Abstract
We isolated oryctin, a 66-residue peptide, from the hemolymph of the coconut rhinoceros beetle Oryctes rhinoceros and cloned its cDNA. Oryctin is dissimilar to any other known peptides in amino acid sequence, and its function has been unknown. To reveal that function, we determined the solution structure of recombinant 13C,15N-labeled oryctin by heteronuclear NMR spectroscopy. Oryctin exhibits a fold similar to that of Kazal-type serine protease inhibitors but has a unique additional C-terminal α-helix. We performed protease inhibition assays of oryctin against several bacterial and eukaryotic proteases. Oryctin does inhibit the following serine proteases: α-chymotrypsin, endopeptidase K, subtilisin Carlsberg, and leukocyte elastase, with Ki values of 3.9 × 10−10 m, 6.2 × 10−10 m, 1.4 × 10−9 m, and 1.2 × 10−8 m, respectively. Although the target molecule of oryctin in the beetle hemolymph remains obscure, our results showed that oryctin is a novel single domain Kazal-type inhibitor and could play a key role in protecting against bacterial infections.
Keywords: Disulfide, Insect, NMR, Peptide Conformation, Protease Inhibitor, Protein Structure, Protein-Protein Interactions, Serine Protease
Introduction
Eggs and larvae of the coconut rhinoceros beetle Oryctes rhinoceros live in compost in warm areas such as Southeast Asia. This beetle has thus developed a self-defense system that includes antimicrobial peptides. Several antimicrobial peptides were isolated from the hemolymph of the beetle: defensin (1), rhinocerosin (2), and scarabaecin (3). Oryctin (GenBankTM accession no. BAA36402), a 66-residue peptide with three intramolecular disulfide bonds, was found as a hemolymph peptide of the beetle during a search for antibacterial peptides. The function of oryctin has been unclear, because no other known peptide or protein is sequentially similar to oryctin. We took a structural approach to reveal the function of oryctin. First, we determined the solution structure of recombinant 13C,15N-labeled oryctin by heteronuclear NMR spectroscopy. Next, we searched for proteins that are structurally similar to oryctin in the Protein Data Bank (PDB).2 The search revealed that oryctin has a fold that is similar in part to that of the turkey ovomucoid third domain (OMTKY3), a serine protease inhibitor. We then performed protease inhibition assays of oryctin and found that oryctin inhibits eukaryotic chymotrypsin-like serine proteases such as α-chymotrypsin and leukocyte elastase and bacterial subtilisin-like serine proteases such as subtilisin Carlsberg and endopeptidase K. Therefore, oryctin is a novel single domain Kazal-type inhibitor despite its unique amino acid sequence. Kazal-type serine protease inhibitors usually consist of multiple Kazal domains, each of which has a characteristic disulfide linkage pattern, CysI-CysV (where CysI, for example, is the first cysteine residue from the N terminus), CysII-CysIV, and CysIII-CysVI, as well as a secondary structure consisting of an α-helix and an antiparallel β-sheet. Here, we discuss the structure and function of oryctin by comparing the sequences, the patterns of the disulfide linkages, and the tertiary structures.
EXPERIMENTAL PROCEDURES
Purification of Oryctin
The hemolymph of the third instar larvae of O. rhinoceros collected on the islands of Okinawa and Ishigaki, Japan, was collected into an ice-cooled 50-ml centrifugation tube containing 1 mg of aprotinin 24 h after injection of heat-killed Escherichia coli. The hemolymph was centrifuged at 39,100 × g for 50 min at 4 °C. The supernatant was heated in boiling water for 10 min and then cooled on ice and centrifuged at 39,100 × g for 50 min at 4 °C. The supernatant was acidified with 0.1% (v/v) trifluoroacetic acid (TFA) and applied onto a Sep-Pak Vac tC18 column (Waters, Milford, MA) equilibrated with 0.1% (v/v) TFA and eluted with 10, 20, 30, 40, 50, and 100% (v/v) acetonitrile containing 0.1% (v/v) TFA. The 40% (v/v) acetonitrile fraction was applied to a PepRPC column (GE Healthcare) equilibrated with 0.05% (v/v) heptafluorobutanoic acid. The adsorbent was eluted with a linear gradient of acetonitrile: 0–20% (v/v) in 5 min, followed by 20–40% (v/v) in 40 min at a flow rate of 0.5 ml/min. The fractions containing oryctin were applied to the same system but using 0.05% (v/v) TFA instead of 0.05% (v/v) heptafluorobutanoic acid.
Peptide Sequencing
Peptide sequencing was performed using LF3400 (Beckman Coulter, Fullerton, CA) and Procise cLC492 (Applied Biosystems, Foster City, CA) protein sequencers.
Matrix-assisted Laser Desorption Ionization Time-of-flight Mass Spectrometry (MALDI-TOF MS)
MALDI-TOF MS was measured on a Voyager Linear spectrometer (Applied Biosystems). About 1 pmol of purified oryctin was dissolved in 1 μl of 0.1% (v/v) TFA. The sample solution was then mixed with saturated sinapinic acid solution in 50% (v/v) acetonitrile containing 0.1% (v/v) TFA directly on the target.
cDNA Cloning
The cDNA encoding oryctin was cloned using the following three-step PCR amplification.
Step 1: Reverse transcriptase-PCR
The fat body was collected 10 h after the injection of heat-killed E. coli. The poly(A)+ RNA was purified from the fat body using a Quick Prep mRNA purification kit (GE Healthcare). The first-strand cDNA was synthesized from 1.32 μg of the poly(A)+ RNA using a First-Strand cDNA synthesis kit (GE Healthcare). Using the first-strand cDNA as a template, RT-PCR was performed using the following degenerate primers: the sense primer O1F (17-mer), whose sequence was deduced from the amino acid sequence of Val1–Asp6 (5′-GTNCCNGTNGGN(AT)(CG)NGA-3′) and the antisense primer O1R (20-mer), whose sequence was deduced from the amino acid sequence of Asn23–Val29 (5′-AC(AGT)ATNCCYTTYTCNGGRTT-3′). A 45-cycle step-down protocol was used: denaturation at 94 °C for 8 min (first cycle) or 30 s (second and following cycles), annealing at 60 °C (initial five cycles), 55 °C (second five cycles), 50 °C (third five cycles), 45 °C (fourth five cycles), or 40 °C (final 25 cycles) for 30 s and polymerization at 72 °C for 30 s. The resultant 86-bp fragment was subcloned into a TA cloning vector (Invitrogen). DNA sequencing was performed using an ABI Prism model 373A DNA sequencer with the dye-terminator protocol (Applied Biosystems).
Step 2: 3′-Rapid Amplification of cDNA End
3′-RACE was done using the first-strand cDNA as the template, the antisense NotI adaptor primer 5′-AACTGGAAGAATTCGCGGC-3′, and the sense primer O2F (22-mer), whose sequence was derived from the result of Step 1: 5′-CTGTGAACCCAAACTATGCACC-3′. A 35-cycle PCR was used: denaturation at 94 °C for 8 min (first cycle) or 30 s (second and following cycles), annealing at 50 °C for 30 s, and polymerization at 72 °C for 30 s. The amplified fragment was subcloned and sequenced as described above.
Step 3: 5′-Rapid Amplification of cDNA End
5′-RACE was done using the 5′-RACE kit (Invitrogen). The first-strand cDNA was synthesized using the antisense primer OGSP1 (16-mer), whose sequence was derived from the result of Step 2: TTATGGACGTGGTGCA. The terminal deoxynucleotidyl transferase reaction was performed on ice for 1 h. The nested PCR was performed using the antisense primer OGSP2 (24-mer) TTTTTGATTGCTTCTTCACACTCG and the abridged anchor primer. A 40-cycle PCR was used: denaturation at 94 °C for 6 min (first cycle) or 1 min (second and following cycles), annealing at 50 °C for 30 s, and polymerization at 72 °C for 1 min. The amplified fragment was subcloned and sequenced as described above.
Determination of Disulfide Linkages
About 1 μg of oryctin was dissolved in 10 μl of 20 mm sodium bicarbonate (pH 8.5) containing 0.1 μg of lysyl endopeptidase (Wako, Tokyo, Japan) and endoprotease Asp-N (Roche Applied Science) and incubated at 37 °C for 4 h. The reaction mixture was applied directly to MALDI-TOF MS. α-Cyano-4-hydroxycinnamic acid was used as the matrix.
Gene Expression Analysis
RT-PCR was carried out as described previously (1) using the following primers: 5′-AGTTGTGAATTTGTGCCGATACTG-3′ (forward) and 5′-CATTTGCGACAATTTATTTAAAAGC-3′ (reverse). A 40-cycle PCR was used: denaturation at 94 °C for 6 min (first cycle) or 30 s (second and following cycles), annealing at 50 °C for 30 s, and polymerization at 72 °C for 30 s.
Expression and Purification of 15N- and 13C,15N-labeled Oryctin
Oryctin with an N-terminal His6 tag that was cleavable by tobacco etch virus (TEV) protease (Invitrogen) digestion was expressed in E. coli BL21-Star(DE3) (Invitrogen) grown in M9 minimal medium using a pET28a-based (Novagen, Madison, WI) expression plasmid by adding a final concentration of 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mm Tris-HCl (pH 8.0), 300 mm NaCl, 10 mm imidazole), and disrupted by sonication. After centrifugation, 15N-labeled and 13C,15N-labeled oryctin with an N-terminal His6 tag was purified with Ni Sepharose 6 Fast Flow (GE Healthcare). The His6 tag was removed by tobacco etch virus (TEV) protease digestion. 15N-labeled and 13C,15N-labeled oryctin were further purified by cation exchange followed by size exclusion chromatographies with Mono S HR 10/10 and Superdex 200 HR 10/30 (GE Healthcare), respectively.
NMR Spectroscopy
Purified 13C,15N-labeled oryctin was dissolved in 50 mm sodium phosphate buffer (pH 6.8), 100 mm NaCl, and 0.02% NaN3 in 90% H2O, 10% D2O (v/v) to a final concentration of 2 mm. All NMR spectra were measured at 25 °C on a Unity Inova 500-MHz spectrometer (Varian, Palo Alto, CA) equipped with a triple-resonance probe. The following NMR data were acquired: 1H-15N HSQC, HNCO, HNCA, HN(CO)CA, CBCA(CO)NH, HNCACB, C(CO)NH, H(CCO)NH, HCCH-correlation spectroscopy (HCCH-COSY), HCCH-total correlation spectroscopy (HCCH-TOCSY), 15N-edited NOESY-HSQC, and 13C-edited NOESY-HSQC. The mixing times for the 15N-edited and 13C-edited NOESY-HSQCs were 75 and 150 ms. All of the acquired data were processed, visualized, and analyzed with the programs NMRPipe (4), NMRDraw (4), and Sparky (NMR Assignment and Integration Software, University of California, San Francisco), respectively.
Dihedral Angle Restraints
The programs CSI (5) and TALOS (6) were used to predict the regions of α-helix and β-strand based on 13Cα, 13Cβ, 13C′, 1Hα, and 15N chemical shifts. The predicted angle ranges were used as the dihedral angle restraints.
Hydrogen Bond Restraints
Hydrogen bonds were detected by the following HgD exchange experiment. First, the 1H-15N HSQC spectrum of 0.2 mm 15N-labeled oryctin dissolved in 50 mm sodium phosphate (pH 6.8), 100 mm NaCl, and 0.02% (w/v) NaN3 in 90% H2O, 10% D2O (v/v) were acquired. Then, the solvent was changed to 50 mm sodium phosphate (pH 6.8), 100 mm NaCl, and 0.02% (w/v) NaN3 in D2O by ultrafiltration using a Vivaspin-20 (molecular weight cutoff 3000; Sartorius Stedim Biotech, Aubagne, France). 1H-15N HSQC spectra were acquired at 6, 12, 24, 36, and 48 h after the solvent exchange. The peak intensities in these 1H-15N HSQC spectra were analyzed. Slowly exchanging amide protons were detected, and their respective carbonyl acceptors were deduced from the NOE data, and the oryctin structures at intermediate stages of the structure calculations. Hydrogen bond distance restraints were set as rHN-O = 1.8–2.0 Å, rN-O = 2.7–3.0 Å.
Structure Calculation and Structural Similarity Search
Interproton distance restraints were derived from peak intensities in the 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra of 13C,15N-labeled oryctin with a mixing time of 75 ms. The cross-peak intensities were translated into interproton distances based on the relationship, NOE ∝ (distance)−6; the standard distance between HNi and HNi+1 in the α-helix was 2.8 Å, and the standard distance between Hαiand Hαj in the antiparallel β-sheet was 2.3 Å. Structure calculations were performed using CYANA (7). Two hundred conformers were annealed in 10,000 steps of torsion angle dynamics calculations, of which 20 conformers with the lowest values in the target function were used to represent the solution structure of oryctin. The conformer with the lowest target function was used as the representative structure of oryctin. The tertiary structure was visualized with the programs MOLMOL (8) and PyMOL (DeLano, W. L.). The five conformers with the lowest target functions were submitted to the Dali server (9) to search for proteins structurally similar to oryctin.
Protease Inhibition Assays
α-Chymotrypsin, leukocyte elastase, subtilisin Carlsberg, endopeptidase K, trypsin, papain, and thermolysin were purchased from Sigma-Aldrich; V8 protease, pepsin, and lysyl endopeptidase were from Wako Pure Chemical Industries (Osaka, Japan); proline-specific endopeptidase was from Seikagaku Corp. (Tokyo, Japan); and endoproteinases Arg-C and Asp-N were from Roche Diagnostics. Peptidyl-MCA substrates (referred to as MCA substrates) and MOCAc/Dnp type fluorescence-quenching substrates (referred to as MOCAc/Dnp substrates) for the above-mentioned proteases (shown in Table 2) were purchased from Peptide Institute (Osaka, Japan). In the case of MCA substrates, the protease activity was measured using fluorescence intensity, where 380 and 460 nm were used as the excitation and detection wavelengths, respectively. In the case of MOCAc/Dnp substrates, the protease activity was measured using fluorescence intensity, where 328 and 393 nm were used as the excitation and detection wavelengths, respectively. The pH-dependent inhibitory activity was assayed as follows. A reaction mixture containing 3.3 nm protease, 10 nm oryctin, and 66, 100, or 130 μm MCA substrate or 6.6, 10, or 13 μm MOCAc/Dnp substrate in 10 mm Tris-HCl (pH 7.2–9.2) was incubated at 37 °C for 10 min, and then HCl was added to stop the reaction. Exceptionally, for a reaction involving pepsin, the pH of the reaction mixture was adjusted to 1.0–3.0 with HCl, and the reaction was stopped by adding 3 m Tris-HCl (pH 8.0). The temperature-dependent inhibitory activity was assayed as follows. A reaction mixture containing 3.3 nm protease, 10 nm oryctin, and 33, 66, 100, or 130 μm MCA substrate or 3.3, 6.6, 10, or 13 μm MOCAc/Dnp substrate in 10 mm Tris-HCl (pH 8.0) was incubated at 10–70 °C for 3 min, and then HCl was added to stop the reaction. The mode of protease inhibition and the inhibition constant (Ki) values were determined with a reaction mixture containing 3.3 nm protease, 83, 170, 250, 330, or 500 μm MCA substrate, and 0.83, 1.7, 2.5, 3.3 or 5.0 nm oryctin (for α-chymotrypsin, endopeptidase K, and subtilisin Carlsberg) or 8.3, 17, 25, 33, or 50 nm oryctin (for leukocyte elastase), respectively. The reaction mixtures were incubated at 37 °C for 5 min, and then HCl was added to stop the reaction. The acquired data were plotted on Lineweaver-Burk plots. The kinetics parameters (Km and kcat) were obtained with the reaction mixture containing 3.3 nm protease, 66, 100, or 130 μm MCA substrate or 6.6, 10, or 13 μm MOCAc/Dnp substrate in 10 mm Tris-HCl (pH 8.0). The reaction mixture was incubated at 37 °C for 3 min, and then HCl was added to stop the reaction. This reaction was performed in triplicate. Exceptionally, for a reaction involving pepsin, the pH of the reaction mixture was adjusted to 1.5 with HCl, and the reaction was stopped by adding 3 m Tris-HCl (pH 8.0).
TABLE 2.
Kinetic parameters, specificity constants and inhibition constants of oryctin and aprotinin for the hydrolysis of MCA and MOCAc/Dnp substrates by eukaryotic and bacterial proteases
The parameters and constants were obtained at 37 °C and pH 8.0, except for pepsin (at 37 °C and pH 1.5). NI, no detectable inhibition.
| Protease (origin) | Substrate | Without inhibitor |
Inhibitor | Ki | ||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | ||||
| μm | s−1 | s−1·mm−1 | nm | |||
| α-Chymotrypsin (bovine) | Suc-AAPF-MCAb | 110 ± 7.7 | 6.2 ± 0.54 | 58 ± 4.7 | Oryctin/Aprotinin | 0.39/1.3 |
| Leukocyte elastase (human) | Suc-AAA-MCAb | 82 ± 6.1 | 0.11 ± 0.007 | 1.4 ± 0.09 | Oryctin/Aprotinin | 12/1300 |
| Subtilisin Carlsberg (Butilus licheniformis) | Suc-LLVY-MCAb | 64 ± 5.8 | 2.5 ± 0.82 | 39 ± 4.5 | Oryctin/Aprotinin | 1.4/NI |
| Endopeptidase K (Tritirachium album) | Suc-AAPF-MCAb | 84 ± 9.5 | 2.6 ± 0.35 | 31 ± 2.8 | Oryctin/Aprotinin | 0.62/NI |
| Trypsin (porcine) | Boc-QAR-MCAb | 57 ± 0.29 | 5.7 ± 0.19 | 100 ± 5.1 | Oryctin | NI |
| V8 protease (Staphylococcus aureus) | Z-LLE-MCAb | 510 ± 52 | 0.43 ± 0.022 | 0.84 ± 0.044 | Oryctin | NI |
| Proline-specific endopeptidase (Flavobacterium meningosepticum) | Suc-GP-MCAb | 130 ± 6.9 | 0.041 ± 0.003 | 0.32 ± 0.015 | Oryctin | NI |
| Lysyl endopeptidase (Lysobacter enzymogenes) | Boc-VLK-MCAb | 15 ± 0.7 | 0.50 ± 0.049 | 32 ± 2.8 | Oryctin | NI |
| Endoproteinase Arg-C (Clostridium histolyticum) | Boc-QAR-MCAb | 190 ± 15 | 0.12 ± 0.009 | 0.63 ± 0.090 | Oryctin | NI |
| Endoproteinase Asp-N (Pseudomonas fragi) | MOCAc-YVADAP-K(Dnp)-NH2b | 12 ± 0.1 | 53 ± 2.0 | 4400 ± 230 | Oryctin | NI |
| Pepsin (porcine) | MOCAc-APAKFFRL-K(Dnp)-NH2b | 11 ± 0.4 | 370 ± 3.1 | 34000 ± 980 | Oryctin | NI |
| Papain (Papaya latex) | Z-FR-MCAb | 230 ± 16 | 0.22 ± 0.003 | 0.99 ± 0.028 | Oryctin | NI |
| Thermolysina (Bacillus thermoproteolyticus rokko) | MOCAc-KPLGL-A2pr(Dnp)-AR-NH2b | 20 ± 2.2 | 380 ± 21 | 19000 ± 1200 | Oryctin | NI |
a The final concentration of 1 μm of zinc ion was added to activate proteolysis.
b Z, benzyloxycarbonyl; Suc, succinyl; Boc, t-butyloxycarbonyl; MOCAc, 7-methoxycoumarin-4-yl; MCA, 4-methylcoumaryl-7-amide; K(Dnp), Nϵ-(2,4-dinitrophenyl)-L-lysine; and A2pr(Dnp), Nβ-(2,4-dinitrophenyl)-L-2,3-diaminopropionic acid.
Chemical Shift Perturbation
1H-15N HSQC spectra of [15N]oryctin only, 15N-labeled oryctin-α-chymotrypsin, and 15N-labeled oryctin-leukocyte elastase were measured to investigate the protease recognition sites of oryctin. The final concentrations of 15N-labeled oryctin and protease were 0.2 and 0.4 mm, respectively. The chemical shift change for each residue was determined quantitatively according to the function, δΔ = (δΔHN2 + 0.25 δΔ15N2)1/2 (10).
RESULTS
Peptide Isolation, cDNA Cloning, and Primary Structure Determination
Oryctin was found and isolated during the purification of antibacterial peptides from the hemolymph of the coconut rhinoceros beetle O. rhinoceros. About 1.0 μg of oryctin was obtained from 1.0 ml of the hemolymph by four steps of reverse-phase HPLC. The N-terminal amino acid sequence was determined using a protein sequencer, and the oryctin cDNA was cloned by three steps of PCR using fat body mRNA from immunized larvae. The deduced amino acid sequence from the nucleotide sequence indicated that oryctin contains an 85-amino acid precursor, and a 66-amino acid mature peptide is assumed to be produced by cleavage of the signal peptide (supplemental Fig. S1). There is no sequence similarity between oryctin and any other known peptides or proteins. The MALDI-TOF MS data indicated that oryctin is a monomeric peptide with three intramolecular disulfide bonds and without any other modifications.
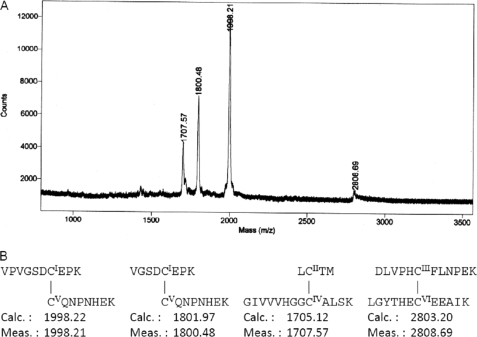
The molecular masses (m/z) of detected ions after the digestion with lysyl endopeptidase and endoproteinase Asp-N were 1707.57, 1998.21, and 2808.69, respectively, using MALDI-TOF MS (Fig. 1). These ions were assigned to the pairs of CysI-CysV, CysII-CysIV and CysIII-CysVI, respectively. An ion with an m/z of 1800.48 was assigned to fragments 3–10 and 42–49, which also indicated the disulfide bond CysI-CysV. Thus, we have concluded that three disulfide bonds are formed in oryctin: Cys7–Cys42, Cys12–Cys35, and Cys20–Cys56 (Fig. 1).
FIGURE 1.
A, MALDI-TOF MS of natural oryctin after the digestion with lysyl endopeptidase and endoprotease Asp-N. B, structure and molecular mass (m/z) of the disulfide-linked fragments. N-terminally truncated oryctin (starting at Val3) also existed in the sample. A site between Ser5 and Asp6 was not digested under this experimental condition.
Gene Expression Analysis
Oryctin was constitutively expressed in the fat bodies, hemocytes, midguts, and Malpighian tubules of the third instar larvae. Injection of E. coli into the larvae did not affect the expression of oryctin (supplemental Fig. S2).
NMR Structural Analysis
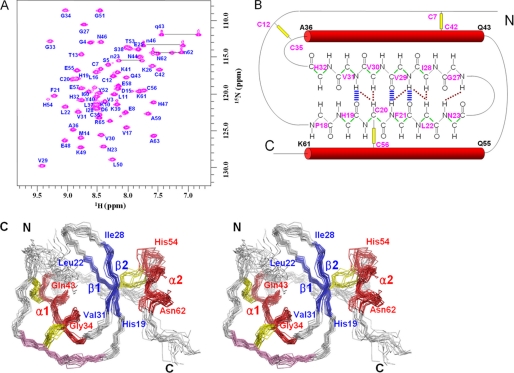
By using the conventional three-dimensional NMR data, 607 atoms of backbone and side chain 1H,13C,15N were assigned. The 1H-15N HSQC spectrum of oryctin is shown in Fig. 2A. 100% (57 residues, except proline and the N-terminal residue) of the backbone 1H,13C,15N atoms were assigned. The secondary structure regions predicted by both CSI and TALOS were restrained with the backbone dihedral angle (ϕ and ψ) restraints in the structure calculations (α, Ala36–Gln43 and Gln55–Lys61; β, His19–Leu22 and Val29–Val31). Three hydrogen bonds (Phe21 HN–Val29 O, Val29 HN–Phe21 O, and Val31-HN–His19 O) were assigned based on the low HgD exchange rates of amide protons and the oryctin structures at intermediate stages of structure calculations. A diagram showing secondary structures, hydrogen bonds, and interstrand NOEs is shown in Fig. 2B.
FIGURE 2.
A, 1H-15N HSQC spectrum of oryctin. B, the secondary structure diagram of oryctin. The hydrogen bonds (blue) and interstrand NOEs (brown) in the antiparallel β-sheet are shown. The three disulfide bonds and the two α-helices are shown as yellow lines and red tubes. The dihedral angle restraints used are shown in green. C, stereoview of the superposition of 20 structures of oryctin. Two α-helices (α1, Gly34–Gln43; α2, His54–Asn62) and two β strands (β1, His19–Leu22; β2, Ile28–Val31) are shown in red and blue, respectively. The disulfide bonds are shown in yellow. The local sequence similarity between oryctin and OMTKY3 is shown in purple.
Tertiary Structure
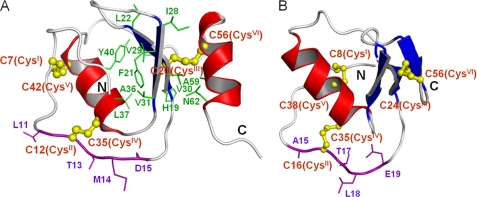
The three-dimensional structure of oryctin was calculated based on 552 NOE-derived distance restraints, three disulfide bond restraints (nine distance restraints), three hydrogen bond restraints (12 distance restraints), and 79 dihedral angle (38ϕ and 41ψ) restraints. A total of 200 structures were calculated, and the 20 structures with the lowest target functions were selected as an ensemble representing the solution structure of oryctin (Fig. 2C). The restraints used and the structural statistics for the final structure are summarized in Table 1. The oryctin molecule consists of a two-stranded antiparallel β-sheet (β1, His19–Leu22; β2, Ile28–Val31) and two α-helices (α1, Gly34–Gln43; α2, His54–Asn62). Three disulfide bonds were formed at Cys7–Cys42, Cys12–Cys35, and Cys20–Cys56. The pairwise root mean square deviations for backbone and all non-hydrogen atoms were 0.79 ± 0.17 Å and 1.43 ± 0.21 Å, respectively, among the 20 structures excluding five N- and five C-terminal residues. A representative structure with the lowest target function value is shown in Fig. 3A. The central β-sheet is rich in hydrophobic residues (His19, Cys20, Phe21, Leu22, Ile28, Val29, Val30, and Val31). The hydrophobic residues on the β-sheet and on the α-helices (α1, Ala36, Leu37, and Tyr40; α2, Cys56, Ala59, and Asn62) form a hydrophobic cluster and contribute to conformational stabilization. The Cys20–Cys56 disulfide bond is included in the hydrophobic cluster.
TABLE 1.
Statistics for the 20 NMR structures of oryctin
| Restraints | |
| Distance restraints derived from NOEs | |
| All | 552 |
| Intraresidual (|i − j| = 0) | 117 |
| Sequential (|i − j| = 1) | 226 |
| Medium range (2 ≤ |i − j| ≤ 5) | 116 |
| Long range (|i − j| > 6) | 103 |
| Hydrogen bond restraintsa | 12 |
| Disulfide bond restraintsb | 9 |
| Dihedral angle restraints | |
| ϕ | 38 |
| ψ | 41 |
| Ramachandran plot (no. of residues/%) | |
| Most favored | 815/76.9 |
| Additionally allowed | 228/21.5 |
| Generously allowed | 15/1.4 |
| Disallowed | 2/0.2 |
| r.m.s.d. of atomic coordinatesc | |
| Backbone atoms (Å)d,e | 0.79 ± 0.17 |
| Non-hydrogen atoms (Å)e | 1.43 ± 0.21 |
| Restraint violations | |
| r.m.s.d. distance restraint violations (Å) | 0.018 ± 0.002 |
| r.m.s.d. dihedral angle restraint violations | 0.45 ± 0.09 ° |
| Target function | |
| CYANA target function (Å2) | 1.3 ± 0.1 |
a Three hydrogen bonds each with four distance restraints.
b Three disulfide bonds each with three distance restraints.
c r.m.s.d., root mean square deviation.
d N, Cα, and C′ of each residue.
e For well defined regions (residues 6–61).
FIGURE 3.
A and B, the ribbon representation of oryctin (A) and of OMTKY3 (B). The residues forming disulfide bonds and hydrophobic cores are shown in yellow and green, respectively. The local sequence similarity between oryctin and OMTKY3 is shown in purple (11LCTMD15 (oryctin) versus 15ACTLE19 (OMTKY3)).
Structural Similarity to the Third Domain of Turkey Ovomucoid
A structural similarity search using the Dali server (9) showed that oryctin is structurally similar to the third domain of OMTKY3 (turkey ovomucoid inhibitor) (Fig. 3B; root mean square deviation = 3.2 and 3.0 Å for 41 and 39 Cα pairs, respectively; PDB codes 1PPF and 1YU6, respectively), which is a Kazal-type serine protease inhibitor. Despite the lack of amino acid sequence similarity between them, their patterns of three disulfide bridges are the same: CysI-CysV, CysII-CysIV and CysIII-CysVI (supplemental Fig. S3A). Oryctin, consisting of 66 residues, has three intramolecular disulfide bonds (Cys7–Cys42, Cys12–Cys35, and Cys20–Cys56), and OMTKY3, consisting of 51 residues, also has three intramolecular disulfide bonds (Cys8–Cys38, Cys16–Cys35, and Cys24–Cys56). In terms of secondary structure, oryctin has two α-helices and two β-strands, whereas OMTKY3 has one α-helix and three β-strands. Both oryctin and OMTKY3 possess a backbone fold similar to those of Kazal-type inhibitors: a central α-helix and an antiparallel β-sheet. The structural difference between oryctin and OMTKY3 is largest in the C-terminal regions, where oryctin has an amphipathic α-helix, whereas OMTKY3 has a short β-strand. The amino acid sequence of the reactive site loop (P4-P3-P2-P1-P1′) including CysII of OMTKY3 is AlaP4-CysP3(II)-ThrP2-LeuP1-GluP1′, whereas that of oryctin is LeuP4-CysP3(II)-ThrP2-MetP1-AspP1′ (shown in purple in Fig. 3, A and B), suggesting that oryctin also could function as a serine protease inhibitor.
Protease Inhibitory Activity
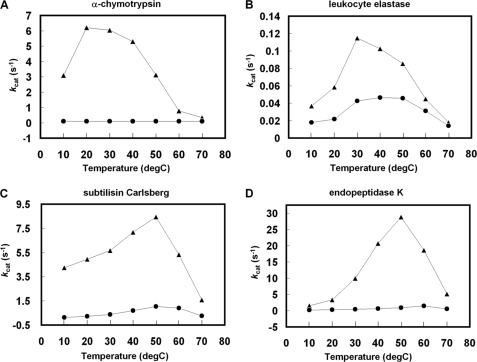
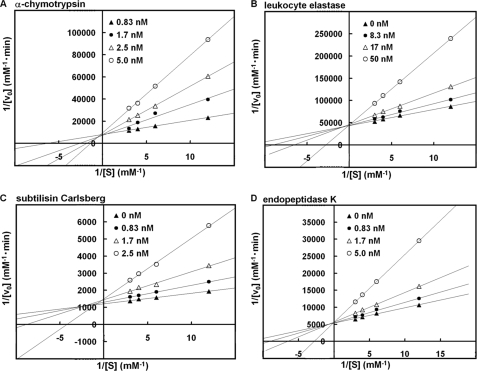
Supplemental Fig. S4 shows the pH-dependent inhibitory activity of oryctin against eukaryotic and bacterial proteases. Oryctin inhibited the eukaryotic serine proteases α-chymotrypsin and leukocyte elastase and the bacterial serine proteases subtilisin Carlsberg and endopeptidase K over a pH range of 7.2–9.2, indicating it acts as a serine protease inhibitor in the beetle hemolymph. Fig. 4 shows the temperature-dependent inhibitory activity of oryctin against the above-mentioned four proteases, demonstrating that oryctin has inhibitory activity in a temperature range of 10–70 °C against the four proteases in 10 mm Tris-HCl (pH 8.0). Based on Lineweaver-Burk plots, oryctin inhibited α-chymotrypsin, endopeptidase K, subtilisin Carlsberg, and leukocyte elastase in a competitive manner with Ki values of 3.9 × 10−10 m, 6.2 × 10−10 m, 1.4 × 10−9 m, and 1.2 × 10−8 m, respectively (Fig. 5).
FIGURE 4.
A–D, the temperature-dependent inhibition activity of oryctin toward α-chymotrypsin (A), leukocyte elastase (B), subtilisin Carlsberg (C), and endopeptidase K (D). The triangles and circles shown represent the turnover number (kcat) in the absence and presence of oryctin (3-fold molar concentration compared with each protease), respectively.
FIGURE 5.
A–D, Lineweaver-Burk plot analysis of the inhibitory activity of oryctin against α-chymotrypsin (A), leukocyte elastase (B), subtilisin Carlsberg (C), and endopeptidase K (D). The inhibition constant (Ki) values against α-chymotrypsin, leukocyte elastase, subtilisin Carlsberg, and endopeptidase K were 3.9 × 10−10 m, 1.2 × 10−8 m, 1.4 × 10−9 m, and 6.2 × 10−10 m, respectively.
Putative Serine Protease Binding Region(s)
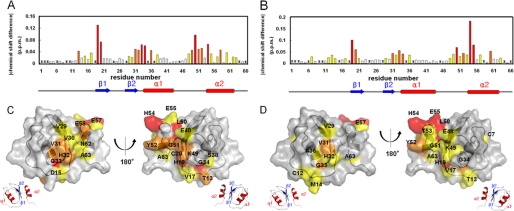
1H-15N HSQC spectra of 15N-labeled oryctin only and 15N-labeled oryctin-α-chymotrypsin showed similar distribution patterns of NMR signals. Chemical shift changes of oryctin by binding α-chymotrypsin and leukocyte elastase were evaluated quantitatively with the formula δΔ = (δΔHN2 + 0.25 δΔ15N2)1/2 (10) (Fig. 6, A and B). In the case of α-chymotrypsin binding (Fig. 6A), His19, Cys20, Gly33, Gly34, Leu50, and His54 exhibited large chemical shift changes (> 0.06 ppm); Thr13, Val31, His32, Ala36, Lys49, Gly51, Tyr52, and Glu58 exhibited moderate chemical shift changes (0.04–0.06 ppm); and Asp15, Val17, Val29, Val30, Ser38, Glu48, Glu55, Glu57, Ile60, Asn62, and Ala63 exhibited small chemical shift changes (0.02–0.04 ppm). The residues with degenerated or invisible signals are not colored in this figure (Val1, Val3, Asp6, Lys10, Ile28, Cys35, and Lys37), nor are prolines (positions 2, 9, 18, 24, 45, 64, and 66). In the case of leukocyte elastase binding (Fig. 6B), His19, Leu50, His54, and Glu55 exhibited large chemical shift changes (> 0.06 ppm); Cys20, Val31, Gly33, Gly34, Tyr52, and Cys60 exhibited moderate chemical shift changes (0.04–0.06 ppm); and Cys7, Cys12, Thr13, Met14, Val17, Phe21, Val29, His32, Ala36, Glu48, Lys49, Gly51, Thr53, Glu57, and Ala63 exhibited small chemical shift changes (0.02–0.04 ppm). Degenerated or invisible signals are not colored in this figure (Val1, Val3, Asp6, Lys10, Asp15, Ile28, Cys35, Lys37, and Glu58), nor are prolines (positions 2, 9, 18, 24, 45, 64, and 66). The amino acid residues whose chemical shifts were perturbed by the addition of α-chymotrypsin and/or leukocyte elastase are mostly common and located in three regions of the sequence (Fig. 6, A and B): (i) the putative reactive site near the N terminus (residues 13–20 and 12–21 in the cases of α-chymotrypsin and leukocyte elastase binding, respectively), (ii) the central part (residues 29–38 and 29–36), and (iii) the C-terminal part (residues 48–63 in both cases). When these residues are mapped on the oryctin structure, they form a circle around the molecule like a belt (Fig. 6, C and D). Because the similar surface areas of oryctin show chemical shift perturbations through the binding of oryctin to α-chymotrypsin or leukocyte elastase, oryctin should recognize these serine proteases in a very similar manner.
FIGURE 6.
A and B, the chemical shift changes of oryctin between the free state and the bound state to α-chymotrypsin (A) and leukocyte elastase (B). Asterisks represent unassigned residues. C and D, mapping of the chemical shift-perturbed residues on the oryctin molecule by binding to α-chymotrypsin (C) and leukocyte elastase (D). The bars in (A and B) and the residues in (C and D) are colored according to the extent of chemical shift change defined by the formula, δΔ = (δΔHN2 + 0.25 δΔ15N2)1/2 (10). White, 0–0.02 ppm; yellow, 0.02–0.04 ppm; orange, 0.04–0.06 ppm; and red, >0.06 ppm.
DISCUSSION
Proposed Inhibition Mode of Oryctin against α-Chymotrypsin and Leukocyte Elastase
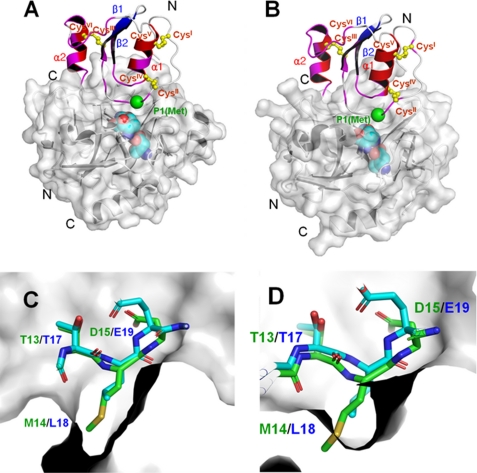
The crystal structures of OMTKY3 bound to α-chymotrypsin and leukocyte elastase have already been solved (PDB codes 1CHO and 1PPF, respectively) (11, 12) (supplemental Fig. S5). OMTKY3 binds α-chymotrypsin and leukocyte elastase through its reactive site loop. The only sequentially similar segment between OMTKY3 and oryctin (15ACTLE19 in OMTKY3 and 11LCTMD15 in oryctin) is located in the active site loop of OMTKY3 and the corresponding loop in oryctin. Because oryctin inhibits chymotrypsin- and subtilisin-like family proteases in a competitive manner (Fig. 5), it should exhibit an inhibition mechanism similar to that of OMTKY3 by using the sequentially similar segment within the putative reactive site loop. Based on the crystal structure of OMTKY3 bound to α-chymotrypsin and leukocyte elastase, we have constructed a binding model of oryctin to α-chymotrypsin and leukocyte elastase (Fig. 7, A and B). In this model, not only the reactive site loop region but also the C-terminal α-helix uniquely found in oryctin is involved in the protease binding, which can cause a change in the orientation of the α-helix. This is consistent with the fact that the chemical shifts of most residues in the C-terminal α-helix are perturbed (> 0.02 ppm) by binding to the target proteases, although the chemical shift of the putative P1 residue, Met14, is not perturbed.
FIGURE 7.
A and B, the binding model of oryctin to α-chymotrypsin (A) and leukocyte elastase (B), predicted based on the complex structures of the OMTKY3–α-chymotrypsin complex (PDB code 1CHO) and OMTKY3-leukocyte elastase complex (PDB code 1PPF) (supplemental Fig. S5). The chemical shift-perturbed residues (>0.02 ppm) are colored in magenta. The Cα atom of the reactive site residue (P1 residue) is shown as a green sphere. The disulfide bonds are shown in yellow. The active site triads of the α-chymotrypsin and leukocyte elastase are shown as cyan spheres. A unique additional α-helix at the C terminus (α2) of oryctin also would be involved in the recognition of α-chymotrypsin and leukocyte elastase and would contribute to the binding affinity. C and D, the crystal structure of the active site loop of OMTKY3 (green) and the model of the corresponding loop of oryctin (blue) in the S1 pockets of α-chymotrypsin (C) and leukocyte elastase (D). Met14, the putative residue at the P1 position in oryctin, can fit into the S1 pocket of α-chymotrypsin, but it is too big to be accommodated by the S1 pocket of leukocyte elastase.
α-Chymotrypsin and leukocyte elastase have large and small hydrophobic substrate-binding (S1) pockets and thus prefer large and small hydrophobic residues at the P1 position, respectively (Fig. 7, C and D). This explains why oryctin with Met at the P1 position inhibits α-chymotrypsin (Ki = 3.9 × 10−10 m) more tightly than leukocyte elastase (Ki = 1.2 × 10−8 m). The large S1 pocket of α-chymotrypsin will fit to the large hydrophobic side chain of Met14 at the P1 position, but the small S1 pocket of leukocyte elastase will be too small to accommodate the Met side chain. In contrast, OMTKY3 with Leu at the P1 position can bind both α-chymotrypsin and leukocyte elastase very tightly, with Kd values of 5.5 × 10−12 m and 2.4 × 10−11 m, respectively (11, 13), because the small side chain of Leu at the P1 position can be accommodated in both the S1 pockets of α-chymotrypsin and leukocyte elastase.
The Number of Residues between CysII and CysIII Provides Insight into the Functional Characterization of Kazal-type Inhibitors
Oryctin and the reported Kazal-type inhibitors that contain three disulfide bonds show no sequence similarity, indicating that oryctin and the other Kazal-type inhibitors have evolved convergently, whereas all of the Kazal-type inhibitors except oryctin have evolved divergently (supplemental Table S1 and supplemental Fig. S3B). In fact, the phylogenetic analysis of all the Kazal-type inhibitors in supplemental Table S1 except oryctin can be performed without error using the Phylogeny.fr server (supplemental Fig. S6) (14), but that with oryctin results in an error. Supplemental Table S1 compares the number of residues among six cysteine residues (CysI-Xna-CysII-Xnb-CysIII-Xnc-CysIV-Xnd-CysV-Xnd-CysVI). The putative reactive site loop (LeuP4-CysP3(II)-ThrP2-MetP1-AspP1′) of oryctin contains CysP3(II). MetP1, probably the most important for the inhibitory activity, is located between CysII and CysIII. The number of residues between CysII and CysIII (seven) is strictly conserved in all the members, including oryctin, with the exception of the second domain of rhodniin (15), which contains eight amino acid residues in that region. The number of residues between CysII and CysIII yields insight into the functional characterization of the Kazal-type inhibitors. Interestingly, there are more residues between CysI and CysII in vertebrate inhibitors than in invertebrate ones (supplemental Table S1). The lymphoepithelial Kazal-type-related inhibitor (LEKTI) isolated from human blood ultrafiltrate, which consists of 15 Kazal domains, contains longer sequence stretches of 12–13 residues between CysI and CysII (16). This may imply that the N-terminal region of the Kazal-type inhibitors in higher animals played some additional role in the course of evolution.
Possible Biological Roles of Oryctin
Proteases and protease inhibitors are involved in the metabolism, immunity, and metamorphosis of insects (17, 18). In particular, hemolymph proteases and protease inhibitors are involved in immune responses in insects, e.g. antimicrobial peptide induction, prophenoloxidase activation, hemolymph coagulation, and protection against virulence-related microbial proteases (17). In the present study, we have shown that oryctin inhibits subtilisin-like serine proteases such as subtilisin Carlsberg and endopeptidase K. This suggests that oryctin is also involved in protection against microbial proteases. In the silkworm Bombyx mori, a protease inhibitor against fungal protease was isolated and is considered to play a role in innate immunity (19). Although further investigation is required to reveal the in vivo function(s) of oryctin, the results obtained in this study indicate that oryctin is a novel and unique member of Kazal-type serine protease inhibitors. Despite the lack of sequence similarity, oryctin exhibits a similar linkage pattern of three disulfide bonds and an inhibition mechanism similar to that of the traditional Kazal-type inhibitors.
Supplementary Material
Acknowledgments
We thank Jun Yang and Masanori Yamamoto for technical assistance.
This work was supported by the Enhancement of Center of Excellence, Special Coordination Funds for Promoting Science and Technology, Science and Technology Agency, Japan; and by the National Project on Protein Structural and Functional Analyses, the Targeted Proteins Research Program, and grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The atomic coordinates and structure factors (code 2KSW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S6, and additional references.
- PDB
- Protein Data Bank
- HSQC
- heteronuclear single-quantum coherence
- MCA
- 4-methylcoumaryl-7-amide
- MOCAc/Dnp
- (7-methoxycoumarin-4-yl)-acetyl/2,4-dinitrophenyl
- RACE
- rapid amplification of cDNA end
- NOESY
- nuclear Overhauser enhancement spectroscopy.
REFERENCES
- 1.Ishibashi J., Saido-Sakanaka H., Yang J., Sagisaka A., Yamakawa M. (1999) Eur. J. Biochem. 266, 616–623 [DOI] [PubMed] [Google Scholar]
- 2.Yang J., Yamamoto M., Ishibashi J., Taniai K., Yamakawa M. (1998) Eur. J. Biochem. 255, 734–738 [DOI] [PubMed] [Google Scholar]
- 3.Tomie T., Ishibashi J., Furukawa S., Kobayashi S., Sawahata R., Asaoka A., Tagawa M., Yamakawa M. (2003) Biochem. Biophys. Res. Commun. 307, 261–266 [DOI] [PubMed] [Google Scholar]
- 4.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 5.Wishart D. S., Sykes B. D. (1994) J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 6.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 7.Jee J., Güntert P. (2003) J. Struct. Funct. Genomics 4, 179–189 [DOI] [PubMed] [Google Scholar]
- 8.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 9.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda M., Terasawa H., Sakakura M., Yamaguchi Y., Kajiwara M., Kawashima H., Miyasaka M., Shimada I. (2003) J. Biol. Chem. 278, 43550–43555 [DOI] [PubMed] [Google Scholar]
- 11.Fujinaga M., Sielecki A. R., Read R. J., Ardelt W., Laskowski M., Jr., James M. N. (1987) J. Mol. Biol. 195, 397–418 [DOI] [PubMed] [Google Scholar]
- 12.Bode W., Wei A. Z., Huber R., Meyer E., Travis J., Neumann S. (1986) EMBO J. 5, 2453–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker B. M., Murphy K. P. (1997) J. Mol. Biol. 268, 557–569 [DOI] [PubMed] [Google Scholar]
- 14.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Nucleic Acids Res. 36, W465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich T., Kröger B., Bialojan S., Lemaire H. G., Höffken H. W., Reuschenbach P., Otte M., Dodt J. (1993) J. Biol. Chem. 268, 16216–16222 [PubMed] [Google Scholar]
- 16.Lauber T., Schulz A., Schweimer K., Adermann K., Marx U. C. (2003) J. Mol. Biol. 328, 205–219 [DOI] [PubMed] [Google Scholar]
- 17.Kanost M. R. (1999) Dev. Comp. Immunol. 23, 291–301 [DOI] [PubMed] [Google Scholar]
- 18.Natori S., Shiraishi H., Hori S., Kobayashi A. (1999) Dev. Comp. Immunol. 23, 317–328 [DOI] [PubMed] [Google Scholar]
- 19.Eguchi M., Itoh M., Nishino K., Shibata H., Tanaka T., Kamei-Hayashi K., Hara S. (1994) J. Biochem. 115, 881–884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.