Abstract
In organs involved in metabolic homeostasis, transmembrane α and βklothos direct FGFR signaling to control of metabolic pathways. Coordinate expression of βklotho and FGFR4 is a property of mature hepatocytes. Genetic deletion of FGFR4 or βklotho in mice disrupts hepatic cholesterol/bile acid and lipid metabolism. The deletion of FGFR4 has no effect on the proliferative response of hepatocytes after liver injury. However, its absence results in accelerated progression of dimethynitrosamine-initiated hepatocellular carcinomas, indicating that FGFR4 suppresses hepatoma proliferation. The mechanism underlying the FGFR4-mediated hepatoma suppression has not been addressed. Here we show that βklotho expression is more consistently down-regulated in human and mouse hepatomas than FGFR4. Co-expression and activation by either endocrine FGF19 or cellular FGF1 of the FGFR4 kinase in a complex with βklotho restricts cell population growth through induction of apoptotic cell death in both hepatic and nonhepatic cells. The βklotho-FGFR4 partnership caused a depression of activated AKT and mammalian target of rapamycin while activating ERK1/2 that may underlie the pro-apoptotic effect. Our results show that βklotho not only interacts with heparan sulfate-FGFR4 to form a complex with high affinity for endocrine FGF19 but also impacts the quality of downstream signaling and biological end points activated by either FGF19 or canonical FGF1. Thus the same βklotho-heparan sulfate-FGFR4 partnership that mediates endocrine control of hepatic metabolism plays a role in cellular homeostasis and hepatoma suppression through negative control of cell population growth mediated by pro-apoptotic signaling.
Keywords: Apoptosis, Bile Acid, Growth Factors, Liver Metabolism, Tumor Suppressor
Introduction
Canonical short range paracrine FGF signaling controls cellular processes associated with tissue remodeling and homeostasis during development and in adult organs (1, 2). These include cell division, motility, death, and differentiation. Tissue matrix heparan sulfate (HS)3 plays a role in sequestration, stability, and release of canonical FGFs prior to reaching cell-associated HS-FGFR complexes (3). Dysfunction in canonical FGF signaling underlies a wide range of pathologies including birth defects in development, repair and response to injury in the adult, and cancer (4).
In contrast to local acting canonical FGFs, FGF19, 21, and 23 have endocrine roles in bile acid, lipid, glucose, and mineral metabolism (5–9). The endocrine FGF subfamily is controlled by metabolite-activated nuclear receptors at sites distal to target cells and tissues (10). Via the farnesoid X receptor, bile acids and other metabolites stimulate FGF19 (FGF15 in mice) in the ileum that regulates bile acid (7) and lipid metabolism in the liver (11). By activation of peroxisome proliferator-activated receptor α, fatty acids and other metabolites stimulate liver FGF21 that impacts metabolism in adipose tissue and liver (12, 13). Vitamin D and the vitamin D receptor regulate mineral balance between the bone and kidney that is mediated through FGF23 (14, 15). Unlike canonical FGFs, the endocrine FGFs have little or no affinity for HS (16). This permits their endocrine circulation and access through tissue to target cells involved in organism level metabolic homeostasis. The impact of FGF signaling on metabolic functions at the cellular level is directed by transmembrane αklotho or βklotho (KLB). αKlotho is involved in the action of FGF23 and the control of mineral metabolism in the kidney (17), whereas KLB is involved in the control of bile acid and lipid and glucose metabolism by FGF19 and FGF21 in liver and adipocytes (18). HS-FGFR complexes have very low affinity for endocrine FGFs in the absence of αklotho or KLB. A major role of αklotho and KLB is to facilitate high affinity binding and activation of FGFR signaling complexes by endocrine FGFs (18, 19).
Of the four FGFR tyrosine kinases, FGFR4 is dominant in mature hepatocytes (20). Genetic deletion and overexpression experiments in mice show that hepatocyte FGFR4 is involved in the control of hepatic bile acid (7, 21) and lipid metabolism (11), restoration of hepatolobular architecture, and the prevention of fibrosis upon liver insult (22). Hepatocytes are the sole site where KLB and FGFR4 are coordinately expressed at high levels (23, 24) (SIB-CleanEx and GeneSapiens databases), and KLB is essential for regulation of hepatic metabolic activities by FGFR4 and endocrine FGF19 (7, 21, 25).
Germ line deletion, knock-in of a common polymorphic variant (26), or targeted overexpression of a hyperactive mutant of FGFR4 in hepatocytes (27–29) indicates that FGFR4 plays no direct role expected of a canonical FGFR in driving cell proliferation in either development (30), liver regeneration (21), or tumors (26, 28, 29, 31). However, the genetic deletion of FGFR4 markedly accelerated hepatocarcinogenesis initiated by a single neonatal exposure to the hepatocyte-activated pro-carcinogen DEN (29). These genetic studies in mice suggested that rather than contribution to the proliferative response of normal hepatocytes to partial hepatectomy, liver injury, or hepatocarcinogenic initiators and promoters, FGFR4 exerts a negative influence on hepatocarcinogenesis concurrent with its KLB-dependent role in endocrine control of hepatic metabolism. The underlying mechanism of this and the role of KLB in cell population dynamics and tumor suppression has not been addressed. Here we report that expression of neither KLB nor FGFR4 alone affected cell population dynamics in KLB- and FGFR4-deficient hepatic or nonhepatic cells. Co-expression of KLB and FGFR4 induced an FGFR4 kinase-dependent restriction on cell population growth via apoptosis that was stimulated by either endocrine FGF19 or canonical FGF1. This confirms that in addition to its role in mediating endocrine control of hepatic metabolism, the KLB-FGFR4 partnership is a negative regulator of hepatocyte proliferation and hepatoma progression through pro-apoptotic mechanisms.
EXPERIMENTAL PROCEDURES
Tissues, Cells, Transfection, and Expression
Normal and FGFR4−/− mouse liver tissue, DEN-initiated hepatomas, and derived hepatoma cell lines were prepared as described (29). Full-length murine KLB (NM_031180) between EcoR1 and Not1 sites in mammalian expression vector pEF1a, a gift from Dr. M. Kuro-o (32), was used in both transient and stable transfections. Tet-on cell lines bearing inducible FGFR4-pcDNA4/TO were prepared by Lipofectamine-mediated transfection of Tet-off T-REx-293 and T-REx-HeLa cells (Invitrogen) as described (33). KLB-expressing AT3 cells (34) bearing inducible FGFR4 cDNA were established with reagents and protocols from Invitrogen. FGFR4 cDNA (AAM13666) (21) was inserted into pcDNA4/TO (Invitrogen) between HindIII and Xho1 sites. Clones stably expressing KLB were transfected and selected with 400 μg/ml neomycin. FGFR4-pcDNA4/TO was transfected into clones stably expressing KLB and selected with 300 μg/ml zeocin. The expression levels were assessed by RT-PCR analysis of mRNA and by immunoblot.
Cell Population Growth
Replicate cultures in each well of 24-well plates were established with 2 × 104 cells for 24 h followed by replacement every other day with medium containing 7% FBS, Tet, and FGF19 or FGF1 at the concentrations indicated in the text. FGF1 was prepared as described (35). Preparation of FGF19 is described in the supplemental materials. Heparin at 1 μg/ml was added with FGF19 or FGF1 in all of the experiments. The cell number was determined daily for 5 days by harvesting with 0.0075% Pronase and 0.02% EDTA followed by direct count using a Coulter Counter (Beckman Coulter, Brea, CA).
Apoptosis and Cell Death
Cell death in hepatoma cultures was assessed by the loss of membrane permeability indicated by import of APOPercentageTM dye after transient expression of KLB. Approximately 6 × 104 cells/well in 24-well plates were transiently transfected with KLB-pEF1a in serum-free DMEM for 6 h followed by incubation in DMEM containing 10% FBS overnight. The cells were then exposed to 100 ng/ml FGF19 for 6 h followed by new medium containing APOPercentageTM dye (Biocolor Ltd., Belfast, Ireland). After 30 min, the cells were washed three times with PBS to remove unbound dye. Imported dye was released by the addition of APOPercentageTM release reagent at 100 μl/well. Dye was measured at 550 nm using a VERSAmax microplate reader.
Apoptosis from membrane inversion to nuclear permeability was assessed by two-channel flow cytometry using anti-annexin V-FITC and propidium iodide. Approximately 3 × 105 cells were cultured for 2 days in each well of 12-well plates (Greiner Bio-one, Monroe, NC) and then subjected to treatment with Tet and FGF19 or FGF1 at the concentrations and times indicated. Unattached cells were combined with attached cells harvested by Pronase/EDTA treatment. Cell number was determined, and samples of 2 × 105 cells were collected by low speed centrifugation. The pellets were washed twice with 0.5 ml of reaction buffer (10 mm Hepes/NaOH, pH 7.4, 0.14 m NaCl, and 2.5 mm CaCl2) and resuspended in 0.1 ml of buffer. Ten μl of anti-annexin V-FITC solution (BD Biosciences, San Jose, CA) (0.5 mg/ml) was mixed with the cell suspension and incubated on ice for 15 min followed by the addition of 10 μl of a solution containing 50 μg/ml propidium iodide and further incubation for 5 min. The apoptotic fractions of the cell population displaying fluorescence for FITC, propidium iodide, and both were determined by sorting in a FACSCalibur flow cytometer (BD Biosciences) and analysis with CellQuest Pro software following the manufacturer's protocols.
The change in mitochondrial membrane potential Δψm was assessed by uptake and retention of the cationic voltage-sensitive lipophilic dye Rhodamine 123 (Sigma-Aldrich). The cells expressing the indicated combinations of KLB and FGFR4 after treatment with 1 μg/ml Tet and 1 μg/ml FGF19 for 36 h were labeled with 1 μm Rhodamine 123 at 37 °C for 30 min in the dark. After washing with PBS, the cells were harvested by Pronase/EDTA, which was then neutralized by 10% FBS. Approximately 2 × 105 cells were suspended in 1 ml of cold PBS and analyzed by the FACSCalibur flow cytometer at FI-2 channel. Cleavage of caspase 3 was assessed by immunoblot analysis. Approximately 6 × 105 cells expressing cKLB bearing inducible FGFR4 cDNA were treated with 1 μg/ml Tet and 1 μg/ml FGF19 for 36 h, lysed in 150 μl of SDS sample buffer. The cleavage of pro-caspase 3 into active fragments was then assessed by antibody for caspase 3 (Cell Signaling Technology, Danvers, MA).
Tests for Activity of Soluble KLB
cDNA coding for the soluble form of KLB missing the transmembrane and intracellular domain was constructed by replacing the sequence coding from 993KPLIFFGC to the C terminus of the full-length KLB with a coding sequence for a His6 tag and cloned into vectors pEF1a and tetracycline-on pcDNA4/TO. Clones of HEK293 cells expressing inducible soluble KLB-His6 were established by transfection and selection as described for FGFR4. The secretion of soluble KLB-His6 into the medium or diffusion through semi-permeable membranes upon the addition of 300 ng/ml Tet was assessed by His probe antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Approximately 4.5 × 104 cells were seeded onto a Transwell insert (Corning Inc., Lowell, MA) with a polycarbonate membrane of 3-μm pores sufficient to allow secreted KLB to pass through into the bottom well. Bottom wells contained 3 × 104 cells expressing FGFR4 alone or control cells co-expressing full-length KLB and FGFR4. Tet and FGF19 treatment was applied, and apoptotic cell death was monitored by flow cytometry after 2 days.
Quantitative PCR
Total RNA was extracted from tissues or cells after treatment with Tet, FGF19, and heparin using the Ultraspec RNA isolation kit (Biotecx Laboratories). The first strand cDNAs were transcribed from the RNA template by the SuperScript II reverse transcriptase (Invitrogen) and random primers according to the manufacturer's protocol. Quantitative PCR analyses were carried out based on the SYBR Green JumpStart Taq Ready Mix protocol (Sigma-Aldrich) by MX3000p (Strategene/Agilent Technologies, Santa Clara, CA). The relative abundances of mRNA were calculated by the comparative threshold cycle method and normalized to the β-actin level as an internal control.
Radiolabeled FGF Binding and Affinity Cross-linking
Human FGF19 cDNA was cloned from colon SW480 cell cDNA, fused with a coding sequence for a His6 tag at the N terminus, and cloned into the bacterial expression vector pET28. FGF19 was then expressed in Escherichia coli BL21 DE3, extracted and refolded from inclusion bodies, and purified on nickel-chelating Sepharose chromatography using an AKTApurifier (GE HealthCare). The purified material exhibited an ED50 of 30 ng/ml in a KLB/FGFR4 ERK1/2 activation assay. The methodological details of preparation and quality and quantitative analysis will be described elsewhere. FGF1 and purified His-tagged FGF19 were iodinated as described previously (36). Iodinated FGF19 was purified by nickel-chelating Sepharose chromatography and eluted by 0.25 m imidazole in PBS instead of heparin-agarose chromatography. The cells expressing cKLB, inducible FGFR4 (iFGFR4), or both cKLB and iFGFR4 after 300 ng/ml Tet induction for 24 h were incubated with 2 ng/ml labeled FGF1 or FGF19 for 30 min at 4 °C as indicated. Unbound or nonspecific binding was eliminated by wash with PBS. Cell surface bound radioactivity was then determined by γ-counter. After binding with 10 ng/ml labeled FGF, the covalent cross-linker disuccinimidyl suberate was added and incubated for 10 min at room temperature. The resultant cross-linked complexes were separated by SDS-PAGE and revealed by autoradiography as described (37).
Immunoblot Analysis
The cells were lysed in SDS or modified cold radioimmune precipitation assay buffer (20 mm Tris-HCl, pH 7.2), 50 mm NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 2 mm sodium orthovanadate, and 2 mm NaF). One tablet (Roche Applied Science) of protease inhibitors/10 ml and one tablet of phosphatase inhibitors/10 ml were added prior to lysis. The lysates were clarified by centrifugation, and supernatants reacted with the indicated antibodies and immunoprecipitates collected with protein A/G-agarose beads. Whole cell lysate supernatants or immunoprecipitates were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were probed with antibodies against FGFR4, KLB, AKT (AK mouse transforming, v-akt murine thymoma viral oncogene homolog 1, protein kinase B), ERK, and phospho-ERK1/2 (Santa Cruz Biotechnology), and phospho-AKTser473, phospho-mTOR2481, caspase 3, and phosphotyrosine (Cell Signaling Technology) as indicated. The bound primary antibodies were then probed with respective secondary antibodies labeled with horseradish peroxidase. Immunolabeled proteins were detected by using the ECL Plus detection kit (GE HealthCare).
Reproducibility and Statistical Analysis
Six independent cell lines bearing the described expression vectors were selected. The cells stably expressing KLB and bearing inducible FGFR4 cDNA were screened for graded response of expression of FGFR4 to Tet concentrations and negligible expression in the absence of Tet. One of three independent clonal cultures was randomly employed for specific experiments. Unless otherwise indicated, each experiment was reproduced at least three times independently in triplicate within each experiment. A representative of three or more experiments is shown in micrographs. Where indicated, the mean and S.D. was determined by Student's t test.
RESULTS
A KLB and FGFR4 Partnership Restricts Cell Population Growth via Apoptosis
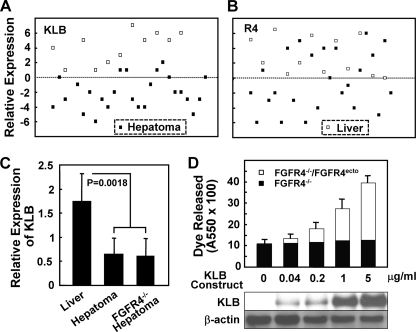
Comparative mRNA expression analyses between normal human liver tissues and hepatomas indicate that KLB is more consistently and significantly reduced in hepatomas than FGFR4 (Fig. 1, A and B) (SIB-CleanEx database: human body index, data set GSE7307; Expression Project for Oncology, data set GSE2109). The expression of KLB in 84% of hepatomas was lower than the lowest level observed in normal samples, whereas only 54% of hepatomas exhibited a similar depression in FGFR4. Analysis by real time PCR revealed that KLB expression was on average 33% of normal liver in both wild type and FGFR4-deficient DEN-induced mouse hepatomas (Fig. 1C), whereas no consistent differences in FGFR4 mRNA levels were observed (29).4 Transient expression of KLB in hepatoma cells from DEN-induced FGFR4-deficient mouse tumors in which FGFR4 had been restored by stable transfection caused a 4-fold increase of cells in the population exhibiting a loss of membrane potential suggestive of apoptotic cell death (Fig. 1D). Basal levels were similar in untransfected cell populations and those expressing either KLB or FGFR4 alone. Together these results suggest that restriction of hepatoma cell population growth by both KLB and FGFR4 together may underlie the observed acceleration of DEN-initiated hepatomas in the absence of FGFR4 (29). They suggest that hepatic KLB may play an essential role in the FGFR4-dependent delay in hepatoma progression and that delay may occur through restraints on cell population growth through induction of apoptosis.
FIGURE 1.
Reduced KLB expression in hepatomas and FGFR4-dependent effect of KLB expression in hepatoma cells. A and B, KLB and FGFR4 expression in human liver and hepatomas. Data from Affymetrix mRNA expression analyses of KLB and FGFR4 in human liver and clinically annotated hepatomas was extracted from the SIB-CleanEx Database and plotted numerically. Closed squares, human hepatoma samples; open squares, normal liver samples. C, reduced KLB expression in mouse hepatomas. The samples were taken from DEN-initiated hepatomas from normal and FGFR4-deficient (FGFR4−/−) mice as described (29). The relative expression of KLB mRNA was determined by quantitative PCR. The indicated data are the means ± S.D. from triplicate analyses of 10 normal liver and 12 hepatoma samples. D, FGFR4-dependent increase in cell death induced by expression of KLB in mouse hepatoma cells. KLB was transiently expressed by the addition of the indicated amounts of the KLB-pEF1a construct to hepatoma cells from FGFR4-deficient mice (FGFR4−/−) or cells (FGFR4−/−/FGFR4ecto) in which FGFR4 had been restored by stable transfection (29). Cell death was assessed by the uptake and release of APOPercentageTM dye. The indicated data are the means ± S.D. of three independent experiments with replicate analyses. FGFR4 expression levels (29) and the indicated KLB expression levels were assessed by immunoblotting of whole cell lysates using β-actin as the loading control.
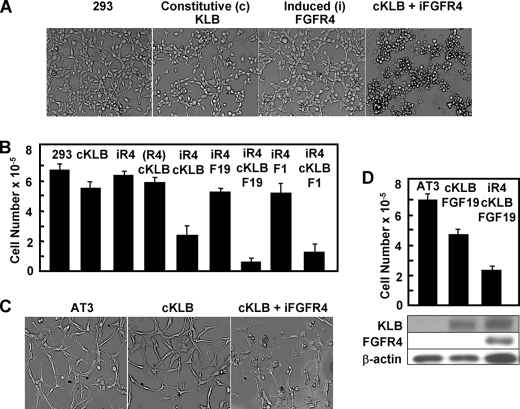
To test the general nature of the negative impact of the KLB-FGFR4 combination on cell population dynamics, we employed the generic nonhepatic nontumorigenic cultured human embryonic kidney cell transfection host (HEK293) whose parent tissue has no documented history of KLB or FGFR4 expression in regard to physiological function. Attempts to select for cell populations stably expressing both KLB and FGFR4 concurrently was severely hampered because of the long time required and the overall low frequency of emergence of co-expressing populations. Concluding that this might reflect restrictions of the KLB-FGFR4 partnership on cell proliferation and gradual selection of highly proliferative clones that become resistant to growth inhibitory signals, we employed an inducible expression system (33). An overnight induction of iFGFR4 in T-REx 293 constitutively expressing KLB (cKLB) caused a dramatic morphological change in the cell population, detachment of anchored cells, and reduction in cell number (Fig. 2, A and B). The effect of expression of KLB or FGFR4 alone was minimal. The FGFR4-KLB partnership resulted in reduction of cell populations to 37% of that of untreated parental 293 cells. FGF19 caused a further reduction to 10% of controls. The inhibitory activity in the absence of added FGF may reflect a level of FGFR4 expression sufficient to overcome membrane context and HS-imposed negative restrictions on transactivation (derepression) of kinase activity within unliganded dimeric FGFR complexes (38). Alternatively, it may reflect the presence of serum FGF19 that can be as high as 600 pg/ml (39) or endogenous cellular FGF1. Similar to FGF19, canonical FGF1, even though its high affinity interaction with HS-FGFR4 is independent on the presence of KLB, also caused a KLB-dependent decrease in cell number to 22% of that in parental cultures (Fig. 2B). This suggests that KLB directs signaling of the HS-FGFR4 complex in addition to simply conferring high affinity for FGF19.
FIGURE 2.
Co-expression of KLB and FGFR4 also restricts nonhepatic cell population growth. A, cell morphology. Untransfected T-Rex-293 cells (293), cells stably transfected with KLB (cKLB), cells transfected with inducible FGFR4 cDNA (iFGFR4), and cKLB cells bearing inducible FGFR4 cDNA were examined by light microscopy after exposure overnight to 1 μg/ml Tet and 300 ng/ml of FGF19. B, population growth rates. The data are cell numbers after 5 days of culture. FGF19 or FGF1 was present at 300 ng/ml where indicated. The data are the means ± S.D. from three independent experiments. (R4), cells bearing inducible FGFR4 cDNA in Tet-free medium. C and D, co-expression of KLB and FGFR4 inhibits malignant prostate tumor cell growth. The induction of FGFR4 and effect on population dynamics of AT3 cells was determined as described for 293 cells. Expression of KLB and FGFR4 were measured by immunoblotting of whole cell lysates. β-Actin was used as the loading control.
Induced expression of FGFR4 in a highly malignant prostate tumor cell line (AT3) constitutively expressing KLB also similarly inhibited cell population growth (Fig. 2, C and D). Unlike 293 cells, FGF19 caused a notable decrease in KLB-expressing cells in the absence of FGFR4. This may be due to the high level of FGFR1 in the AT3 tumor cells (34). Separate experiments indicate that KLB similarly causes inhibition of cell population growth via apoptosis in partnership with FGFR1. The induction of FGFR4 in KLB-expressing cells also inhibited the anchorage-independent growth of the 293 cells and cervical cancer-derived HeLa cells (supplemental Fig. S1).
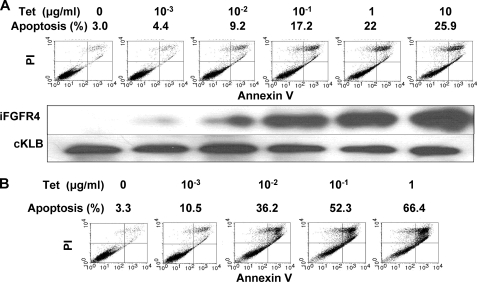
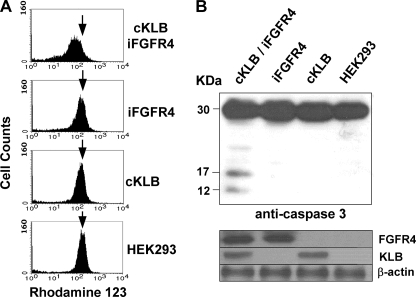
The morphological changes that occurred concurrent with the inhibition of cell population growth in the presence of both KLB and FGFR4 suggested a rate of cell death sufficient to offset the intrinsically high rate of cell division characteristic of cell lines that have been highly selected for proliferation in culture. To determine whether the cell population loss in 293 cells occurred by stepwise progression through the apoptotic cell death pathway, we tracked early perturbation of the cell membrane indicated by exposure of phospholipids to a late loss of nuclear envelope permeability. The results indicated that apoptosis in cell populations constitutively expressing KLB was proportional to expression of FGFR4 in respect to both level and time of induction (Fig. 3). Increasing levels of FGFR4 induced for 24 h resulted in a 9-fold increase in the apoptotic fraction at the plateau of induction (Fig. 3A). Induction of FGFR4 alone at the Tet levels shown in Fig. 3A resulted in only 2.5, 3.0, 3.8, 4.6, 5.8, and 8.7% apoptotic cells, respectively (supplemental Fig. S2). Extending the period of induction of FGFR4 at 1 μg/ml Tet to 3 days resulted in 66% of the population in apoptosis, an increase in apoptotic cells 20-fold that of cells expressing KLB alone (Fig. 3B). Six days of FGFR4 induction at 1 μg/ml Tet in cells expressing KLB resulted in the death of nearly 100% of cells (Fig. 2B). Basal levels of apoptosis in untransfected 293 populations and those expressing KLB alone were 2–5% and independent of Tet at the dose and times indicated in Fig. 3. The loss of mitochondrial membrane permeability indicated by the loss of retention of Rhodamine 123 (40) (Fig. 4A) and the cleavage of 30-kDa proenzyme caspase-3 into active 12- and 17-kDa fragments, a marker of the execution phase of apoptosis (Fig. 4B), were observed only in cell populations expressing both KLB and FGFR4. This further confirmed that the KLB and FGF4 partnership restricts cell population growth rates via the apoptotic pathway from initiation through loss of membrane permeability.
FIGURE 3.
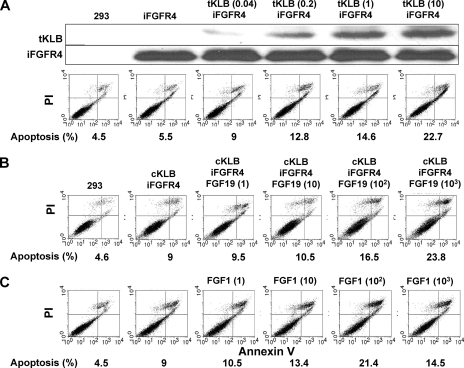
KLB-dependent apoptosis is proportional to FGFR4 expression. A, apoptotic cell death after induction of FGFR4 for 24 h in 293 cells. FGFR4 was induced in KLB-expressing cells by the indicated concentrations of Tet. Apoptosis was monitored by flow cytometric analysis of membrane phospholipid exposure and propidium iodide uptake. Expression of FGFR4 and KLB were assessed by immunoblotting. B, rates of apoptosis in KLB-expressing 293 cells after induction of FGFR4 for 3 days.
FIGURE 4.
Loss of mitochondrial transmembrane potential and induction of caspase cleavage by co-expression of KLB and FGFR4. A, mitochondrial membrane potential. Uptake and retention of the cationic voltage-sensitive lipophilic dye Rhodamine 123 in 293 cells expressing the indicated combinations of KLB and FGFR4 and treated with Tet and FGF19 were assessed by flow cytometry. B, cleavage of caspase 3. The lysates from 293 cells co-expressing KLB and FGFR4 after treatment with Tet and FGF19 were separated by SDS-PAGE, and the cleavage of caspase 3 into active fragments was then assessed by immunoblot analysis. The levels of FGFR4 and KLB were shown by immunoblotting of whole cell lysates as in Fig. 2.
FGFR4-mediated Apoptotic Cell Death Is Dependent on Full-length KLB and Stimulated by Either FGF19 or FGF1
To further confirm that FGFR4-dependent increases in the rates of apoptosis were dependent on KLB, increasing amounts of KLB were expressed by transient transfection of cells expressing a constant amount of FGFR4 induced at 10 ng/ml Tet. The rates of apoptosis in the co-expressing 293 cell populations increased with increasing levels of KLB expression (Fig. 5A). This recapitulates the effect of transiently expressed KLB in FGFR4−/−/FGFR4ecto hepatoma cells (Fig. 1D).
FIGURE 5.
FGFR4-dependent apoptotic cell death increases with increasing KLB expression and is stimulated by either FGF19 or FGF1. A, transient expression of KLB in FGFR4-expressing cells. 293 cells harboring inducible FGFR4 cDNA were transiently transfected (tKLB) overnight by the amount of KLB-pEF1a in μg/ml indicated in parentheses. FGFR4 was then induced by the addition of 10 ng/ml of Tet for 24 h followed by analysis of apoptosis and expression. B and C, stimulation of KLB-FGFR4-induced apoptosis by either FGF19 or FGF1. FGF19 (B) or FGF1 (C) was added at the concentrations in ng/ml indicated in parentheses to cells co-expressing constitutive KLB and FGFR4 induced by 10 ng/ml Tet.
Substitution of the vector for full-length KLB with one coding for a truncated KLB missing the transmembrane and intracellular domains failed to similarly increase apoptosis and cell death when it was stably co-expressed with induced FGFR4 (“Experimental Procedures”). Co-culture between cells expressing the induced soluble ectodomain of KLB and cells expressing FGFR4 separated by semi-permeable membranes (3 μm) also failed to induce apoptosis-associated changes in morphology and inhibition of cell population growth comparable with cells co-expressing full-length KLB and inducible FGFR4 (Fig. 2). We conclude that intact membrane-anchored KLB containing both extracellular and intracellular domains is required to support the pro-apoptotic KLB-FGFR partnership.
Apoptosis initiated by the FGFR4 and KLB partnership was responsive to FGF19 in a dose-dependent mode over a range of 1–1000 ng/ml (Fig. 5B). FGF1 similarly enhanced the rates of KLB-dependent apoptosis in the same concentration range as FGF19 (Fig. 5C). This confirms that KLB directs signaling of the activated FGFR4 kinase complex that is in addition to the role of KLB in facilitating high affinity binding and activation by FGF19.
Direct Participation of KLB in the FGFR4 Signaling Complex and Dependence of KLB-induced Apoptosis on the FGFR Tyrosine Kinase
Protein bands at 130 and 90 kDa from immunoprecipitates of 293 cells co-expressing mouse KLB and human FGFR4 harvested with anti-FGFR4 antibody that specifically targets the C-terminal of FGFR4 were identified as mouse KLB (supplemental Fig. S3A) and human FGFR4 (supplemental Fig. S3B) by nano-flow HPLC coupled with nano-electrospray tandem MS analysis (33). No KLB was detected in the pull-down with control mouse IgG or beads alone (not shown). This was direct evidence of the presence of KLB-FGFR4 complexes in co-expressing cells under conditions that induce apoptosis.
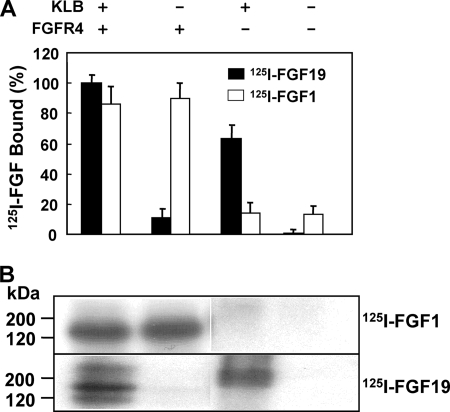
To further understand how KLB forms a functional partner with FGFR4, we performed ligand binding analysis on the surface of 293 cells expressing KLB, FGFR4, and their combination. As expected (20), FGF1 binds to cells expressing FGFR4 independent of KLB (Fig. 6A). In contrast, the binding of FGF19 to cells was largely KLB-dependent. The cells expressing KLB alone bound radiolabeled FGF19 at 60% that of cells expressing the KLB and FGFR4 combination. Covalent affinity cross-linking analysis with radiolabeled FGF1 indicated a single band consistent with a complex of FGF1 and FGFR4 without direct interaction with KLB (Fig. 6B). The same analysis with radiolabeled FGF19 revealed additional radiolabeled bands indicative of interactions with KLB. Bands were present that correlated with the estimated apparent molecular masses of cross-linked complexes of KLB-FGF19 and FGFR4-KLB-FGF19 in addition to FGF19-FGFR4. The results indicate that KLB bound FGF19 with sufficient affinity to be covalently cross-linked in absence of FGFR4 and also concurrently to FGFR4. A complete biochemical and kinetic analysis of the interaction of FGF1 and FGF19 to KLB-HS-FGFR complexes, competition binding, and the resultant covalent affinity cross-linked species will be the subject of a subsequent report.
FIGURE 6.
Binding and complex formation among KLB, FGFR4, FGF19, and FGF1. A, differential binding of 125I-labeled FGF1 and FGF19. 293 cells expressing cKLB, iFGFR4, or their combination after Tet induction for 24 h were incubated with labeled FGF1 (white bar) or FGF19 (black bar). Cell surface bound radioactivity was determined by γ-counter. The data are the means ± S.D. of three independent experiments. B, covalent affinity cross-linked complexes. After binding with 10 ng/ml labeled FGF, the covalent affinity cross-linker disuccinimidyl suberate was used to cross-link the formed complex as described (37).
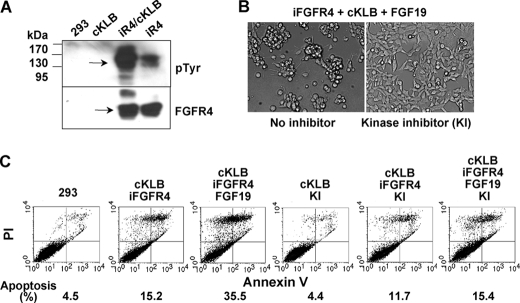
KLB enhanced FGF19-stimulated tyrosine phosphorylation of FGFR4 (Fig. 7A). An FGFR tyrosine kinase inhibitor (Calbiochem 341608) rescued cells from the KLB-FGF19-FGFR4-induced restrictions on the population expansion of 293 cells (Fig. 7B) and apoptosis (Fig. 7C). Separate experiments indicated that a depression of tyrosine phosphorylation of FGFR4 was coincident with the decreased rates of cell death in presence of the inhibitor. Taken together, these experiments indicate that KLB participates directly in the HS-FGFR signaling complex to facilitate activating autophosphorylation of the FGFR4 tyrosine kinase. The apoptosis and cell death-promoting effect of the KLB-FGFR4 partnership is proportional to FGFR4 kinase activity.
FIGURE 7.
Activation and dependence of apoptotic cell death on activity of the FGFR4 tyrosine kinase. A, activation of the FGFR4 kinase by KLB. 293 cells co-expressing constitutive KLB and FGFR4 induced by 300 ng/ml Tet overnight were maintained in serum-free medium for 6 h (33) and then exposed to 300 ng/ml of FGF19 for 10 min followed by immunoblot analysis of lysates with anti-phosphotyrosine (pTyr) and anti-FGFR4 antibodies. B and C, rescue of KLB-FGFR4-induced apoptosis and cell death by inhibition of tyrosine kinase activity. The FGFR kinase inhibitor 1-(2-amino-6-(3,5-dimethoxyphenyl) pyrido [2,3-d] pyrimidin-7-yl)-3-tert-butyl urea (341608; Calbiochem, San Diego, CA) at 1 μm was added during the 24-h induction of FGFR4 by 300 ng/ml Tet in cells expressing KLB followed by analysis of cell morphology (B) and apoptosis and cell death (C). FGF19 was present at 1 μg/ml.
Suppression of Activated AKT and mTOR by the KLB-FGFR4 Partnership
Activation of the Ras/MAPK pathway indicated by phosphorylation of ERK1/2 and the PI3K/AKT pathway indicated by phosphorylation of AKT is a response associated with canonical FGF signaling (2). Both are generally associated with a positive influence on cell population growth rates. Activation of AKT and mTOR that can be but is not solely a downstream target of AKT activation (41–43) elicits pro-survival and anti-apoptotic activities (44, 45).
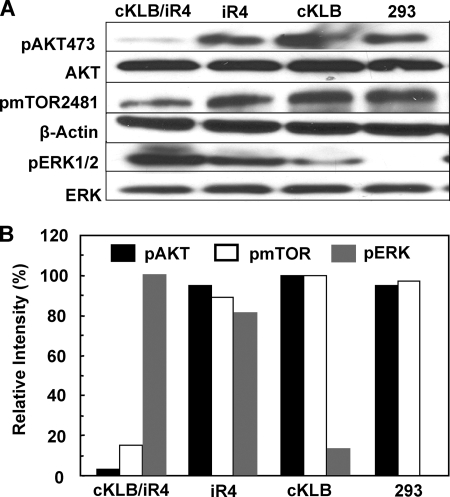
Activation of AKT occurs by phosphorylation of Thr308 and Ser473 (46). Activation of mTOR is associated with phosphorylation of Ser1261, Thr2446, Ser2448, and Ser2481 (47). Using phosphorylation site-specific antibodies, we compared the impact of the FGF19-stimulated KLB-FGFR partnership on activated AKT and mTOR to ERK1/2 in 293 cells. Under conditions that induced ∼20% apoptotic cells in the population, the KLB-FGFR4 partnership reduced pAKT473 to less than 5% of that in cells expressing only FGFR4, KLB, or parental untransfected cells (Fig. 8). The KLB-FGFR4 partnership reduced pmTOR2481 to ∼15% of that of control cells. Inhibition of PI3K activity further increased the rate of apoptosis by ∼40%, but the percentage of increase above controls caused by the inhibitor appeared independent on the FGF19-KLB-FGFR4 combination (supplemental Fig. S4). This indicates that PI3K has effects on apoptotic rate that are independent of the KLB-FGFR4 partnership and suggests the corollary. The partnership may have specific effects on AKT and mTOR that are not strictly mediated by PI3K. The depression of AKT and mTOR was in marked contrast to pERK1/2 that was elevated under the same conditions. Notably the depression in AKT and mTOR was completely dependent on the KLB-FGFR4 partnership compared with the elevation of pERK1/2, which was significant in the presence of KLB and iFGFR4 alone.
FIGURE 8.
Selective depression of AKT and mTOR activity by the KLB-FGFR4 partnership. A, 293 cells were treated with 300 ng/ml Tet overnight followed by addition of 1 μg/ml FGF19 for 6 h and analysis of lysates by immunoblot with the indicated antibodies. B, quantification of pAKT, pmTOR, and pERK. The relative levels of pAKT to total AKT, pmTOR to β-actin, and pERK to total ERK were calculated from a densitometric scan of band intensities in A. 100% was the most intense band from each protein among samples.
DISCUSSION
Although genetic deletion of FGFR4 and the targeted overexpression of hyperactive FGFR4 in mice indicate a role of FGFR4 in hepatic metabolism (7, 21, 25), they fail to indicate a direct role for FGFR4 expected for canonical FGFRs in driving cell division. This includes development (30), liver regeneration (21), and tumors (26, 28, 29, 31). Genetic ablation of FGFR4 accelerates progression of carcinogen-initiated hepatomas, suggesting a negative controlling role for FGFR4 in cell proliferation (29). Here we investigated for the first time the mechanism of this tumor suppressive effect of FGFR4 in partnership with KLB that has been implicated in control of hepatic metabolic pathways. Consistent with a negative influence on proliferation of hepatic cell populations and hepatoma progression, we showed that KLB is more consistently depressed than FGFR4 in both human and mouse hepatomas. Restoration of KLB by transient expression that does not select for expressing cells increased the rate of cell death within mouse hepatoma cell populations but only in cells expressing FGFR4. We observed no stimulatory effects of FGFR4 when it was introduced into cultured hepatic cells and a variety of nonhepatic cells. This is consistent with reports that relative to other FGFR isotypes, FGFR4 introduced into a variety of nonhepatic cells exhibits little or no activity in response to canonical FGF1 and FGF2 in respect to autophosphorylation, tyrosine phosphorylation of downstream signal mediators, and stimulation of the cell cycle and mitogenesis (48–51). Similarly we observed no positive effects on cell population dynamics when FGFR4 and KLB were co-expressed. Transient expression of FGFR4 or KLB in the presence of the other revealed that the partnership has a negative influence on cell population growth.
Therefore to study the effects of the transmembrane KLB-FGFR4 partnership in negative control of cell population growth, we necessarily employed an inducible gene expression system. This prevented selection for proliferative cell populations that bypass KLB-FGFR4-induced growth restrictive pathways or possibly become dependent on one or both for survival and growth in culture. This revealed that restriction of cell population growth rates by the co-expression of the activated KLB-FGFR4 partnership is a general phenomena extending across both hepatic and nonhepatic cell lines of both nonmalignant and highly tumorigenic character. We show here that the restriction on cell population growth caused by the KLB-FGFR4 partnership occurs by induction of apoptosis resulting in cell death. Neither KLB nor FGFR4 alone had the effect. Thus our results are consistent with the genetic experiments indicating a suppressive role for FGFR4 in hepatoma progression. They are inconsistent with reports suggesting that FGFR4 is a direct hepatocyte proliferation and hepatoma promoter through mediation of effects of chronic or pharmacological levels of FGF19 administered in vivo (52, 53). Conceivably, chronic stimulation of the hepatic KLB-FGFR4 partnership results in damaging cholesterol/bile acid and lipid metabolism and hepatocyte death that triggers foci of regenerating hepatocytes. Genetic screens otherwise indicate that both endogenous FGFR4 (54) and FGF19 (55) genes have properties of tumor suppressors. Our results suggest that similar to its essential role in partnership with FGFR4 in regulating hepatic metabolic pathways (7, 21, 25), KLB is essential in FGFR4-dependent negative control of hepatic cell proliferation and hepatocarcinogenesis (29) and therefore qualifies as a hepatoma suppressor.
αKlotho, the homolog of KLB, which partners with FGFR1 to mediate endocrine FGF23 control of mineral metabolism in kidney cells, also exhibits context-dependent properties of a tumor suppressor (56, 57). Inconsistent with this, it has been reported that soluble αklotho together with FGF23 elicits DNA synthesis in both renal and nonrenal cells in culture (58), although it was not demonstrated that FGFR mediates the effect. Because soluble αklotho confers high affinity on FGFR1 for FGF23 (17, 59), it may promote activation of the canonical growth-promoting signaling profile expected of FGFR1 without redirection of downstream signaling caused by membrane αklotho. Coincidental with expected effects on mineral metabolism, chronic systemic expression of FGF23 in mice failed to elicit DNA synthesis in kidney where the αklotho-FGFR1 partnership elicits its metabolic effects (58). The relative role of soluble and membrane-bound αklotho in directing metabolic and cell population growth end points both in vitro and in vivo remains to be established.
In contrast to αklotho, full-length membrane-anchored KLB with FGFR4 is required to form an FGF-independent complex competent for high affinity binding and activation by endocrine FGF19 (24). In line with this, we found that full-length KLB with an intact transmembrane domain is required for apoptosis and resultant cell population growth restrictions elicited by the KLB-FGFR4 partnership. In addition, we show that FGF1 is equally as effective as FGF19 in directing the KLB-dependent apoptotic effect of FGFR4. This is despite the fact that KLB neither binds FGF1 nor is required for high affinity binding of FGF1 to the HS-FGFR4 complex. This suggests that KLB forms a composite signaling complex with HS-FGFR4 that affects the quality of downstream signaling of the FGFR4 kinase in addition to facilitating high affinity binding of FGF19. In contrast to endocrine FGF19, the access of tissue matrix FGF1 to the HS-FGFR4 complex and the specificity of FGF1 for hepatic HS-FGFR4 is set by hepatocyte-specific HS (3, 20). The release of FGF1 from tissue matrix depots that occur in response to injury may serve to maintain hepatocyte metabolic functions. In addition FGF1 may contribute to feedback control via the resident hepatocyte KLB-HS-FGFR4 complex on the extent of the normal hepatocyte proliferative response or chronic proliferative stimuli that can lead to hepatocarcinogenesis.
Our results show that, similar to HS, KLB is a direct participant in oligomeric FGFR kinase signaling complexes. Similar to HS, KLB participates in FGFR signaling by interaction with both FGF and FGFR. Tissue-specific motifs within HS set affinity and specificity for locally acting paracrine FGFs that have diverse roles in cellular homeostasis (3, 20, 60–64). In addition, the interaction of HS with self-associated unliganded transmembrane FGFR kinase oligomers prevents FGF-independent activation of HS-FGFR complexes (3, 20, 38, 64). The requirement of HS in KLB-directed FGFR signaling (65) indicates that KLB interacts with and modifies the HS-FGFR signaling complex rather than replacing HS and its role in the complex. Similar to the role of specific oligosaccharide motifs of HS in canonical FGFR signaling (3, 20), αklotho and KLB interact directly with FGFR to enable high affinity binding and activation of the FGFR complex by specific endocrine FGFs. Our results indicate that similar to HS, the interaction of membrane KLB with the HS-FGFR complex impacts the structure of the signaling complex sufficiently to alter the qualitative nature of downstream signaling from it. This has not yet been demonstrated for HS.
The current models of control and activation of the oligomeric FGF-HS-FGFR signaling complex are in contention with respect to the stoichiometry and order of assembly of subunits as well as the mode of activation (3, 38, 64, 66, 67). Collectively static structural studies and dynamic structure-function and biochemical analyses point to a symmetrical two-FGFR and two-HS chain complex where conformational restriction imposed by unliganded membrane-anchored self-associated FGFR dimers, HS and divalent cations limit a transphosphorylating kinase-substrate relationship between intracellular domains (3, 38, 64). Conformational restrictions are overcome by changes caused by the entry of two FGFs either free or bound to an oligosaccharide that are then transmitted to the intracellular domain of the complex (68). The requirement for full-length membrane-anchored KLB for high affinity association of KLB with HS-FGFR4, activation, and the qualitative nature of FGFR4 signaling is consistent with the notion that KLB is an integral component of the inactive unliganded membrane-anchored HS-FGFR4 signaling complex that is poised for activation by endocrine FGF19 or tissue matrix FGF1.
In this study, we showed that the KLB-directed proliferation and tumor suppressive effects are dependent on the FGFR4 kinase activity, and this correlates with depression of the AKT and mTOR pathways. AKT and mTOR pathways have major impact on net expansion of cell populations through their well characterized pro-survival and anti-apoptotic effects. The observed depression of activated AKT and mTOR occurs concurrently with activation of ERK1/2. Activation of ERK1/2 is the most commonly observed indicator of canonical FGF-stimulated HS-FGFR (2) as well as klotho-dependent endocrine FGF signaling (19, 32, 69). It is unclear whether the concurrent activation of ERK1/2 cooperates with the depression of pAKT and pmTOR to elicit the pro-apoptotic effects of the KLB-FGFR4 partnership. Although it is most commonly associated with growth stimulation, activation of ERK1/2 has been implicated in growth suppression, cell death, and other biological functions (70, 71). The specific inhibition of anti-apoptotic and cell growth-stimulating AKT and mTOR without effect on activation of ERK1/2 is consistent with the idea that membrane KLB specifically modifies the overall quality of signals elicited from the HS-FGFR complex at the intracellular membrane interface. Activation of the three pathways is thought to diverge from the same single FGFR-associated adapter and signal mediator FRS2α (2). Recently, mTOR has been shown to form a complex with activated FGFR1-FRS2α apparently independently and upstream of AKT (43).
In conclusion, our results show that the same KLB-FGFR4 partnership that mediates endocrine control of hepatic metabolism plays a role in cellular homeostasis and hepatoma suppression through negative control of cell population growth mediated by pro-apoptotic signaling. This insures that a major transiently activated endocrine metabolic circuit that is of benefit to the organism does not contribute to a proliferative stimulus in the liver or to promotion of hepatocarcinogenesis after it is initiated. Given the strong inhibitory effect of the KLB-FGFR4 partnership relative to FGFR4 alone on cell population growth, it is predicted that reduction and restoration of hepatocyte KLB levels may play a role in normal compensatory regeneration of liver. The coordinate expression of KLB and FGFR4 appears limited to hepatocytes in vivo. However, the KLB-FGFR4 partnership elicits controlling effects on cell population growth via pro-apoptotic signaling in diverse nonhepatic cell types including highly malignant tumor cells. Thus ectopic expression of the partnership in tumor cells in general in addition to restoration in deficient hepatomas may have possibilities for limiting diverse types of malignancies. It will be of interest to determine whether the cell growth controlling effects of the KLB-FGFR4 partnership demonstrated here extend to the αklotho-FGFR1 partnership in kidney and a KLB-FGFR partnership in adipocytes. Clarification of the mechanism by which membrane KLB participation in the HS-FGFR complex modifies FGFR-associated intracellular membrane signal transduction complexes is a subject for future study. To what extent KLB-dependent FGFR4 signaling pathways overlap or diverge for regulation of metabolism and control of hepatocyte proliferation and hepatoma suppression is also a subject for future investigation.
Supplementary Material
This work was supported by United States Public Health Service Grants DK56338 (to Y. L.) and P50 CA140388 (to W. L. M. and F. W.), the Susan Komen Breast Cancer Foundation, and the John S. Dunn Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Y. Luo, C. Yang, and W. L. McKeehan, unpublished results.
- HS
- heparan sulfate
- DEN
- dimethynitrosamine
- FGFR
- FGF receptor tyrosine kinase
- KLB
- klotho beta, betaklotho, or βklotho
- cKLB
- constitutively expressed KLB
- mTOR
- mammalian target of rapamycin
- iFGFR4
- induced FGFR4.
REFERENCES
- 1.McKeehan W. L., Wang F., Kan M. (1998) Prog. Nucleic Acid Res. Mol. Biol. 59, 135–176 [DOI] [PubMed] [Google Scholar]
- 2.Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 3.Luo Y., Ye S., Kan M., McKeehan W. L. (2006) J. Biol. Chem. 281, 21052–21061 [DOI] [PubMed] [Google Scholar]
- 4.Turner N., Grose R. (2010) Nat. Rev. Cancer. 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 5.Shimada T., Kakitani M., Yamazaki Y., Hasegawa H., Takeuchi Y., Fujita T., Fukumoto S., Tomizuka K., Yamashita T. (2004) J. Clin. Invest. 113, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., Gromada J., Brozinick J. T., Hawkins E. D., Wroblewski V. J., Li D. S., Mehrbod F., Jaskunas S. R., Shanafelt A. B. (2005) J. Clin. Invest. 115, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Cell. Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 8.Kuro-o M. (2008) Trends Endocrinol Metab. 19, 239–245 [DOI] [PubMed] [Google Scholar]
- 9.Kurosu H., Kuro-o M. (2008) Curr. Opin. Nephrol. Hypertens. 17, 368–372 [DOI] [PubMed] [Google Scholar]
- 10.Moore D. D. (2007) Science 316, 1436–1438 [DOI] [PubMed] [Google Scholar]
- 11.Huang X., Yang C., Luo Y., Jin C., Wang F., McKeehan W. L. (2007) Diabetes 56, 2501–2510 [DOI] [PubMed] [Google Scholar]
- 12.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., Elmquist J. K., Gerard R. D., Burgess S. C., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. (2007) Cell. Metab. 5, 415–425 [DOI] [PubMed] [Google Scholar]
- 13.Badman M. K., Pissios P., Kennedy A. R., Koukos G., Flier J. S., Maratos-Flier E. (2007) Cell. Metab. 5, 426–437 [DOI] [PubMed] [Google Scholar]
- 14.Barthel T. K., Mathern D. R., Whitfield G. K., Haussler C. A., Hopper H. A., 4th, Hsieh J. C., Slater S. A., Hsieh G., Kaczmarska M., Jurutka P. W., Kolek O. I., Ghishan F. K., Haussler M. R. (2007) J. Steroid Biochem. Mol. Biol. 103, 381–388 [DOI] [PubMed] [Google Scholar]
- 15.Razzaque M. S. (2009) Nat. Rev. Endocrinol. 5, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz R., Beenken A., Ibrahimi O. A., Kalinina J., Olsen S. K., Eliseenkova A. V., Xu C., Neubert T. A., Zhang F., Linhardt R. J., Yu X., White K. E., Inagaki T., Kliewer S. A., Yamamoto M., Kurosu H., Ogawa Y., Kuro-o M., Lanske B., Razzaque M. S., Mohammadi M. (2007) Mol. Cell. Biol. 27, 3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosu H., Ogawa Y., Miyoshi M., Yamamoto M., Nandi A., Rosenblatt K. P., Baum M. G., Schiavi S., Hu M. C., Moe O. W., Kuro-o M. (2006) J. Biol. Chem. 281, 6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurosu H., Choi M., Ogawa Y., Dickson A. S., Goetz R., Eliseenkova A. V., Mohammadi M., Rosenblatt K. P., Kliewer S. A., Kuro-o M. (2007) J. Biol. Chem. 282, 26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. (2006) Nature 444, 770–774 [DOI] [PubMed] [Google Scholar]
- 20.Kan M., Wu X., Wang F., McKeehan W. L. (1999) J. Biol. Chem. 274, 15947–15952 [DOI] [PubMed] [Google Scholar]
- 21.Yu C., Wang F., Kan M., Jin C., Jones R. B., Weinstein M., Deng C. X., McKeehan W. L. (2000) J. Biol. Chem. 275, 15482–15489 [DOI] [PubMed] [Google Scholar]
- 22.Yu C., Wang F., Jin C., Wu X., Chan W. K., McKeehan W. L. (2002) Am J. Pathol. 161, 2003–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S., Kinoshita S., Shiraishi N., Nakagawa S., Sekine S., Fujimori T., Nabeshima Y. I. (2000) Mech Dev. 98, 115–119 [DOI] [PubMed] [Google Scholar]
- 24.Lin B. C., Wang M., Blackmore C., Desnoyers L. R. (2007) J. Biol. Chem. 282, 27277–27284 [DOI] [PubMed] [Google Scholar]
- 25.Ito S., Fujimori T., Furuya A., Satoh J., Nabeshima Y. (2005) J. Clin. Invest. 115, 2202–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seitzer N., Mayr T., Streit S., Ullrich A. (2010) Cancer Res. 70, 802–812 [DOI] [PubMed] [Google Scholar]
- 27.Yu C., Wang F., Jin C., Huang X., McKeehan W. L. (2005) J. Biol. Chem. 280, 17707–17714 [DOI] [PubMed] [Google Scholar]
- 28.Huang X., Yu C., Jin C., Kobayashi M., Bowles C. A., Wang F., McKeehan W. L. (2006) Cancer Res. 66, 1481–1490 [DOI] [PubMed] [Google Scholar]
- 29.Huang X., Yang C., Jin C., Luo Y., Wang F., McKeehan W. L. (2009) Mol. Carcinog. 48, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein M., Xu X., Ohyama K., Deng C. X. (1998) Development 125, 3615–3623 [DOI] [PubMed] [Google Scholar]
- 31.Olson D. C., Deng C., Hanahan D. (1998) Cell Growth & Differ. 9, 557–564 [PubMed] [Google Scholar]
- 32.Ogawa Y., Kurosu H., Yamamoto M., Nandi A., Rosenblatt K. P., Goetz R., Eliseenkova A. V., Mohammadi M., Kuro-o M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y., Yang C., Jin C., Xie R., Wang F., McKeehan W. L. (2009) Cell. Signal. 21, 1370–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng S., Wang F., Matsubara A., Kan M., McKeehan W. L. (1997) Cancer Res. 57, 5369–5378 [PubMed] [Google Scholar]
- 35.Luo Y., Gabriel J. L., Wang F., Zhan X., Maciag T., Kan M., McKeehan W. L. (1996) J. Biol. Chem. 271, 26876–26883 [DOI] [PubMed] [Google Scholar]
- 36.Kan M., Shi E. G., McKeehan W. L. (1991) Methods Enzymol. 198, 158–171 [DOI] [PubMed] [Google Scholar]
- 37.Wang F., Kan M., Xu J., Yan G., McKeehan W. L. (1995) J. Biol. Chem. 270, 10222–10230 [DOI] [PubMed] [Google Scholar]
- 38.Kan M., Wang F., To B., Gabriel J. L., McKeehan W. L. (1996) J. Biol. Chem. 271, 26143–26148 [DOI] [PubMed] [Google Scholar]
- 39.Lundåsen T., Gälman C., Angelin B., Rudling M. (2006) J. Intern Med. 260, 530–536 [DOI] [PubMed] [Google Scholar]
- 40.Ferlini C., Scambia G. (2007) Nat. Protoc. 2, 3111–3114 [DOI] [PubMed] [Google Scholar]
- 41.Holz M. K., Blenis J. (2005) J. Biol. Chem. 280, 26089–26093 [DOI] [PubMed] [Google Scholar]
- 42.Chiang G. G., Abraham R. T. (2005) J. Biol. Chem. 280, 25485–25490 [DOI] [PubMed] [Google Scholar]
- 43.Chen P. Y., Friesel R. (2009) Biochem. Biophys. Res. Commun. 382, 424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhaskar P. T., Hay N. (2007) Dev. Cell. 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 46.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 47.Foster K. G., Fingar D. C. (2010) J. Biol. Chem. 285, 14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J. K., Gao G., Goldfarb M. (1994) Mol. Cell. Biol. 14, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaoul E., Reich-Slotky R., Berman B., Ron D. (1995) Oncogene 10, 1553–1561 [PubMed] [Google Scholar]
- 50.Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 51.Kwiatkowski B. A., Kirillova I., Richard R. E., Israeli D., Yablonka-Reuveni Z. (2008) J. Cell. Physiol. 215, 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desnoyers L. R., Pai R., Ferrando R. E., Hötzel K., Le T., Ross J., Carano R., D'Souza A., Qing J., Mohtashemi I., Ashkenazi A., French D. M. (2008) Oncogene 27, 85–97 [DOI] [PubMed] [Google Scholar]
- 53.Wu X., Ge H., Lemon B., Vonderfecht S., Weiszmann J., Hecht R., Gupte J., Hager T., Wang Z., Lindberg R., Li Y. (2010) J. Biol. Chem. 285, 5165–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S., Kang J., Cho M., Seo E., Choi H., Kim E., Kim J., Kim H., Kang G. Y., Kim K. P., Park Y. H., Yu D. Y., Yum Y. N., Park S. N., Yoon D. Y. (2009) Int J. Oncol. 34, 161–172 [PubMed] [Google Scholar]
- 55.Lopez-Serra L., Ballestar E., Ropero S., Setien F., Billard L. M., Fraga M. F., Lopez-Nieva P., Alaminos M., Guerrero D., Dante R., Esteller M. (2008) Oncogene 27, 3556–3566 [DOI] [PubMed] [Google Scholar]
- 56.Wolf I., Levanon-Cohen S., Bose S., Ligumsky H., Sredni B., Kanety H., Kuro-o M., Karlan B., Kaufman B., Koeffler H. P., Rubinek T. (2008) Oncogene 27, 7094–7105 [DOI] [PubMed] [Google Scholar]
- 57.Lee J., Jeong D. J., Kim J., Lee S., Park J. H., Chang B., Jung S. I., Yi L., Han Y., Yang Y., Kim K. I., Lim J. S., Yang I., Jeon S., Bae D. H., Kim C. J., Lee M. S. (2010) Mol. Cancer 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medici D., Razzaque M. S., Deluca S., Rector T. L., Hou B., Kang K., Goetz R., Mohammadi M., Kuro-O M., Olsen B. R., Lanske B. (2008) J. Cell. Biol. 182, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goetz R., Nakada Y., Hu M. C., Kurosu H., Wang L., Nakatani T., Shi M., Eliseenkova A. V., Razzaque M. S., Moe O. W., Kuro-o M., Mohammadi M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guimond S. E., Turnbull J. E. (1999) Curr. Biol. 9, 1343–1346 [DOI] [PubMed] [Google Scholar]
- 61.McKeehan W. L., Wu X., Kan M. (1999) J. Biol. Chem. 274, 21511–21514 [DOI] [PubMed] [Google Scholar]
- 62.Ye S., Luo Y., Lu W., Jones R. B., Linhardt R. J., Capila I., Toida T., Kan M., Pelletier H., McKeehan W. L. (2001) Biochemistry 40, 14429–14439 [DOI] [PubMed] [Google Scholar]
- 63.Kan M., Uematsu F., Wu X., Wang F. (2001) In Vitro Cell. Dev. Biol. Anim. 37, 575–577 [DOI] [PubMed] [Google Scholar]
- 64.Luo Y., Ye S., Kan M., McKeehan W. L. (2006) J. Cell. Biochem. 97, 1241–1258 [DOI] [PubMed] [Google Scholar]
- 65.Wu X., Ge H., Gupte J., Weiszmann J., Shimamoto G., Stevens J., Hawkins N., Lemon B., Shen W., Xu J., Veniant M. M., Li Y. S., Lindberg R., Chen J. L., Tian H., Li Y. (2007) J. Biol. Chem. 282, 29069–29072 [DOI] [PubMed] [Google Scholar]
- 66.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 67.Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 68.Uematsu F., Jang J. H., Kan M., Wang F., Luo Y., McKeehan W. L. (2001) Biochem. Biophys. Res. Commun. 283, 791–797 [DOI] [PubMed] [Google Scholar]
- 69.Wu X., Ge H., Lemon B., Weiszmann J., Gupte J., Hawkins N., Li X., Tang J., Lindberg R., Li Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14379–14384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mebratu Y., Tesfaigzi Y. (2009) Cell. Cycle 8, 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cagnol S., Chambard J. C. (2010) FEBS J. 277, 2–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.