), the transcriptional profile of P. bryantii B14 cultured either with soluble wheat arabinoxylan (WAX) or with a mixture of xylose and arabinose (XA) was investigated. The studies allowed us to assemble the enzymes that likely constitute the xylan-degrading machinery of P. bryantii B14. In addition, we have assigned biochemical function to two hypothetical proteins that were up-regulated in cells metabolizing wheat arabinoxylan compared with a mixture of its monosaccharide components. Genes encoding similar polypeptides, which invariably exhibited endoxylanase activity, were identified in several members of the Bacteroidetes, suggesting that this group of enzymes is critical to the capture of energy from xylan in this phylum. More importantly, our analyses have led to the discovery of a gene cluster, composed of an invariant core of six genes flanked by either biochemically characterized or putative glycoside hydrolases and carbohydrate esterases in P. bryantii B14. The genes within the cluster and their collective response during wheat arabinoxylan utilization suggest that the cluster is critical to xylan utilization in this bacterium. Furthermore, the conservation of the gene cluster in other xylanolytic Prevotella and Bacteroides spp. derived from the bovine rumen and the human colonic microbiomes suggests a conserved mechanism for xylan utilization by xylanolytic Bacteroidetes.
EXPERIMENTAL PROCEDURES
Materials
P. bryantii B14 (DSM 11371) was initially isolated by Bryant et al. (13) and was obtained from our culture collection in the Department of Animal Sciences at the University of Illinois, Urbana-Champaign. Bacteroides intestinalis 341 (DSM 17393) was a kind gift from Jeffrey I. Gordon (Washington University, St. Louis) and was originally obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). Escherichia coli JM109 and E. coli BL21-CodonPlusTM (DE3) RIL competent cells and the PicoMaxx high fidelity DNA polymerase were acquired from Stratagene (La Jolla, CA). The pET-46 Ek/LIC vector kit was obtained from Novagen (San Diego). The RNAprotect bacteria reagent, RNeasy mini kit, RNase-free DNase set, DNeasy blood and tissue DNA purification kit, and QIAprep spin miniprep kit were all obtained from Qiagen (Valencia, CA). Xylo-oligosaccharides and wheat arabinoxylan (medium viscosity, 20 centistokes) were obtained from Megazyme (Bray, Ireland). The Superscript double-stranded cDNA synthesis kit and the RiboMinus transcriptome isolation kit were obtained from Invitrogen. The One-Color DNA labeling kit, hybridization kit, Sample Tracking control kit, and Wash Buffer kit were all obtained from Roche NimbleGen (Madison, WI). All other reagents were of the highest possible purity and were purchased from Sigma.
Growth of P. bryantii B14 and Isolation of RNA
P. bryantii B14 was grown anaerobically in a modified chemically defined medium (4) with either wheat arabinoxylan (0.15% w/v) or a mixture containing both xylose (0.0885% w/v) and arabinose (0.0615% w/v) as the sole carbohydrate sources. The concentrations of XA in the mixture were equivalent to that in the WAX polysaccharide (59:41, xylose/arabinose). Cells were subcultured two times consecutively in triplicate with the respective growth media to ensure complete adaptation to the carbohydrate growth source. At mid-log phase of growth (A600 nm = 0.2), 30 ml of each culture was removed and immediately combined with 60 ml of RNAprotect bacteria reagent, and RNA was purified using the RNeasy kit with the optional on-column DNase treatment step. The quality of the RNA was assessed by using a Bioanalyzer 2100 with a RNA 6000 Nano Assay reagent kit from Agilent (Santa Clara, CA).
Gene Expression Analysis Using DNA Microarrays
DNA microarray slides were fabricated by Roche NimbleGen (Madison, WI) using the partial genome sequence of P. bryantii B14. Four arrays were printed on each slide, with each array containing 68,877 total probes. For each array, a total of 2551 open reading frames (ORFs) were analyzed each with nine individual probes and with three replicates per probe. The estimated genome size for P. bryantii B14 is 3.6 megabases, and the total number of identified features (RNAs and ORFs) is 2710. Thus, the microarrays covered ∼94% of the genome features for P. bryantii B14. The purified RNA was converted to double-stranded cDNA by using the Superscript cDNA synthesis kit. The cDNA was then labeled with Cy3 by using the One-Color DNA labeling kit, which employs Cy3-labeled random nonamer primers. Sample tracking controls were added to each of the labeled cDNA samples, and hybridization was performed at 42 °C using the hybridization kit and a MAUI® 4-bay hybridization system from BioMicro Systems (Salt Lake City, UT). After 18 h, the slides were removed, washed using the Wash Buffer kit, and scanned using a GenePix 4000B microarray scanner with the laser tuned to a wavelength of 532 nm and a photomultiplier tube gain of 680. The files were then analyzed using the NimbleScan version 2.5 software. The gene expression values were determined using ArrayStar version 3.0 from DNASTAR, Inc. (Madison, WI).
Whole Transcriptome Analysis by RNA Sequencing
For RNA-Seq analyses, the RNAs isolated from two individual experiments were used for each growth condition. Bacterial 16S and 23S ribosomal RNAs were subtracted from ∼10 μg of total RNA with the RiboMinusTM bacteria transcriptome isolation kit. The enriched mRNA fraction was converted to an RNA-Seq library using the mRNA-Seq kit from Illumina Inc. (San Diego) with multiplexing adaptors that tag each library with a unique identifier. Fragments 200–400 bp long were size-selected for the final library. Each library was hybridized onto a single lane of an eight-lane flow cell and sequenced with a Genome Analyzer IIx according to the manufacturer's instructions (Illumina, Inc.). The sequences derived from each library were 73 bp long, and the overall error rate of the control lane was 0.83%. Each library yielded the following number of reads: WAX1, 11.2 million reads; WAX2, 7.1 million reads; XA1, 8.8 million reads; and XA2, 6.8 million reads.
The RNA-Seq data were analyzed using CLC genomics workbench version 3.7 from CLC Bio (Cambridge, MA). The partial genome sequence for P. bryantii B14 was uploaded onto the Rapid Annotation using Subsystem Technology (RAST) server (14), and the annotated genome was exported as a GenBank file (.gbk). The P. bryantii B14 GenBank file was then uploaded onto the CLC software and used as a reference genome for RNA-Seq analyses for each of the four samples. Reads were only assembled if the fraction of the read that aligned with the reference genome was greater than 0.9 and if the read matched other regions of the reference genome at less than 10 nucleotide positions. The RNA-Seq output files were then analyzed for statistical significance by using the proportion-based test of Baggerly et al. (15).
De Novo Assembly of the P. bryantii B14 Transcriptome
To analyze the entire transcriptome of P. bryantii B14, a de novo assembly of the RNA-Seq data was performed with 82 million 60-bp single end, nucleotide reads from a total of seven illumina lanes. These data included the four illumina libraries previously mentioned and included three additional illumina libraries. These three additional libraries were constructed with a mixture of RNA from WAX- and XA-grown cultures, in which the multiplexing adaptor reaction had failed, and the individual RNA populations could not be separated based upon the cognate growth substrate. Thus, these libraries were not useful for analyzing differential gene expression, but they did contain information on transcript coverage and were included in the transcriptome assembly. The reads were filtered for adaptor sequences and assembled into contiguous sequences (contigs) using ABySS version 1.0.12 with k-mer lengths of 25, 30, 35, 40, 45, and 50. Twenty five 2.83-GHz cores, on a cluster with 200 processors with 16 gigabytes per 8 cores, were used for each k-mer assembly. The six different k-mer assemblies were merged, and any contig that shared 100% identity to a larger contig and was completely contained within the larger contig was filtered out. The merged Abyss assembly was reassembled using the 64-bit phrap version 1.080721 with the revise greedy option on a 16-core Intel Xeon 2.93 GHz server with 128 gigabytes DDR2 RAM. The phrap assembly resulted in a total of 1927 contigs with a median length of 2340 bp, and maximum and minimum lengths of 47,199 and 100 bp, respectively. The N50 contig length was 6593 bp, and 292 contigs were larger than this size. The contigs were matched to the reference P. bryantii genomic sequence using BLAT (16) at 99% identity. Transcriptome contigs, which matched more than one contig from the reference genome, were considered as potentially closing gaps in the partial genome sequence and were confirmed by PCR.
Cloning, Expression, and Purification of P. bryantii B14 ORF0150, P. bryantii B14 ORF0336, Bacteroides eggerthii ORF1299, B. intestinalis ORF1125, and B. intestinalis ORF4213
Genomic DNA was isolated from P. bryantii B14 (13) and B. intestinalis DSM 17393 (17) using the DNeasy blood and tissue DNA purification kit. B. eggerthii DSM 20697 genomic DNA was a kind gift from Abigail A. Salyers (Department of Microbiology, University of Illinois, Urbana-Champaign). The DNA sequences for the five genes were amplified from genomic DNA using the PicoMaxx high fidelity PCR system with the oligonucleotide primers listed in supplemental Table S1. Putative signal sequences with corresponding cleavage sites were predicted for each of the five genes (PbORF0150, MNKKIIIVCLACAISLSSMA; PbORF0336, MKKILAFIGSLLLLPMAALA; BeORF1299, MKNMKNTVSILLFLFVFLSACS; BiORF4213, MKNNMRKIIYLLTLLLGVSLAACS; and BiORF1125, MKNITNVFYEFLIALCCLMSSSTLWA) with the SignalP version 3.0 on-line server (18). To ensure that proteins accumulated within the cytoplasm of E. coli cells, the primers were engineered to clone the respective gene beginning with the amino acid just downstream of the predicted cleavage site.
The five genes were cloned into pET-46b by ligation-independent cloning as described previously (4). The recombinant hexahistidine proteins were expressed in E. coli BL21-CodonPlus (DE3) RIL cells, and different purification schemes were used for each protein. For PbORF0150, the recombinant protein was purified by metal affinity chromatography using an ÄKTAxpress instrument equipped with a HisTrap FF nickel column and a HiPrep 26/10 desalting column as described previously (4). For BeORF1299 and BiORF4213, the proteins were purified by metal affinity chromatography (HisTrap FF) followed by size exclusion chromatography (HiLoad 16/60 Superdex 200 column) with protein storage buffer (50 mm Tris-HCl, 150 mm NaCl (pH 7.5)) as the mobile phase. For PbORF0336 and BiORF1125, the proteins were purified by metal affinity chromatography using Talon resin from Clontech as described by the manufacturer followed by anion exchange chromatography (HiTrap Q XL) using the following binding and elution buffer pairs: PbORF0336, binding buffer, 50 mm Na2HPO4/NaH2PO4 (pH 8.0), and elution buffer, 50 mm, Na2HPO4/NaH2PO4, 1 m NaCl, (pH 8.0); BiORF1125, binding buffer, 50 mm Na2HPO4/NaH2PO4 (pH 7.0) and elution buffer, 50 mm, Na2HPO4/NaH2PO4, 1 m NaCl (pH 7.0). Fractions were analyzed by SDS-PAGE followed by staining with Coomassie Blue G-250. The purified protein concentrations were calculated using the method of Gill and von Hippel (19) with the following extinction coefficients: PbORF0150, 152.53 cm−1 mm−1; PbORF0336, 192.52 cm−1 mm−1; BeORF1299, 137.17 cm−1 mm−1; BiORF4213, 137.17 cm−1 mm−1; and BiORF1125, 294.17 cm−1 mm−1.
Evaluation of Xylanase Activity on Agar Plates
The hydrolytic activity of PbXyn5A with plant polysaccharides as substrate was assessed by using a Congo red-based assay adapted from Teather and Wood (20). The substrates were WAX, carboxymethyl cellulose, locust bean gum, or lichenan, each at 1% w/v.
Hydrolysis of Xylohexaose and Soluble Wheat Arabinoxylan
PbXyn5A (2 μm, final concentration) was incubated with xylohexaose (1% w/v, final concentration) in citrate buffer (50 mm sodium citrate, 150 mm NaCl (pH 5.5)) at 37 °C. At regular time intervals (0, 0.5, 1, 3, 5, 10, 20, 30, and 60 min). 1-μl aliquots were removed, and the products of hydrolysis were analyzed by TLC as described below.
To evaluate the capacity of PbXyn5A, BeXyn5A, and BiXyn5A to hydrolyze a longer polysaccharide chain such as xylan, each enzyme (0.5 μm, final concentration) was incubated with soluble wheat arabinoxylan (1% w/v) in citrate buffer (500 μl, final volume) at 37 °C. After 15 h, the concentration of reducing ends was estimated using the para-hydroxybenzoic acid hydrazide assay with glucose as the standard (21). For qualitative identification of the hydrolysis products, the reactions were resolved by TLC as described previously (9). For more quantitative analysis of the products of hydrolysis, the wheat arabinoxylan hydrolysates were analyzed by high performance anion exchange chromatography (HPAEC) as described earlier (4).
Site-directed Mutagenesis
Mutagenesis was performed by use of the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). All the methods were as described in our earlier report (4). Furthermore, the expression and purification of the mutant recombinant proteins were performed as described above for the wild-type (WT) PbXyn5A, BeXyn5A, and BiXyn5A.
GenBankTM Accession Numbers
The genes listed in Fig. 1 and Table 3 are available on GenBankTM with the following accession numbers: ORF0150, HM454201.1; ORF0315, HM454202.1; ORF0316, HM454203.1; ORF0336, HM454200.1; ORF1114, HM454204.1; ORF1131, HM454205.1; ORF1893, CAB01855.1; ORF1906, HM454206.1; ORF1908, CAA89208.1; ORF1909, CAA89207.1; ORF1911, CAD21013.1; ORF1912, HM454207.1; ORF1917, ADD92015.1; ORF2001, HM454208.1; ORF2002, HM454209.1; ORF2003, HM454210.1; ORF2004, HM454211.1; ORF2350, HM454212.1; and ORF2351, HM454213.1.
FIGURE 1.
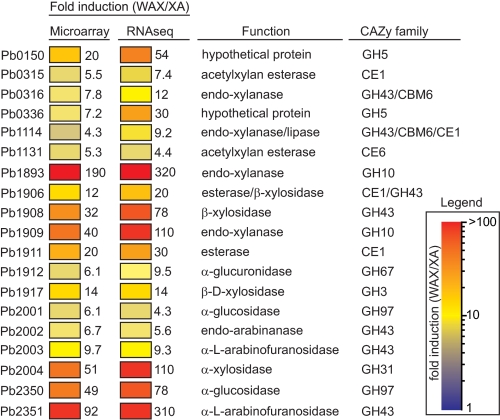
Carbohydrate esterase and glycoside hydrolase genes up-regulated on soluble WAX compared with XA. Each gene that was overexpressed at least 4-fold was used as a query in a BLASTp search of the nonredundant (nr) database at GenBankTM. Genes were assigned to CAZy (43) families if they exhibited significant similarity (E-value <1 × 10−5) to biochemically characterized proteins already catalogued in a CAZy family. The GenBankTM accession numbers for these genes are listed under “Experimental Procedures.”
TABLE 3.
Domain architecture for ORF0150 from P. bryantii B14 and its top BLAST hits
a Functional domains were assigned utilizing the Conserved Domain Database on the NCBI database (www.ncbi.nlm.nih.gov). The regions of the polypeptides were selected as belonging to a domain if the expected value (E-value) for the protein query compared with the consensus sequence generated for the particular domain was less than or equal to 0.01.
b Amino acid identities are shown for each open reading frame derived from alignments with P. bryantii ORF0150. For B. intestinalis ORF1125 and B. cellulosilyticus ORF2048, the amino acid sequence alignments with P. bryantii ORF0150 were discontinuous with similarity occurring at the N and C termini but with no significant similarity at the regions predicted to code for the GH 43 domain. Correspondingly, two amino acid identity values are provided that correspond to alignment with P. bryantii ORF0150 at the N and C termini, respectively.
RESULTS
Transcriptional Analysis of P. bryantii B14 Using Microarrays and RNA-Seq
To assess the reproducibility of the microarray and RNA-Seq analyses, the expression levels for all annotated genes in the partial genome sequence of P. bryantii B14 were compared for the two biological replicates using both technologies. These analyses revealed a high correlation in the expression level for each gene in the two biological replicates derived from either DNA microarray or RNA-Seq technologies (supplemental Fig. S1). These results suggest that the biological reproducibility in the gene expression levels was very high for both the microarray (WAX, R2 = 0.969; XA, R2 = 0.977) and RNA-Seq (WAX, R2 = 0.972; XA, R2 = 0.932) analyses.
To evaluate how well data derived from DNA microarrays and RNA-Seq analyses agree, the expression level for each gene was compared for each sample using both technologies. These results revealed that there was a positive correlation in the expression of genes identified using the two different techniques (supplemental Fig. S2); however, this correlation was moderate (WAX1, R2 = 0.649; WAX2, R2 = 0.662; XA1, R2 = 0.663; and XA2, R2 = 0.568). The moderate correlation between microarray and RNA-Seq analyses has been reported before, and it is predicted that this may arise due to the fact that microarrays lack sensitivity for genes expressed at low and high levels (22). Because of the reported higher dynamic range for gene expression using RNA-Seq, the subsequent analyses of transcriptional regulation focused on the results from the RNA-Seq experiments rather than the DNA microarray data.
The Illumina reads were assembled onto the reference genome for P. bryantii B14, and a transcriptome map was generated for the bacterium during growth with each carbohydrate source. A representative transcriptome map is shown in supplemental Fig. S3 where the number of assembled reads for each nucleotide position within the genome is plotted on the inner circle. The most abundant reads mapped onto ribosomal RNA genes (16S and 23S), indicated by the most prominent peak on the transcriptome map and five of the smaller peaks. The high abundance of reads aligning to rRNA genes demonstrates that the removal of ribosomal RNA from the sample using the RiboMinusTM kit was incomplete. In addition to the ribosomal RNA genes, the other most highly expressed genes included genes important for housekeeping functions in the cell such as fatty acid biosynthesis (acyl carrier protein, ORF0076; FabF, ORF0077), ribosome biogenesis (small and large subunit ribosomal proteins as follows: ORF2542, ORF2216, ORF1370, ORF1137, ORF1214, ORF1368, ORF0740, ORF2553, ORF0177, ORF1421, and ORF1369), and glycolysis (fructose-bisphosphate aldolase, ORF2552; and glyceraldehyde-3-phosphate dehydrogenase, ORF2095) (supplemental Table S2).
Identification of Xylanolytic Genes Up-regulated at the Transcriptional Level during Growth of P. bryantii B14 with WAX Compared with XA
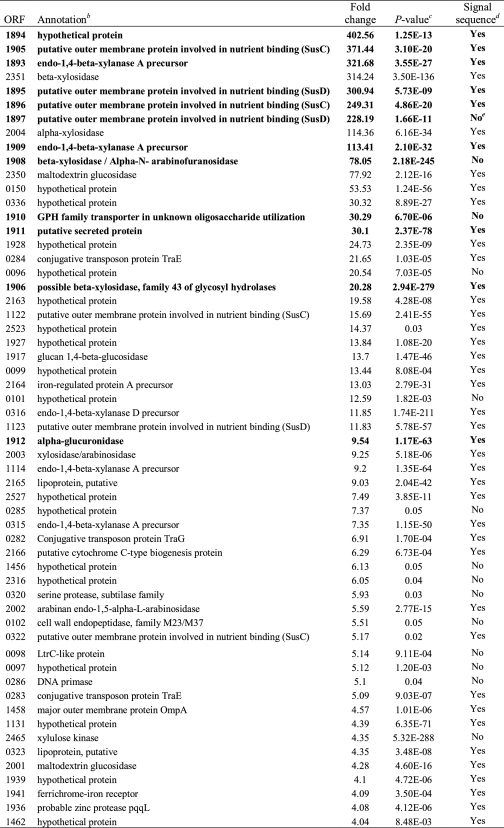
During growth with WAX relative to XA, ∼57 genes were induced greater than 4-fold, and 32 genes were repressed greater than 4-fold (Baggerly's test (15), p < 0.05) using RNA-Seq analysis (Tables 1 and 2). Annotation, based on predicted function, of the induced genes revealed that the majority code for hypothetical proteins, although GHs and membrane transporters, including starch utilization system (Sus) homologues, were also highly enriched. Additionally, a few genes with predicted functions in conjugative transposition, protein degradation, and carbohydrate ester hydrolysis were also identified (Table 1). The GH genes that were highly induced had predicted functions associated with the hydrolysis of xylan, including endoxylanases (five genes), β-xylosidases (four genes), and arabinofuranosidases (two genes). These results suggest that P. bryantii B14 elaborates a specific transcriptional response in the presence of xylan leading to induced expression of a subset of genes related to xylan degradation and utilization. Of the 57 induced genes, 77% code for proteins with putative signal sequences as predicted by using the SignalP server (Table 1) (18), suggesting that the metabolic repertoire induced during growth of P. bryantii B14 on WAX is polarized toward the periplasmic space or outer membrane.
TABLE 1.
Genes induced greater than 4-fold during growth of P. bryantii B14 with WAX as compared with XA assessed by RNA-Seq and listed by magnitude of inductiona
a P. bryantii B14 was cultured in a synthetic medium with either soluble WAX or XA as the sole carbohydrate source. RNA was then extracted, and RNA-Seq experiments were performed as described under “Experimental Procedures.”
b Open reading frames were predicted, and annotations were assigned to the putative gene products using the RAST server (14). Genes located within the major xylanolytic gene cluster shown in Fig. 2 are indicated in boldface type.
c Two-sided p values derived from Baggerly's test of the differences in means for two independent experiments are reported (15).
d Signal peptides were predicted using the SignalP version 3.0 on-line server (18).
e The predicted start codon of ORF1897 is just downstream of the beginning of a contig, and BLASTp analyses revealed that close homologs of this protein possess N-terminal extensions, which suggested that this protein is truncated at the N terminus. Thus, the assignment of the absence of a signal peptide for this protein may not be accurate.
TABLE 2.
Genes repressed greater than 4-fold during growth of P. bryantii B14 with WAX as compared with XA assessed by RNA-Seq and listed by magnitude of repressiona
a P. bryantii B14 was grown in a synthetic medium with either WAX or XA as the sole carbohydrate source. RNA was then extracted, and RNA-Seq experiments were performed as described under “Experimental Procedures.”
b Open reading frames were predicted, and annotations were assigned to the putative gene products using the RAST server (14).
c Two-sided p values derived from Baggerly's test of the differences in means for two independent experiments are reported (15).
d Signal peptides were predicted using the SignalP version 3.0 on-line server (18).
e The predicted start codon of ORF0667 is just downstream of the beginning of a contig, and BLASTp analyses revealed that close homologs of this protein possess N-terminal extensions, which suggest that this protein may be truncated at the N-terminus. Thus, the assignment of the absence of a signal peptide for this protein may not be accurate.
To improve annotation of genes, which may encode xylanolytic enzymes, the 57 highly induced genes were analyzed using BLASTp to catalogue genes into different carbohydrate active enzyme (CAZy) families. Of the 57 highly induced genes, 19 clustered into CAZy families. Fourteen of the genes grouped into GH families, three grouped into carbohydrate esterase families, and two genes were predicted to encode polypeptides harboring both GH and carbohydrate esterase domains (Fig. 1). The most highly represented CAZy family in the subset of induced genes was GH43 and the top six most highly induced genes included members from GH10 (ORF1893, ORF1909), GH31 (ORF2004), GH43 (ORF1908, ORF2351), and GH97 (ORF2350). The trends in these gene expression patterns were confirmed by both DNA microarray and RNA-Seq methods (Fig. 1).
The genes that were repressed during growth on WAX compared with XA were highly enriched for hypothetical genes and transporters (Table 2). The subset of glycoside hydrolase genes that were repressed had predicted functions associated with the depolymerization of polysaccharides such as arabinan, galacturonan, cellulose, and starch. Furthermore, SusC and SusD homologues were also found to be among the group of repressed genes. These results suggest that during growth on WAX, a transcriptional program is initiated that represses the expression of genes involved with degradation of non-xylan-containing polysaccharides.
Identification of the Major Xylanolytic Gene Cluster in P. bryantii B14
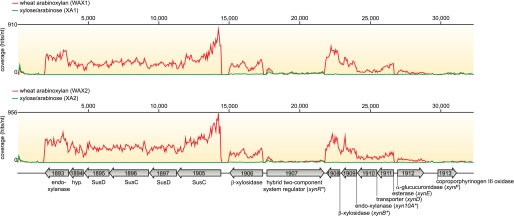
Five of the most highly induced genes clustered together near the edge of a single contiguous DNA sequence (contig 17) in the partial genome sequence of P. bryantii B14. The gene that was positioned closest to the end of the contig (ORF1897) appeared to be truncated at the N-terminal region relative to other homologous proteins within the GenBankTM database (data not shown). To evaluate whether the transcriptomic data may aid in completing the sequence for this highly expressed gene, a de novo assembly of the RNA-Seq data was carried out. Within the expressed sequence tag assembly, a large contig (expressed sequence tag contig 1388, 12,646 bp) was identified that exhibited overlap with the ends of two genomic DNA contigs (contig 17, 8656-bp overlap; contig 19, 3875-bp overlap). These data suggested that contigs 17 and 19 are closely positioned on the P. bryantii B14 chromosome and are separated by a 104-bp gap (supplemental Fig. S4). To test the results from this assembly, an oligonucleotide primer set was designed to amplify a 746-bp fragment that spans the predicted gap between the two genomic DNA contigs. Cloning and sequencing of the PCR fragment revealed that it indeed joins contigs 17 and 19, thus confirming the presence of a contiguous xylanolytic gene cluster on the P. bryantii B14 genome (Fig. 2).
FIGURE 2.
RNA-Seq coverage for the major xylanolytic gene cluster in P. bryantii B14. P. bryantii B14 was grown with either WAX or a XA as the sole carbohydrate source, and the transcriptomes were analyzed by RNA-Seq as described under “Experimental Procedures.” A detailed view of the nucleotide coverage for the major xylanolytic gene cluster in P. bryantii B14 during growth on WAX (red) or XA (green) is shown for two biological replicates (WAX1, WAX2; XA1, XA2). The portion of the gene cluster including ORF1907–1912 has been studied previously by Flint and co-workers (7). Asterisks denote genes for which biochemical activities have been demonstrated for their cognate gene products: xynB and xynA (7) and xynR (5).
Of the 20 most highly induced genes in P. bryantii B14, 12 of them mapped to this major xylanolytic gene cluster (Table 1 and Fig. 2). Annotation of the genes identified a central gene (xynR, ORF1907) predicted to encode a hybrid two-component system regulator flanked by two groups of xylanolytic genes (Fig. 2). The genes upstream of xynR are divergently oriented and include a putative β-xylosidase/esterase (ORF1906) and an operon, which contained two pairs of SusC-SusD (ORF1905-ORF1897 and ORF1896-ORF1895) genes in tandem followed by a hypothetical protein (ORF1894), and then an endoxylanase gene (ORF1893, xyn10C). The genes downstream of xynR are also divergently oriented relative to xynR and include a putative β-xylosidase encoding gene (ORF1908, xynB) preceded by an endoxylanase gene (ORF1909, xyn10A), a sugar transporter encoding gene (ORF1910, xynD), and a putative esterase gene (ORF1911, xynE). Further downstream from this cluster relative to xynR is a putative α-glucuronidase gene (ORF1912, xynF), which is in the same orientation as xynR. The RNA coverage or expression of each gene in the cluster was higher for the WAX-grown cultures relative to the XA-grown cultures except for xynR and ORF1913 (Fig. 2).
The RNA coverage was continuous throughout ORF1911-ORF1908, which is highly suggestive that these four genes are co-transcribed within a single messenger RNA molecule (Fig. 2). Furthermore, the RNA coverage is continuous throughout ORF1905-ORF1893, which suggests that these six genes are also co-transcribed within a single mRNA molecule (Fig. 2). The observed differences in the RNA coverage, e.g. ORF1908 and ORF1909 versus ORF1910 and ORF1911, may also indicate that there are multiple promoter elements present within this cluster and that the RNA coverage represents the presence of mRNA molecules with different 5′ ends. Another notable fluctuation in coverage occurred within the genes, ORF1906 and ORF1905. These fluctuations were observed in both of the biological replicates (WAX1 and WAX2), which provides evidence that the observed patterns in RNA coverage were reproducible. The intragenic variations in coverage, such as that observed with ORF1905, may reflect different susceptibilities of regions within the transcripts to degradation by native endoribonuclease enzymes.
P. bryantii B14 ORF0150 Encodes a Glycoside Hydrolase Family 5 Endoxylanase
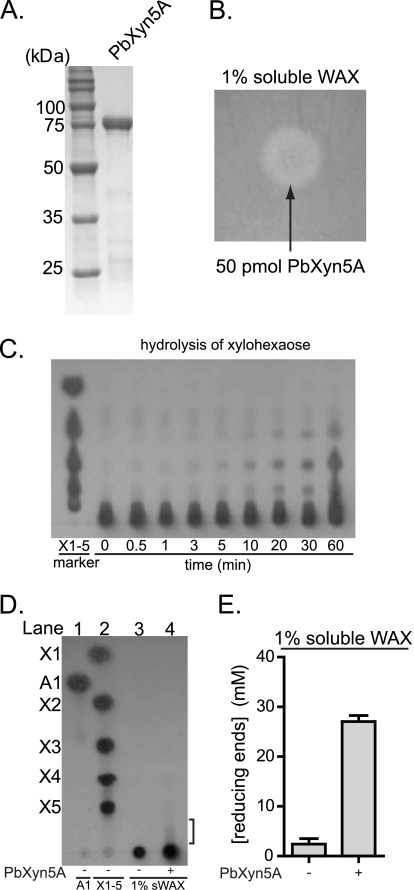
Of the 57 genes that were induced greater than 4-fold during growth on WAX relative to XA, 18 were predicted to code for hypothetical proteins, according to the RAST server and a GenBankTM database search (Table 1). Further analysis showed that two of the genes (ORF0150 and ORF0336) exhibited homology at the amino acid level to each other (50% identity, 415 residues aligned) and also low homology to enzymes from the glycoside hydrolase family 5. The gene for ORF0150 was selected for further analysis, because this gene was more highly induced on WAX relative to XA in the transcriptional study. For ORF0150, the most similar enzyme with a biochemically defined function in the GenBankTM database is the alkaline endoglucanase enzyme from Bacillus sp. KSM-635 (23), although the amino acid conservation between these two proteins is low (27% identity over 207 amino acids aligned). These results suggested that ORF0150 encodes a glycoside hydrolase, and the likely group is GH family 5. To test this possibility, the recombinant hexahistidine fusion protein, with a predicted molecular mass of 79 kDa, was made in E. coli cells and purified (Fig. 3A). Enzymatic activity was then tested with WAX, carboxymethyl cellulose, locust bean gum, or lichenan as substrates using an agar plate assay. A zone of Congo red exclusion, representing hydrolysis of WAX was observed (Fig. 3B). In contrast, clearing zones were not seen on the plates containing carboxymethyl cellulose, locust bean gum, or lichenan. These results suggested that the gene product of ORF0150 possesses xylanase activity but not endoglucanase, mannanase, or lichenenase activities. Because this gene encodes the first GH family 5 endoxylanase identified in P. bryantii B14, it was designated xyn5A.
FIGURE 3.
P. bryantii B14 ORF0150 encodes an enzyme with endoxylanase activity. A, purification of recombinant PbXyn5A. ORF0150 was cloned into an expression vector and expressed heterologously as a hexahistidine fusion protein in E. coli. The protein (PbXyn5A) was purified using cobalt affinity chromatography, and the eluate was analyzed by 12% SDS-PAGE, followed by Coomassie Brilliant Blue G-250 staining. B, depolymerization of soluble wheat arabinoxylan. PbXyn5A was assessed for its capacity to depolymerize soluble WAX by incubating the protein on an agar plate infused with WAX followed by staining and destaining with Congo red and 1 m NaCl, respectively. C, hydrolysis of xylohexaose. PbXyn5A-catalyzed hydrolysis of xylohexaose was assessed by incubating the enzyme with the substrate, removing aliquots at the indicated time points, and then resolving the products by thin layer chromatography followed by staining with methanolic orcinol. D, thin layer chromatography of products released from WAX by PbXyn5A. PbXyn5A (0.50 μm) was incubated with WAX (1% w/v), and the products were resolved by thin layer chromatography followed by staining with methanolic orcinol. Xylo-oligosaccharide standards X1–X5 and arabinose (A1) were spotted on the plate in lanes 2 and 1, respectively, to serve as markers for the identification of hydrolysis products. In lane 4, PbXyn5A was incubated with WAX at 37 °C for 15 h, and 2.5 μl of the reaction mixture were resolved on the TLC plate. E, reducing sugars released from WAX by PbXyn5A. Wild-type Xyn5A was incubated with WAX (1% w/v), and the amounts of reducing sugars released were determined by the para-hydroxybenzoic acid hydrazide assay. The reducing sugar concentrations were calculated from the absorbance at 410 nm by comparison to a standard curve generated with known concentrations of glucose. E, the values are reported as the means ± S.D. from three independent experiments.
To further verify the endoxylanase activity of PbXyn5A, the enzyme was incubated with xylohexaose, and aliquots of end products were removed at specific time intervals and analyzed by TLC. Products corresponding to xylobiose, xylotriose, and xylotetraose accumulated in the reaction mixture (Fig. 3C), thus supporting assignment of endoxylanase activity to PbXyn5A. The sizes of products released from WAX by Xyn5A were resolved by TLC, and the amounts of reducing ends released were also determined. In the absence of the enzyme, no depolymerization of the substrate was evident (Fig. 3D, lane 3). However, when the enzyme was incubated with the substrate, a smear of oligosaccharides was apparent near the bottom of the TLC plate (Fig. 3D, lane 4). Furthermore, the concentration of reducing sugars in the reaction mixture increased ∼9-fold following the addition of PbXyn5A (Fig. 3E), indicating depolymerization of the xylan substrate. The results therefore suggested that PbXyn5A releases long oligosaccharides from WAX and that the products are not clearly resolved by TLC.
To evaluate whether PbXyn5A exhibits synergistic activity with other xylanolytic enzymes, it was incubated independently or in combination with an α-l-arabinofuranosidase (Ara43A) and a β-d-xylosidase (Xyl3B) in the presence of WAX, and the products of hydrolysis were resolved by HPAEC. Injection of xylo-oligosaccharide (X2–X6) standards allowed determination of retention times for identification of end products of hydrolysis. The concentrations of xylose and arabinose in the reaction mixtures were determined by comparison of the peak areas with standard curves generated with known concentrations of xylose or arabinose. The enzyme, Ara43A, is a GH family 43 arabinoxylan arabinofuranohydrolase from P. bryantii B14, encoded by ORF2351, and was cloned and expressed in our laboratory.3 Xyl3B is a GH family 3 β-d-xylosidase from P. bryantii B14, and it releases xylose from xylo-oligosaccharides (4). In the absence of all enzymes, neither monomers nor oligosaccharides were detectable in the substrate (Fig. 4). When WAX was incubated with Ara43A, a sizeable amount of arabinose was detected (Fig. 4A). When WAX was incubated with Xyl3B, small amounts of arabinose and xylose were detected (Fig. 4A). The release of both xylose and arabinose by Xyl3B is consistent with previous results which demonstrated activity with both para-nitrophenyl (pNP) α-l-arabinofuranoside and pNP-β-d-xylopyranoside (4). When WAX was incubated with PbXyn5A, no monosaccharides were detected (Fig. 4A); however, several peaks that eluted after the retention times for xylobiose were observed (Fig. 4B), which suggests that these peaks represent xylo-oligosaccharide fragments that are either longer than our standards or are decorated with arabinose. Incubation of WAX with both PbXyn5A and Xyl3B resulted in an increase in the release of xylose and arabinose over either PbXyn5A or Xyl3B alone (Fig. 4A). When PbXyn5A and Ara43A were incubated together with WAX, the concentrations of xylose and arabinose were similar as compared with Ara43A alone (Fig. 4A); however, the pattern of xylo-oligosaccharides was different compared with PbXyn5A alone with peaks appearing that exhibited similar retentions times to the xylo-oligosaccharide standards (X2–X6) (Fig. 4B). These data revealed that PbXyn5A and Ara43A do not exhibit synergism in the release of xylose or arabinose, but they do exhibit synergism in the release of xylo-oligosaccharides. When all three enzymes were incubated together, the amount of xylose released was increased relative to each of the other enzyme combinations, whereas the level of arabinose was similar to Ara43A alone. Moreover, no peaks corresponding to the xylo-oligosaccharide standards were identified. These experiments revealed that PbXyn5A is an endoxylanase that functions synergistically with a β-d-xylosidase/α-l-arabinofuranosidase and an arabinofuranohydrolase from the same bacterium to release xylose from WAX.
FIGURE 4.
PbXyn5A functions synergistically with β-xylosidase and α-l-arabinofuranosidase enzymes. A and B, hydrolysis of WAX was assessed by incubating the respective enzymes with the substrate and then resolving the products by HPAEC followed by detection with a pulsed amperometric detector. The products of hydrolysis were identified by comparison of peaks with retention times of purified substrates. Abbreviations are as follows: FT, flow-through; A1, arabinose; X1–X6, xylose through xylohexaose. The concentration of xylose and arabinose in each of the reactions was estimated by comparison with a calibration curve constructed with known concentrations of each sugar. The same chromatograms are depicted in A and B; however, the scale is adjusted in B to reveal changes in the oligosaccharide patterns between different reactions. These experiments were performed three times, and single representative curves are shown. The concentrations of xylose and arabinose are reported as means ± S.D.
B. eggerthii ORF1299 and B. intestinalis ORF4213 Each Encode Glycoside Hydrolase Family 5 Endoxylanases
A BLASTp search of the GenBankTM nonredundant (nr) database using PbXyn5A as the query revealed that the most closely related proteins derive from members of the human colonic Bacteroidetes (Table 3). These proteins do not have biochemically defined functions; therefore, to assess whether they also encode endoxylanases, the B. eggerthii DSM20697 ORF1299 and B. intestinalis DSM17393 ORF4213 were expressed for biochemical characterization. The recombinant hexahistidine fusion proteins were purified by metal affinity chromatography. The predicted molecular masses for his-BeXyn5A and his-BiXyn5A were 72 and 73 kDa, respectively, and the sizes of the purified proteins, estimated by SDS-PAGE, were in agreement (Fig. 5A). Both the TLC and reducing sugar assays showed that the two enzymes can depolymerize WAX. In the absence of either enzyme, no depolymerization of the substrate was evident (Fig. 5B, lane 3); however, when the enzymes were independently incubated with the substrate, oligosaccharides were released (Fig. 5B, lanes 4 and 5). For both BeXyn5A and BiXyn5A, several spots, including those that migrated to similar distances as xylotriose and xylotetraose, were observed on the TLC plate (Fig. 5B, lanes 4 and 5). Furthermore, these two enzymes released large amounts of reducing sugars from WAX (Fig. 5C). Taken together, these results revealed that both BeXyn5A and BiXyn5A are endoxylanases.
FIGURE 5.
B. eggerthii ORF1299 and B. intestinalis ORF4213 encode endoxylanases. A, purification of recombinant BeXyn5A and BiXyn5A. B. eggerthii ORF1299 and B. intestinalis ORF4123 were cloned into expression vectors and expressed heterologously as hexahistidine fusion proteins in E. coli. The proteins were purified using cobalt affinity chromatography and gel filtration, and the elution fractions were pooled and analyzed by 12% SDS-PAGE, followed by Coomassie Brilliant Blue G-250 staining. B, thin layer chromatography of products released from WAX by BeXyn5A and BiXyn5A. BeXyn5A or BiXyn5A (0.50 μm each) was incubated with WAX (1% w/v) for 15 h at 37 °C, and the products were resolved by thin layer chromatography followed by staining with methanolic orcinol. Xylo-oligosaccharide standards X1–X5 and arabinose (A1) were spotted on the plate in lanes 2 and 1, respectively, to serve as markers for the identification of hydrolysis products. C, reducing sugars released from sWAX by BeXyn5A and BiXyn5A. BeXyn5A or BiXyn5A (0.50 μm each) was incubated with WAX (1% w/v) for 15 h at 37 °C, and the reducing sugars were detected by using the para-hydroxybenzoic acid hydrazide assay. The reducing sugar concentrations were calculated from the absorbance at 410 nm by comparison with a standard curve generated with known concentrations of glucose. C, the values are reported as the means ± S.D. from three independent experiments.
To provide insight into the products that are released from WAX by BeXyn5A and BiXyn5A, and to evaluate whether these enzymes also function synergistically with Ara43A and Xyl3B, the enzymes were incubated alone or in combination with Ara43A and Xyl3B, and the hydrolysates were analyzed by HPAEC. The patterns of hydrolysis for Ara43A and Xyl3B were identical to that described above (Fig. 4A). Following incubation of WAX with BeXyn5A, arabinose was released (Fig. 6A), and a mixture of oligosaccharides, including xylobiose and xylotetraose, was also detected (Fig. 6B). Incubation of WAX with both BeXyn5A and Xyl3B resulted in an increase in the concentration of xylose over either BeXyn5A or Xyl3B alone (Fig. 6A), and the oligosaccharide pattern was different from BeXyn5A alone (Fig. 6B). Incubation of both BeXyn5A and Ara43A with WAX released arabinose at a concentration that was higher than the sum of each enzyme acting alone (Fig. 6A), and xylo-oligosaccharides (X2–X6) were clearly visible (Fig. 6B). These results demonstrate that BeXyn5A functions synergistically with Xyl3B to release xylose from WAX. In addition, BeXyn5A and Ara43A function synergistically to release arabinose and xylo-oligosaccharides from WAX. The data also show that BeXyn5A releases both oligosaccharides and arabinose from WAX, indicating that this enzyme has both endoxylanase and arabinofuranosidase activities.
FIGURE 6.
BeXyn5A and BiXyn5A release long xylo-oligosaccharides from WAX. A–D, hydrolysis of wheat arabinoxylan was assessed by incubating the enzymes with WAX (1% w/v) and then resolving the products by HPAEC followed by detection with a pulsed amperometric detector. The products of hydrolysis were identified by comparison of peaks with retention times of purified substrates. Abbreviations are as follows: FT, flow-through; X2, xylobiose, X3, xylotriose; X4, xylotetraose; X5, xylopentaose; X6, xylohexaose,. The concentration of xylose and arabinose in each of the reactions was estimated by comparison with a calibration curve constructed with known concentrations of each sugar. The same chromatograms are depicted in A and B as well as in C and D; however, the scale is adjusted in B and D to reveal changes in the oligosaccharide patterns between different reactions. These experiments were performed three times, and single representative curves are shown. The concentrations of xylose and arabinose are reported as means ± S.D.
When BiXyn5A was incubated with WAX, no monosaccharides were released (Fig. 6C). However, larger oligosaccharides were detected in the hydrolysates, and these included xylobiose and xylotetraose (Fig. 6D). Incubation of WAX with both BiXyn5A and Xyl3B resulted in an increase in the concentration of xylose over either BiXyn5A or Xyl3B alone (Fig. 6C), and the oligosaccharide pattern was different from BiXyn5A alone, with no xylobiose or xylotetraose being detected (Fig. 6D). The level of arabinose released by BiXyn5A and Ara43A in combination was similar to Ara43A alone (Fig. 6C); however, in the presence of both enzymes, xylo-oligosaccharides (X2–X6) were released (Fig. 6D). This indicated that BiXyn5A functions synergistically with Ara43A to improve the release of unbranched xylo-oligosaccharides; however, no synergism was observed between the two enzymes in the release of arabinose. These results confirmed that B. intestinalis ORF4213 encodes an endoxylanase.
Although both BeXyn5A and BiXyn5A have endoxylanase activity, the oligosaccharide patterns in their hydrolysates differed. Moreover, BeXyn5A releases almost twice the amount of arabinose and xylose when co-incubated with Ara43A and Xyl3B relative to BiXyn5A. This observation clearly showed that although the two enzymes encode endoxylanases within the same GH family and also have similar domain organization (Table 3), they release different products from soluble wheat arabinoxylan.
Mutational Analyses Reveal Conserved Active Site Residues in PbXyn5A, BeXyn5A, and BiXyn5A
To further test the prediction that these three enzymes are GH family 5 enzymes and that they employ a similar mechanism for hydrolysis as other biochemically characterized GH5 enzymes, we probed for conserved amino acid sequence motifs by using position-specific iterative BLAST (PSI-BLAST) (24). This analysis revealed the presence of two amino acid sequence motifs that contained the putative glutamic acid catalytic acid/base (supplemental Fig. S5A, motif 2) and the putative glutamic acid catalytic nucleophile (supplemental Fig. S5A, motif 3). An additional motif was identified (supplemental Fig. S5A, motif 1) that contained an aspartic acid in PbXyn5A, BeXyn5A, and BiXyn5A that aligned with a conserved tyrosine residue that was shown to make contact with the cellobiose ligand in the crystal structure for the alkaline cellulase K from Bacillus sp. strain KSM-635 (25). On mutating each of the three residues (PbXyn5A D104A, PbXyn5A E203A, PbXyn5A E308A, BeXyn5A D129A, BeXyn5A E229A, BeXyn5A E349A, BiXyn5A D127A, and BiXyn5A E227A and E347A), only the mutations that targeted the glutamic acid residues abolished catalysis (supplemental Fig. S5, B and C). This finding was further evidence that the three proteins belong to GH family 5, because the data suggest that they employ similar amino acid residues in catalysis as described for GH5 enzymes (25).
DISCUSSION
Xylans are an abundant group of plant polysaccharides that are hydrolyzed and fermented by commensal microbes within the rumen (26–28) and the human gut (29–35). Despite the abundance of xylanolytic bacteria in these microbiomes, relatively little is known about the mechanisms underlying the degradation and utilization of xylan by these bacteria.
In this study, a whole genome transcriptomic approach was employed to evaluate the repertoire of genes that the rumen hemicellulolytic bacterium P. bryantii B14 employs to degrade xylan. The regulation of xylanase activity has been studied previously in P. bryantii B14, and it was found that xylanase activity is not induced by glucuronic acid, arabinose, xylose, or small xylo-oligosaccharides (X2–X5) (12). Rather, the major inducers of xylanase activity were found to be medium to large sized xylo-oligosaccharides. Our data confirmed the results from this previous study and reveal that the two previously studied endoxylanase genes (xyn10A and xyn10C) are highly induced during growth on soluble WAX compared with the component monosaccharides, XA. Although these genes were among the top 10 most highly induced genes, a large number of additional genes were also induced under these conditions. This observation underscores the complexity in the transcriptional response of P. bryantii B14 during growth on this arabinoxylan polysaccharide.
The major xylanolytic gene cluster in P. bryantii B14 identified in this study contains a genomic DNA fragment that was previously cloned by Gasparic et al. (7). The genes previously identified include open reading frames 1907 (xynR), 1908 (xynB), 1909 (xyn10A), 1910 (xynD), 1911 (xynE), and 1912 (xynF), although our data suggest that two genes (xynR and xynF) on either end of the DNA fragment reported by Gasparic et al. (7) were truncated in their study. A translation start site (ATG) that occurs 1602 bp upstream of the start site predicted for this gene by Gasparic et al. (7) was identified for xynR (ORF1907), and the assignment of this alternative start site was corroborated by the RNA-Seq coverage, which is continuous throughout the entire length of ORF1907 (Fig. 2). This result suggests that the xylan catabolism regulator (XynR) in P. bryantii B14 is 534 amino acids longer than originally reported. Analysis of the domain architecture for this protein revealed three functional domains, including a putative N-terminal periplasmic sensing domain, followed by a histidine kinase domain and a response regulator domain (supplemental Fig. S6). Thus, this hybrid two-component system regulator contains all of the regulatory elements found in “classical” two-component regulatory systems, but rather than encoding the sensor and effector domains in separate genes as in the classical system, xynR sencodes all of these functions within a single polypeptide. The domain organization for XynR is analogous to that for a hybrid two-component system regulator (BT3172) characterized from the related bacterium, Bacteroides thetaiotaomicron VPI-5482 (36); however, the sequence similarity across the entire polypeptide is relatively low (23%, 1405 amino acids aligned). The gene xynR was shown to be important for coordinating the transcription of a xylan utilization gene cluster in response to growth on xylan in P. bryantii B14 (5); thus, this protein may be the primary regulator responsible for the transcriptional response identified in this study. This hypothesis can be tested by constructing a P. bryantii B14 strain carrying a deletion in this gene and examining its growth on xylan. However, the lack of a genetic system for manipulating P. bryantii B14 precludes this analysis at the current time. The gene xynR is conserved in the human colonic xylan utilizing bacterium Bacteroides ovatus, which is amenable to genetic manipulation. Thus this bacterium can be used as a model to investigate the role of the hybrid two-component system in the catabolism of xylan.
The xylan utilization gene cluster previously characterized by Gasparic et al. (7) was found to be part of a larger xylanolytic gene cluster that includes the previously characterized endoxylanase gene, xyn10C (10). The gene for Xyn10C (ORF1893) occurs in a group of six genes that the RNA-Seq data suggest are co-transcribed within a single polycistronic mRNA molecule. This operon includes six of the seven most highly induced genes during growth on WAX relative to XA, which indicates that it is likely to be of critical importance to xylan utilization by P. bryantii B14. Contained within this operon are two tandem repeats of genes predicted to encode outer membrane proteins that share homology to the starch utilization system (Sus) components, SusC and SusD, followed by a hypothetical gene that is then followed by xyn10C. This arrangement of genes is similar in certain respects to the starch utilization system identified by Salyers and co-workers (37) in B. thetaiotaomicron. SusC is predicted to encode an outer membrane porin that transports oligosaccharides into the periplasm in a TonB-dependent fashion. SusD harbors a signal peptidase II cleavage site that may facilitate the tethering of this protein onto the outer leaflet of the outer membrane where it may play a role in oligosaccharide binding (38). The hypothetical protein (ORF1894) and Xyn10C both possess putative signal peptidase II cleavage sites, which suggests that these two proteins are also tethered on the outer surface of the cell. The function of the hypothetical protein (ORF1894) has yet to be determined; however, its location within this highly induced xylan utilization operon suggests that it is involved in the degradation and utilization of xylan. The gene product has been made and purified, and when tested it exhibited a zone of Congo red exclusion following incubation on a WAX-infused agar plate (data not shown). Analysis of the activity of this protein is an ongoing focus in our laboratory. These observations suggest that this cluster of six proteins may be critical for the binding, depolymerization, and transport of extracellular xylan fragments into the periplasmic space, although further functional studies must be performed to verify this hypothesis.
The arrangement of this operon is conserved among a number of bacteria for which genome sequences are available, including Prevotella ruminicola, Prevotella copri, Prevotella buccae, Prevotella bergensis, B. intestinalis, Bacteroides cellulosilyticus, Bacteroides sp. 2_2_4, B. ovatus, B. eggerthii, Bacteroides plebeius, and an additional Bacteroidetes member, Spirosoma linguale that is commonly isolated from soil or freshwater (Fig. 7 and supplemental Fig. S7). These organisms are members of the phylum Bacteroidetes and, with the exception of S. linguale, they were isolated from the bovine rumen or the human alimentary tract (supplemental Fig. S7). This is the first evidence of a xylan utilization cluster that is strictly conserved across xylan degrading members of the rumen Prevotella and the human-associated Bacteroides spp. The conservation of this cluster is highly suggestive that xylanolytic members from these two bacterial genera employ a conserved mechanism to degrade and utilize xylan. Apart from the high level of conservation in the orientation of genes within this operon, there are no other strictly conserved arrangements of genes in the regions nearby on the chromosome (Fig. 2). It has been proposed that the Sus system, initially identified by Salyers and co-workers (37, 39), represents a paradigm for oligo- and polysaccharide utilization by Bacteroidetes (40), and the core xylan utilization cluster identified in this study provides further support for this prediction.
FIGURE 7.
Core xylan utilization system is conserved among certain species within the phylum Bacteroidetes. The P. bryantii B14 endoxylanase, Xyn10C, was used as the query sequence in a BLASTp search of the GenBankTM database. The genomic context is shown for each of the top BLASTp hits. Only the region that contains genes with predicted roles associated with xylan deconstruction are shown. Genes are color-coded based on their predicted roles as indicated in the legend. ORF numbers are indicated within each of the genes as derived from the genome project for each organism in the GenBankTM database.
The majority of the genes that were induced by P. bryantii B14 during growth on WAX compared with XA code for proteins that are currently designated hypothetical proteins. This observation underscores the fact that there are large gaps in our current understanding of how polysaccharides are metabolized by P. bryantii B14 and other gut-associated bacteria. Whole genome transcriptional profiling represents a useful approach to gain insight into the potential roles of genes with unassigned functions. In this study, a subset of genes with low homology to glycoside hydrolases has been shown to belong to GH family 5 based on biochemical data. These genes were only found within certain members of the Bacteroidetes phylum, which are resident within the alimentary tract of humans or ruminants. Most of these GH5 genes occur near the conserved xylan utilization cluster in the genome (Fig. 7; P. copri, ORF6092; P. buccae, ORF0844, Bacteroides sp. 2_2_4, ORF3750; B. intestinalis, ORF4213; B. cellulosilyticus, ORF3415; B. eggerthii, ORF1299), which suggests that they may be directly involved in the degradation of xylan. BeXyn5A and BiXyn5A contain putative signal peptidase II cleavage sites, raising the possibility that the two proteins are anchored on the outside of the cell and perhaps function coordinately with the core xylan utilization cluster.
All of the GH5 proteins identified in this study possess N- or C-terminal stretches of amino acids that did not align with known domains in either the Pfam or the NCBI conserved domains database (Table 3). Whether or not these regions represent functional domains is currently unclear. The two Bacteroides proteins (BeXyn5A and BiXyn5A) share 78% identity at the amino acid level and also share similar domain organizations with both proteins possessing a C-terminal bacterial Ig2-like domain. The function of this C-terminal extension is currently unknown; however, the Ig2-like domain is predicted to be involved in cell surface adhesion (41). Many xylan-degrading enzymes are associated with carbohydrate-binding modules within a single polypeptide, and it is possible that the Ig2-like domain serves a carbohydrate binding function. Further studies in our laboratory will focus on delineating the functional role of this domain in BeXyn5A and BiXyn5A.
Despite the fact that the three enzymes characterized in this study are clearly related to each other at the amino acid sequence level, they exhibited differences in the products released from WAX. The observation that BeXyn5A (ORF1299, Table 3) synergizes with Ara43A, whereas BiXyn5A (ORF4213, Table 3) and PbXyn5A (ORF0150, Table 3) do not exhibit synergy with Ara43A, clearly indicates that there is a fundamental difference in the enzymatic activities among these enzymes, most likely originating from the GH5 active site domain.
Two additional genes (P. bryantii B14 ORF0336 and B. intestinalis ORF1125, Table 3), encoding similar GH5 modules as those found in the enzymes described above, were expressed. Both proteins exhibited endoxylanase activity, and the gene products were named PbXyn5B and BiXyn5B (supplemental Fig. S8). In addition to the GH5 module, BiXyn5B also contains a GH43 domain and is therefore different from BiXyn5A.
Recently, a large number of genome sequences have been made available for human colonic bacteria through the human microbiome project (42). These genome sequences provide a wealth of information on the genome contents of commensal microbes within the human gut microbiome; however, proper annotation of these genes is critical to interpreting the genomic data in terms of the metabolic repertoire of the microbial community. In this study, transcriptional profiling of a rumen bacterium led to the assignment of function to a group of hypothetical proteins within the rumen Prevotella spp. and human colonic Bacteroides spp. Furthermore, this study has provided insights that suggest a conserved mechanism for xylan utilization among members of the phylum Bacteroidetes.
Supplementary Material
Acknowledgments
We thank Shosuke Yoshida, Yejun Han, Michael Iakiviak, and Xiaoyun Su of the Energy Biosciences Institute for valuable scientific discussions and Hiroshi Miyagi for technical assistance. We also thank members of the North American Consortium for Genomics of Fibrolytic Ruminal Bacteria for access to the partial genome sequence of P. bryantii B14, Alvaro Hernandez and Chris Wright of the W. M. Keck Center for Comparative and Functional Genomics for assistance with Illumina sequencing, and Mark Band from the same center for assistance with microarray analyses.
This work was supported by the Energy Biosciences Institute. D. D. was partially supported by National Institutes of Health Fellowship 1F30DK084726 from NIDDK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Tables S1 and S2.
S. Kiyonari, Y.-H. Moon, D. Dodd, R. I. Mackie, and I. K. O. Cann, manuscript in preparation.
- GH
- glycoside hydrolase
- WAX
- wheat arabinoxylan
- HPAEC
- high performance anion exchange chromatography
- XA
- xylose and arabinose.
REFERENCES
- 1.Dodd D., Cann I. O. (2009) GCB Bioenergy 1, 2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevenson D. M., Weimer P. J. (2007) Appl. Microbiol. Biotechnol. 75, 165–174 [DOI] [PubMed] [Google Scholar]
- 3.Edwards J. E., McEwan N. J., Travis A. J., Wallace R. J. (2004) Antonie van Leeuwenhoek 86, 263–281 [DOI] [PubMed] [Google Scholar]
- 4.Dodd D., Kiyonari S., Mackie R. I., Cann I. K. (2010) J. Bacteriol. 192, 2335–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki K., Miyamoto H., Mercer D. K., Hirase T., Martin J. C., Kojima Y., Flint H. J. (2003) J. Bacteriol. 185, 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazaki K., Martin J. C., Marinsek-Logar R., Flint H. J. (1997) Anaerobe 3, 373–381 [DOI] [PubMed] [Google Scholar]
- 7.Gasparic A., Martin J., Daniel A. S., Flint H. J. (1995) Appl. Environ. Microbiol. 61, 2958–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasparic A., Marinsek-Logar R., Martin J., Wallace R. J., Nekrep F. V., Flint H. J. (1995) FEMS Microbiol. Lett. 125, 135–141 [DOI] [PubMed] [Google Scholar]
- 9.Dodd D., Kocherginskaya S. A., Spies M. A., Beery K. E., Abbas C. A., Mackie R. I., Cann I. K. (2009) J. Bacteriol. 191, 3328–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint H. J., Whitehead T. R., Martin J. C., Gasparic A. (1997) Biochim. Biophys. Acta 1337, 161–165 [DOI] [PubMed] [Google Scholar]
- 11.Morrison M., Daugherty S. C., Nelson W. C., Davidsen T., Nelson K. E. (2010) Microb. Ecol. 59, 212–213 [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki K., Hirase T., Kojima Y., Flint H. J. (2005) Microbiology 151, 4121–4125 [DOI] [PubMed] [Google Scholar]
- 13.Bryant M. P., Small N., Bouma C., Chu H. (1958) J. Bacteriol. 76, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., Formsma K., Gerdes S., Glass E. M., Kubal M., Meyer F., Olsen G. J., Olson R., Osterman A. L., Overbeek R. A., McNeil L. K., Paarmann D., Paczian T., Parrello B., Pusch G. D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. (2008) BMC Genomics 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baggerly K. A., Deng L., Morris J. S., Aldaz C. M. (2003) Bioinformatics 19, 1477–1483 [DOI] [PubMed] [Google Scholar]
- 16.Kent W. J. (2002) Genome Res. 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakir M. A., Kitahara M., Sakamoto M., Matsumoto M., Benno Y. (2006) Int. J. Syst. Evol. Microbiol. 56, 151–154 [DOI] [PubMed] [Google Scholar]
- 18.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 19.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 20.Teather R. M., Wood P. J. (1982) Appl. Environ. Microbiol. 43, 777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lever M. (1972) Anal. Biochem. 47, 273–279 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Gerstein M., Snyder M. (2009) Nat. Rev. Genet. 10, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozaki K., Shikata S., Kawai S., Ito S., Okamoto K. (1990) J. Gen. Microbiol. 136, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 24.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirai T., Ishida H., Noda J., Yamane T., Ozaki K., Hakamada Y., Ito S. (2001) J. Mol. Biol. 310, 1079–1087 [DOI] [PubMed] [Google Scholar]
- 26.Hespell R. B., Wolf R., Bothast R. J. (1987) Appl. Environ. Microbiol. 53, 2849–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehority B. A. (1967) Appl. Microbiol. 15, 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehority B. A. (1966) J. Bacteriol. 91, 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salyers A. A., Balascio J. R., Palmer J. K. (1982) J. Food Biochem. 6, 39–56 [Google Scholar]
- 30.Chassard C., Goumy V., Leclerc M., Del'homme C., Bernalier-Donadille A. (2007) FEMS Microbiol. Ecol. 61, 121–131 [DOI] [PubMed] [Google Scholar]
- 31.Salyers A. A., Gherardini F., O'Brien M. (1981) Appl. Environ. Microbiol. 41, 1065–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chassard C., Bernalier-Donadille A. (2006) FEMS Microbiol. Lett. 254, 116–122 [DOI] [PubMed] [Google Scholar]
- 33.Chassard C., Delmas E., Lawson P. A., Bernalier-Donadille A. (2008) Int. J. Syst. Evol. Microbiol. 58, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 34.Mirande C., Kadlecikova E., Matulova M., Capek P., Bernalier-Donadille A., Forano E., Bera-Maillet C. (2010) J. Appl. Microbiol. 109, 451–460 [DOI] [PubMed] [Google Scholar]
- 35.Weaver J., Whitehead T. R., Cotta M. A., Valentine P. C., Salyers A. A. (1992) Appl. Environ. Microbiol. 58, 2764–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenburg E. D., Sonnenburg J. L., Manchester J. K., Hansen E. E., Chiang H. C., Gordon J. I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8834–8839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shipman J. A., Berleman J. E., Salyers A. A. (2000) J. Bacteriol. 182, 5365–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koropatkin N. M., Martens E. C., Gordon J. I., Smith T. J. (2008) Structure 16, 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Elia J. N., Salyers A. A. (1996) J. Bacteriol. 178, 7180–7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martens E. C., Koropatkin N. M., Smith T. J., Gordon J. I. (2009) J. Biol. Chem. 284, 24673–24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly G., Prasannan S., Daniell S., Fleming K., Frankel G., Dougan G., Connerton I., Matthews S. (1999) Nat. Struct. Biol. 6, 313–318 [DOI] [PubMed] [Google Scholar]
- 42.Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007) Nature 449, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.