Abstract
Elucidating the regulatory mechanism for tissue-specific gene expression is key to understanding the differentiation process. The chondromodulin-I gene (ChM-I) is a cartilage-specific gene, the expression of which is regulated by the transcription factor, Sp3. The binding of Sp3 to the core-promoter region is regulated by the methylation status of the Sp3-binding motif as we reported previously. In this study, we have investigated the molecular mechanisms of the down-regulation of ChM-I expression in mesenchymal stem cells (MSCs) and normal mesenchymal tissues other than cartilage. The core-promoter region of cells in bone and peripheral nerve tissues was hypermethylated, whereas the methylation status in cells of other tissues including MSCs did not differ from that in cells of cartilage, suggesting the presence of inhibitory mechanisms other than DNA methylation. We found that a transcriptional repressor, YY1, negatively regulated the expression of ChM-I by recruiting histone deacetylase and thus inducing the deacetylation of associated histones. As for a positive regulator, we found that a transcriptional co-activator, p300, bound to the core-promoter region with Sp3, inducing the acetylation of histone. Inhibition of YY1 in combination with forced expression of p300 and Sp3 restored the expression of ChM-I in cells with a hypomethylated promoter region, but not in cells with hypermethylation. These results suggested that the expression of tissue-specific genes is regulated in two steps; reversible down-regulation by transcriptional repressor complex and tight down-regulation via DNA methylation.
Keywords: DNA Methylation, Histone Modification, p300, Sp1, Stem Cell, Transcription Regulation, Chondromodulin-I, YY1, Mesenchymal Stem Cell
Introduction
The expression of cell lineage-specific genes is a key to initiating the differentiation of stem cells into a particular cell lineage, and the down-regulation of such genes in cells of other lineages is also critical to maintain a normal cellular physiology. The chondromodulin-I (ChM-I)2 gene is a specific gene for cartilage tissue (1). We have found that the basal promoter activity of ChM-I is driven by a ubiquitous transcription factor, Sp3, and chondrocyte-specific expression is regulated by the methylation status of the Sp3-binding motif in the core-promoter region (2). Demethylation treatment in vitro restored the expression of ChM-I in cells of the osteogenic lineage (2, 3). A similar result was obtained with cells of the adipogenic lineage, in which the expression of an adipocyte-specific gene was restored in non-adipogenic cells by the elimination of methylated DNA in a regulatory region (4). Because DNA methylation is considered a tight epigenetic change under physiological conditions, it is a suitable mechanism for cells to inhibit the expression of unnecessary genes. It is, however, still to be investigated whether cells in tissues other than cartilage share the same inhibitory mechanism. It is also important to know how the expression of lineage-specific genes is down-regulated in tissue stem cells before differentiation is initiated. Mesenchymal stem cells (MSCs) in bone marrow are tissue stem cells, which can differentiate into multiple mesenchymal cell lineages including chondrogenic cells (5, 6). Because three-dimensional cultures supplemented with growth factors such as TGF-β can induce the chondrogenic differentiation of MSCs (6), there should be a mechanism other than DNA methylation to down-regulate the gene expression of ChM-I in undifferentiated MSCs. Modification of the histone tail is another mechanism regulating gene expression. The acetylation of histone H3 and H4 promotes gene expression, whereas deacetylation inhibits the expression (7). The dimethylation of histone H3 at lysine 9 (H3K9) in particular is correlated with DNA methylation and markedly inhibits gene expression (8, 9). These modifications of the histone tail and methylation status determine differentiation (10), and are regulated by several intrinsic histone modifiers including p300 and YY1 (11–13). p300 possesses intrinsic histone acetyltransferase (HAT) activity (11, 12). YY1 is a member of the polycomb group of transcription factors, which establish and maintain transcriptional silencing by recruiting histone deacetylase (HDAC) (13, 14). These intrinsic factors regulate the epigenetic status and regulate gene expression.
Here we demonstrated that the down-regulation of ChM-I expression by DNA methylation is restricted in particular cell types, whereas other cells including MSCs are free from the methylation, and found that expression of the ChM-I gene in these cells is reversibly dependent on histone modifications, which are regulated by the net activity of intrinsic histone modifiers, YY1 and p300.
EXPERIMENTAL PROCEDURES
Tissue Specimens and Primary Cultured Cells
Mesenchymal (cartilage, bone, fat, muscle, ligament, and tendon) and non-mesenchymal tissues (nerve, artery, and skin) were obtained from the lower limb of a 56-year-old male who underwent above-knee amputation. The tissues were frozen by dry ice and kept at −80 °C until nucleic acid extraction. Human primary cultured chondrocytes (hPCs) was isolated from same patient and cultured as previously mentioned (15). MSCs were isolated from the iliac bone of healthy donor as described (16). Normal human osteoblasts (NHOSTs) and human primary pre-adipocytes (hPAs) were obtained from TaKaRa (TaKaRa Bio, Shiga, Japan). All the primary cells were maintained in DMEM (Sigma-Aldrich) with 10% fetal bovine serum (Thermo Fisher Scientific Inc., Waltham, MA), 100 units/ml penicillin, and 100 mg/ml streptomycin, in 5% CO2 at 37 °C. The Ethics Committee of the Faculty of Medicine, Kyoto University, approved the procedure and informed consent was obtained.
Cell Lines and Culture Conditions
The human cell lines, Saos2, were obtained from American Type Culture Collection (ATCC; Manassas, VA). The human osteosarcoma cell lines TAKAO and ANOS were established in our laboratory (2). All the cell lines used in this study were maintained in DMEM (Sigma-Aldrich) with 10% fetal bovine serum (Thermo Fisher Scientific Inc.), 100 units/ml penicillin, and 100 mg/ml streptomycin, in 5% CO2 at 37 °C.
Antibodies and Expression Vectors
The following antibodies were used; anti-YY1 (sc-7341, Santa Cruz Biotechnology, Santa Cruz, CA), anti-p300 (05–257, Millipore Corp, Billerica, MA), anti-Sp3 (sc-644, Santa Cruz Biotechnology), anti-acetylated H3K9 (06–942, Millipore Corp), anti-dimethylated H3K9 (07–212, Millipore Corp), anti-pan H3 (07–690, Millipore Corp), and anti-HDAC2 (51–5100, Zymed Laboratory Inc., San Francisco, CA). Expression vectors for YY1 (pCEP-YY1) and p300 (pcDNA3-p300) were kindly provided by Drs. E. Seto and K. Miyazono, respectively. The Sp3 expression vector (pCMV-Sp3) was previously described elsewhere (17).
Reverse Transcription (RT)-PCR and Quantitative RT-PCR
RNA was isolated using the Rneasy kit (Qiagen KK, Tokyo, Japan) from frozen tissues and the cultured cell lines. All RT reactions were performed using 1 μg of total RNA with a Super Script First Strand Synthesis System for RT-PCR kit (Invitrogen, Carlsbad, CA). The relative amount of ChM-I mRNA was assessed by TaqMan real-time PCR with the ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA) (2). A75-bp fragment from +411 (exon 4) to +485 (exon 5) of the ChM-I cDNA (GenBankTM accession number XM_007132) was amplified using specific primers (sense, 5′-GAAGGCTCGTATTCCTGAGGTG-3′; antisense, 5′-TGGCATGATCTTGCCTTCCAGT-3′) and labeled with a TaqMan probe (5′-FAM-CGTGACCAAACAGAGCATCTCCTCCA-3′-TAMRA). 18 S rRNA was used as the internal control, and all reactions were run in duplicate. The ratio of ChM-I/18 S in each sample was calculated, and the expression level of ChM-I genes was demonstrated as a relative value using the ChM-I/18 S ratio in human articular cartilage as a standard (1.0) (2).
Drug Treatment
Cells (1 × 105) were seeded on 60-mm dishes in DMEM with 10% FBS. After they had attached to the dish, the cells were treated with either 5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich) (1 μm) for 96 h or MS-275 (Nihon Scherring K.K., Chiba, Japan) (1 μm) for 24 h.
Bisulfite Genomic Sequencing
The bisulfite modification of DNA samples was performed using the EpiTect bisulfite kit (Qiagen). DNA (1 μg) was digested by BamHI for 12 h and subjected to sodium bisulfite treatment. Bisulfite-modified DNA-spanning residues −297 to −104 relative to the transcription start point (2) was amplified, cloned into the TA-vector (Invitrogen), and sequenced using an ABI 377 semiautomatic sequencer (Applied Biosystems).
Electrophoresis Mobility Shift Assay (EMSA)
Double-stranded DNA fragments corresponding to the sequence from −357 to −333 and from −86 to −44 were synthesized by annealing two single-stranded oligonucleotides (OND) (5′-CTTCACCTTCCATGAGCCATCTTC-3′ and 5′-GGGGGAAGATGGCTCATGGAAGGT-3′; 5′-GGGCATCCGGGAGTGCAGGACGAGCTTCCCGCGGCGGGA-3′; and 5′-TCTCTCCCGCCGCGGGAAGCTCGTCCTGCACTCCCGGAT-3′, respectively) and filling in by DNA polymerase I (TOYOBO, Osaka, Japan). These fragments were designated GR3 and GR4 (Fig. 2B). For the formation of the complex, 5 μg of nuclear extract from cell lysate was incubated with 32P end-labeled ONDs for 20 min at room temperature. The mixtures were electrophoresed in 5% polyacrylamide gel in 0.5× Tris borate EDTA at 45 volts for 3 h, and the gel then was dried and autoradiographed. For the competition assay, the OND-protein complex was produced in the same way in the presence of given amounts of non-labeled OND. In the supershift assay, nuclear extracts were incubated with 1 μg of anti-YY1 and anti-p300 antibody for 1 h on ice before being mixed with labeled DNA.
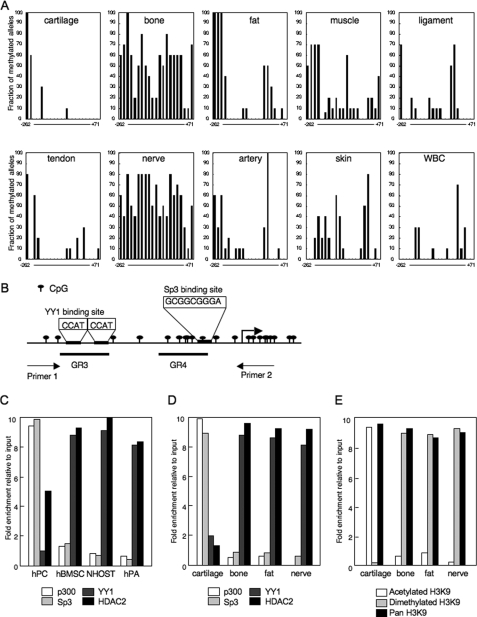
FIGURE 2.
Binding of YY1 and p300 determined the modification of H3K9 and the mRNA expression of ChM-I in mesenchymal tissues. A, methylation status of the core-promoter region of the ChM-I. DNA extracted from normal tissue was analyzed by bisufite genomic sequencing. B, genomic structure of the core-promoter region of ChM-I. CpG sites from −262 to +71 were marked as indicated, and the transcription start site is indicated by an arrow. Two overlapping YY1-binding motifs (−344 to −347 and −342 to −346), and an Sp3-binding motifs (−56 to −48) are also indicated. A DNA fragment for the ChIP assay was amplified by primer 1 (−446 to −425) and primer 2 (+70 to +91). GR3 (−357 to −337) and GR4 (−86 to −44) were OND probes used in the EMSA for YY1 and p300, respectively. ChIP-qPCR assay for the binding of transcriptional regulators in primary cultured cells (C) and cells of normal tissues (D). The y-axis represents fold enrichment relative to input. E, ChIP-qPCR assay for the modification of histones in normal tissues.
Luciferase Assay
The 533-bp fragment from −446 to +86 and 383-bp fragment from −296 to +86 relative to the transcription initiation site of the ChM-I gene was amplified by PCR, and cloned into a TA-vector using the TOPO cloning kit (Invitrogen). These fragments were subcloned into the luciferase reporter plasmid, PGV-B (Toyo Ink, Tokyo, Japan), yielding PGV-B-f1 and PGV-B-f1-del. Two tandem binding motifs of YY1 (CCAT) was mutated to (TTAT) by PCR, cloned into PGV-B, and designated PGV-B-f1-mt. One microgram of each reporter plasmid was co-transfected with 1 μg of pCEP-YY1 or pCMV-p300. Transfection efficiency was standardized by the co-transfection of 1 ng of pRL-TK control vector (Toyo Ink). Cells were harvested 24 h after transfection, and luciferase assays were performed with the PicaGene Dual SeaPansy system (Toyo Ink). Firefly-luciferase activity and SeaPansy-luciferase activity were measured as relative light units with a luminometer (STRATEC Biomedical Systems, Birkenfeld, Deutschland). The fold increase was calculated based on empty vector activity. Each experiment was performed in triplicate.
Chromatin Immunoprecipitation-Quantitative Polymerase Chain Reaction
The suitability of each antibody for the ChIP assay was confirmed by immunoprecipitation-Western blotting (data not shown). Tissue samples were treated using an EpiQuik tissue ChIP kit (Epigentek Group Inc. Brooklyn, NY). Cells were harvested and mixed with formaldehyde at a final concentration of 1.0% for 10 min at 37 °C to cross-link protein to DNA. Cells then were suspended in 0.2 ml of SDS lysis buffer and settled on ice for 10 min. DNA cross-linked with protein was sonicated into fragments of 200–1,000 bp. One-tenth of the sample was set aside as an input control, and the rest was precleared with salmon sperm DNA protein A-Sepharose beads (Millipore Corp) for 30 min with agitation. The soluble chromatin fraction was collected with each antibody at 4 °C overnight with rotation. Immune complexes were collected with salmon sperm DNA protein A-Sepharose beads and washed with the manufacturer's low salt, high salt, and LiCl buffers and then washed twice with TE buffer (10 mm Tris-HCl and 1 mm EDTA). The chromatin-antibody complexes were eluted with elution buffer (1% SDS and 0.1 m NaHCO3). Protein DNA cross-links were reversed with 5 m NaCl at 65 °C for 4 h, proteinase K treatment and phenol-chloroform extraction were carried out, and then the DNA was precipitated in ethanol. The DNA pool from ChIP, input control and negative control was used for quantitative PCR. PCR amplification was performed on an ABI 7700 real-time PCR (Applied Biosystems). PCR amplification was performed using primers specific for the ChM-I regulatory region (sense, 5′-GAATGCAGGCCAGTGAGAAGGT-3′;1 antisense, 5′-GCACCCTGGGATCTGTCCCGCT-3′, Fig. 2B). The PCR conditions were an initial step of 5 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 10 s at 64 °C and 60 s at 72 °C. Primers were designed according to the selected genes for evaluating ChIP. To generate a standard curve for each amplicon, threshold cycle (CT) values of serially diluted input DNA, which were extracted in the ChIP experiment, were determined. The status of histone modification and binding of HDAC2, p300, YY1, and Sp3 changes were determined using the 2(-Delta Delta C(T)) method (18). They were demonstrated as a relative value using the enrichment of IP DNA/input DNA. A melting curve analysis was performed for each reaction to ensure a single peak. Each experiment was performed in triplicate, with the values averaged to obtain 1 datum per sample.
siRNAs
Luciferase siRNA duplex (GL2RN1, Dharmacon) was used as a negative control. 40 μm siRNA for YY1 (GeneSolution siRNA; Hs-YY1–5, Qiagen), p300 (p300 Pub. siRNA, Duplex1, Qiagen), and Sp3 (previously described in (2)) were transfected by Lipofection LTX (Invitrogen). RNA was prepared 48 h after transfection and used for the RT-PCR.
RESULTS
DNA Methylation and Histone Deacetylation Down-regulate the Expression of ChM-I in a Cell Type-specific Manner
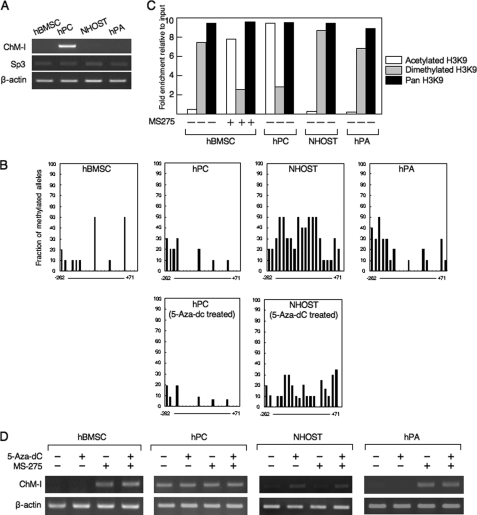
The expression of ChM-I was analyzed by RT-PCR in primary-cultured mesenchymal cells (hMSCs, hPCs, NHOSTs, and hPAs), among which only hPCs expressed the gene (Fig. 1A). We have previously shown the expression of ChM-I to be induced by the binding of Sp3, which was regulated by the methylation status of the binding motif in the core-promoter region (2). Expression levels of the Sp3 gene did not differ among the four types of cells (Fig. 1A). The core-promoter region of ChM-I was hypomethylated in hPCs and hypermethylated in NHOSTs (Fig. 1B), which was consistent with the positive and negative expression of ChM-I in each cell. The methylation status of the core-promoter region of hMSCs or hPAs, however, was not significantly different from that of hPCs in spite that the expression of ChM-I was not detected in these cells (Fig. 1B). We have also shown that the acetylation of histone H3 at lysine 9 (H3K9) is necessary to induce the binding of Sp3 to the core-promoter region of ChM-I. ChIP analyses showed that H3K9 associated with the core-promoter region was acetylated in hPCs, but dimethylated in hMSCs, NHOSTs, and hPAs (Fig. 1C). Treatment with a demethylation reagent (5-aza-dC) induced the expression of ChM-I in NHOSTs (Fig. 1D), which was associated with demethylatiion in the promoter region of the ChM-I gene (Fig. 1B, lower panel). 5-aza-dC treatment, however, showed no effects in hMSCs or hPAs (Fig. 1D). On the other hand, treatment with a HDAC inhibitor (MS-275) induced the expression of ChM-I gene in hMSCs and hPAs, but not in NHOSTs (Fig. 1D). The induction of ChM-1 gene expression in hMSC was associated with the acetylation of H3K9 (Fig. 1C). These results suggested two mechanisms for the down-regulation of ChM-I expression in primary-cultured cells; methylation of the core-promoter region as found in NHOSTs, and histone deacetylation and methylation without DNA methylation as found in hMSCs and hPAs.
FIGURE 1.
DNA methylation and histone deacetylation down-regulate the expression of ChM-I in a cell type-specific manner. A, expression of the ChM-I and Sp3 genes in primary cultured mesenchymal cells. B, methylation status of the core-promoter region of ChM-I. The methylation of each CpG site was analyzed in 10 alleles by bisulfite genomic sequencing. The y-axis indicates the fraction of methylated alleles and the x-axis indicates the position of each CpG site relative to the transcription start site. Methylation status in hPC and NHOST treated with a demethylating reagent (5-aza-dC, 1 μm for 96 h) were also shown. C, ChIP-qPCR assay for the modification of histones in primary cultured mesenchymal cells. Open box, acetylated H3K9; closed box, dimethylated H3K9; gray box, Pan H3. Histone modification in hBMSC treated with an HDAC inhibitor (MS-275, 1 μm for 24 h) were also shown. The y-axis represents fold enrichment relative to input. D, expression of ChM-I after treatment with 5-aza-dC and/or MS-275.
The Binding of YY1 and p300 Correlates to the Expression of ChM-I in Normal Mesenchymal Tissues
Among the normal mesenchymal tissues examined, the expression of ChM-I was observed only in cartilage (supplemental Fig. S1). The methylation status of the core-promoter region, however, differed significantly among tissues (Fig. 2A). DNA extracted from cells in cartilage and fat tissues showed a hypomethylated state in the core-promoter region, which was similar to those found in hPCs and hPAs (Fig. 1B). DNA extracted from cells in bone and nerve tissues showed that the core-promoter region was hypermethylated, which was similar to those that found in NHOSTs (Fig. 1B). In other tissues, the core-promoter region was hypomethylated. These results further suggested a mechanism other than DNA methylation to down-regulate the expression of ChM-I.
A search for factors regulating the chromatin structure revealed two tandem repeats (−344 to −347 and −342 to −346) of the binding motif of YY-1, which represses gene expression by recruiting HDAC to target regions (13, 14). As for factors with HAT activity, we focused on p300, which is known to relieve the transcriptional repression by YY1 (19). p300 also plays a role as a transcriptional adaptor recruiting transcription factors such as Sp3 (20). ChIP-qPCR assay showed that YY1 and HDAC2 bound to the core-promoter region in ChM-I-negative hMSCs, NHOSTs, and hPAs, whereas p300 and Sp3 bound in ChM-I-positive hPC (Fig. 2C). Consistent with the results obtained with primary-cultured cells, p300 and Sp3 bound to the core-promoter region in cells of cartilage, but not bone, fat or nerve tissue (Fig. 2D). On the other hand, the binding of YY1 and HDAC2 was observed in cells of bone, fat, and nerve, but not cartilage (Fig. 2D). ChIP-qPCR assay of the histone tail associated with the core-promoter region demonstrated that H3K9 was acetylated in cells of cartilage tissue, and dimethylated in those of bone, fat, and nerve tissues (Fig. 2E), which corresponded with the expression of ChM-I in each tissue. These results indicated that the expression of ChM-I correlated positively with the binding of p300 and negatively with that of YY1.
YY1 Binds to the Core Promoter Region and Inhibits Transcription
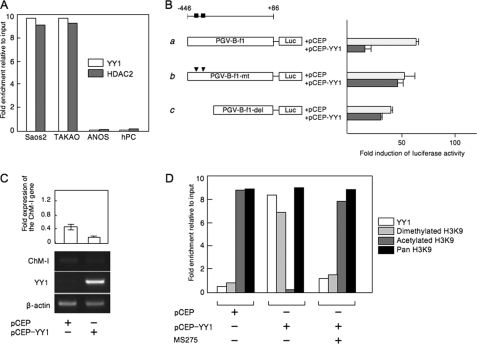
To further analyze the involvement of YY1 and p300 in the regulation of ChM-I, we used an osteosarcoma cell line, ANOS, as a ChM-I positive cell line, which we have previously investigated. The core-promoter region of the gene was hypomethylated in ANOS cells (2). As ChM-I-negative cell lines, two other osteosarcoma cell lines, Saos2 and TAKAO, were used, in which the core-promoter region was hypermethylated (2).
ChIP-qPCR assays showed that YY1and HDAC2 bound to the regulatory region of Saos2 and TAKAO cells, but not ANOS cells and hPCs (Fig. 3A). The binding of YY1 was further confirmed by an EMSA using an oligonucleotide (OND) (GR3) containing putative YY1-binding motifs (−342 to −347) (supplemental Fig. S2A). A shifted band was detected in the protein-OND complex from Saos2 cells, which disappeared on competition with unlabeled OND (supplemental Fig. S2A, left panel). The shifted band was detected also in the protein-OND complex from TAKAO cells, but not in ChM-I-positive cells (ANOS cells and hPCs) (supplemental Fig. S2A, middle panel). The shifted band was further shifted by the pretreatment with anti-YY1 antibody (supplemental Fig. S2A, right panel). These results indicated that YY1 bound to the core-promoter region of ChM-I in ChM-I-negative cells. To analyze the functional involvement of YY1, a reporter assay using the promoter fragment (−446 to +86) was performed (Fig. 3B), which contained the basal transcriptional activity of ChM-I (2). When the YY1 expression vector was co-transfected with the reporter plasmid, the promoter activity was significantly inhibited in ANOS cells (Fig. 3B, a). This inhibitory effect of YY1 was not observed when the reporter vector was replaced with one containing mutations in the YY1 motif (Fig. 3B, b) or lacking the motif (Fig. 3B, c). Forced expression of YY1 inhibited the expression of the endogenous ChM-I gene in hPCs (Fig. 3C), which was associated with the deacetylation and dimethylation of H3 (Fig. 3D). Co-treatment with MS275 inhibited the effect of forced expression of YY-1, rescuing the acetylation of H3K9 (Fig. 3D). These results suggested that YY1 inhibits the transcriptional activity of ChM-I by binding to a putative binding motif in the core-promoter region.
FIGURE 3.
YY1 bound to the regulatory region of ChM-I and decreased the promoter activity of ChM-I. A, ChIP-qPCR assay for YY1 and HDAC2. B, luciferase reporter assay. a, DNA fragment encompassing −446 to +86 was cloned into a reporter vector containing the luciferase gene (PGV-B-f1). The black box indicates the location of the consensus sequence for the YY1-binding motif. b, PGV-B-f1-mt contains mutations in the YY1-binding motif (arrowhead), and PGV-B-f1-del lacked the YY1-binding motifs (c). Each reporter vector was co-transfected with empty vector (pCEP) or the YY1 expression vector (pCEP-YY1) into ANOS. The fold-increase was calculated based on empty vector activity. C, expression of endogenous ChM-I in hPCs transfected with the YY1 expression vector. The expression of ChM-I was semi-quantified taking the value for endogenous expression as 1.0 and is demonstrated at the top. D, ChIP-qPCR assay of hPCs transfected with the YY1 expression vector with or without MS275 treatment (1 μm for 24 h). Forced expression of YY1 in hPCs changed the modification of the H3 tail from acetylation to dimethylation.
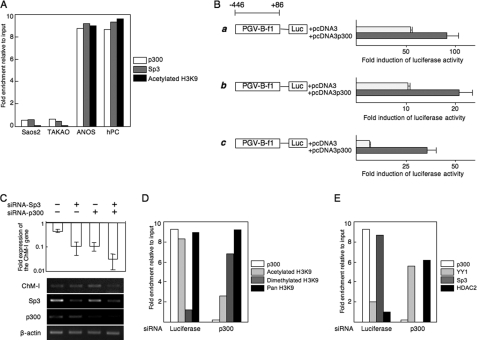
p300 Binds to the Core Promoter Region and Enhances Transcription
ChIP-qPCR assays showed that p300 as well as Sp3 bound to the core-promoter region of ChM-I in ANOS cells and hPCs, but not Saos2 and TAKAO cells (Fig. 4A). H3K9 associated with this region was acetylated in ANOS cells and hPCs, but not Saos2 or TAKAO cells (Fig. 4A). The binding of p300 was further confirmed by an EMSA using an OND (GR4)-containing putative p300 and Sp3-binding motifs (−56 to −48) (supplemental Fig. S2B). A shifted band was observed in extracts from ANOS cells, but not Saos2 or TAKAO cells (supplemental Fig. S2B, middle panel). The specificity of the band was confirmed by addition of a cold OND (supplemental Fig. S2B, left panel). The shifted band was supershifted when cell extracts were pretreated with anti-Sp3 antibody, and the same band disappeared when cell extracts were pretreated with anti-p300 antibody (supplemental Fig. S2B, right panel), suggesting that the protein-OND complex contained both Sp3 and p300. The functional involvement of p300 was analyzed with a promoter assay. Promoter activity was increased by co-transfection of the p300 expression vector in both ChM-I-positive (ANOS, Fig. 4B, a) and negative (Saos2, Fig. 4B, b; TAKAO, Fig. 4B, c) cells. Inhibition of p300 or Sp3 expression by the siRNA for each gene (siRNA-p300 and siRNA-Sp3) reduced the expression of ChM-I in hPCs, while the two siRNAs combined had an additive effect (Fig. 4C). ChIP-qPCR assays showed that the siRNA for p300 changed H3K9 from an acetylated to dimethylated form (Fig. 4D). These results suggest that p300 positively regulates the transcriptional activity of ChM-I by inducing the acetylation of H3K9 associated with this region.
FIGURE 4.
p300 binds to the core promoter region and enhances transcription. A, ChIP-qPCR assay of p300, Sp3 and acetylated H3. B, luciferase reporter assay. The reporter construct was described in the legend for Fig. 3B, and co-transfected with empty vector (pcDNA3) or the p300 expression vector (pcDNA3-p300) into ANOS (a), Saos2 (b), or TAKAO (c). The fold-increase was calculated based on empty vector activity. C, down-regulation of ChM-I gene expression by siRNA for Sp3 and/or p300. siRNAs for Sp3 and/or p300 were transfected into ANOS, and the expression of ChM-I, Sp3, and p300 was analyzed by RT-PCR. The expression of ChM-I was semi-quantified taking the value for endogenous expression as 1.0 and is demonstrated at the top. D, ChIP-qPCR assay for the modification of H3K9 (D) and for the binding of transcription regulators (E): cross-linked DNA-protein complexes were prepared from ANOS treated with or without siRNA for p300 and used for ChIP-qPCR assay.
Involvement of YY1 and p300 in Primary Mesenchymal Cells
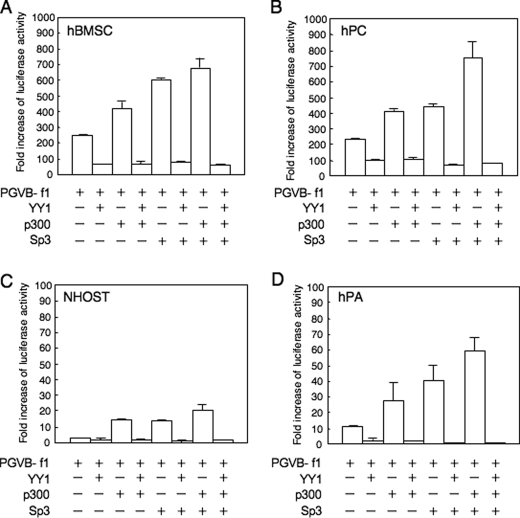
The effect of YY1 and p300 on promoter activity was further analyzed in primary-cultured mesenchymal cells (Fig. 5). Surprisingly, the transcriptional activity of the basal promoter fragment in hMSCs (Fig. 5B) was as strong as those in hPCs (Fig. 5A), although the expression of endogenous ChM-I was weak in hMSCs. In contrast, the basal activity level was low in NHOSTs and hPAs (Fig. 5, C and D). When the YY1 expression vector was co-transfected with the reporter vector, promoter activity was significantly inhibited in all strains (Fig. 5, A–D). Co-transfection with the p300 or Sp3 expression vector enhanced the activity, and simultaneous transfection of the two vectors increased it in an additive manner (Fig. 5, A–D). The enhancement of ChM-I expression by the p300 and/or Sp3 expression vectors was completely inhibited by the co-transfection of YY1 in all strains (Fig. 5, A–D). These results confirmed that YY1 and p300 are involved in the regulation of ChM-I transcription in mesenchymal cells, and that because this luciferase reporter system has no relationship with histone modifications, YY1 may directly inhibit the function of p300 or p300/Sp3 complex in addition to the modification of chromatin structure.
FIGURE 5.
Promoter activity of ChM-I was promoted by Sp3 and p300, but completely inhibited by YY1 in primary cultured cells. The luciferase reporter vector containing the core-promoter fragment of the ChM-I gene (PGV-B-f1) was co-transfected with YY1, p300 and/or Sp3 expression vectors into hMSCs (A), hPCs (B), NHOSTs (C), and hPAs (D). The fold-increase was calculated based on empty vector activity.
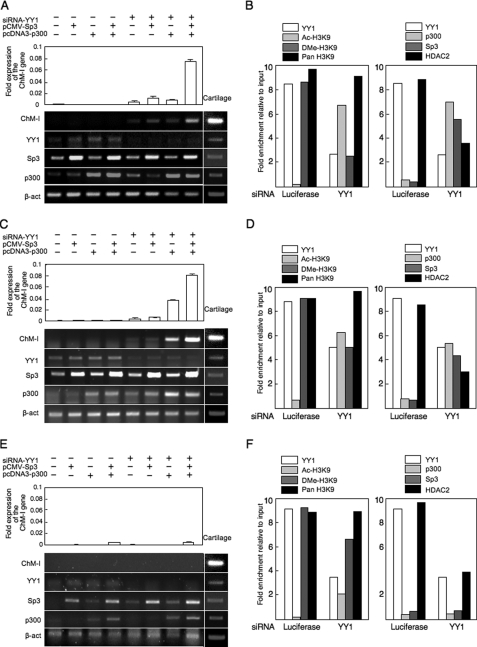
Cell-specific Effects of YY1 Inhibition on the Expression of ChM-I
Finally, the effect of YY1 expression on endogenous ChM-I expression was evaluated using siRNA for the gene (siRNA-YY1). In hMSCs (Fig. 6A), the inhibition of YY1 expression slightly induced the expression of ChM-I gene. The introduction of the p300 and/or Sp3 expression vectors had little effect on the expression of ChM-I, but significantly up-regulated it when combined with siRNA-YY1. Similar results were obtained in hPAs (Fig. 6C). In both cell types, siRNA-YY1 treatment induced the acetylation of H3K9 along with reductions in the dimetylation of H3K9 (Fig. 6, B and D, left panels). At the same time, siRNA-YY1 treatment dissociated HDAC2 and recruited p300 and Sp3 in the core-promoter region of ChM-I (Fig. 6, B and D, right panels). In NHOSTs (Fig. 6E), however, the combination the inhibition of YY1 and over-expression of p300 and Sp3 showed little effect for the induction of ChM-I expression. siRNA-YY1 treatment failed to induce the acetylation of H3K9 (Fig. 6F, left panel). Interestingly, although p300 was successfully recruited to the promoter region, no binding of Sp3 was observed in NHOSTs (Fig. 6F, right panel). These results suggested that Sp3 and p300 independently bind to the promoter region, and the binding of Sp3 was inhibited by the methylation of target DNA, but that of p300 was not. Therefore, the expression of a cartilage-specific gene, ChM-I, can be induced in some types of mesenchymal cells including MSCs by the modification of repressors (YY1) and activators (p300), but not in other cells, in which the expression is irreversibly inhibited by DNA methylation.
FIGURE 6.
Induction of ChM-I expression in ChM-I-negative primary cultured cells by modification of regulators. A–C, primary-cultured cells were transfected with a combination of the siRNA for YY1, p300 expression vector, and Sp3 expression vector, and mRNA level of ChM-1, YY1, p300, and Sp3 gene were analyzed by semi-quantitative RT-PCR (lower panel). The expression of the ChM-I was further analyzed by quantitative RT-PCR and digitalized (upper panel). A, hMSCs; C, hPAs; E, NHOSTs. ChIP-qPCR assay: B, hMSCs; D, hPAs; F, NHOSTs. Cross-linked DNA-protein complexes were prepared from primary cultured cells treated with or without siRNA for YY1 and used for ChIP-qPCR assay for the modificationf H3K9 (left panels) and for the binding of transcription regulators (right panels).
DISCUSSION
Numerous studies support the importance of epigenetic status for the regulation of differentiation, based on experiments involving chemical modifications of genome and the winding protein histone (21, 22). DNA methylation at CpG dinucleotides is a major epigenetic modification of the genome and associated with gene silencing (23). Because no intrinsic DNA-demethylating enzyme has been found, the inhibition by DNA methylation is tight under physiological conditions. Genomic DNA of embryonic stem (ES) cells is hypomethylated, and the total amount of methylated DNA increases with development (22, 24). Thus DNA methylation is a key mechanism to regulate and maintain the expression of cell type-specific genes. Unexpectedly, however, the methylation in the core-promoter region played a role in inhibiting the expression of ChM-I only in particular types of mesenchymal cells; cells in bone (mainly osteocytes and osteoblasts) and peripheral nerve (mainly Schwann cells and perineural cells) (Fig. 2A). The hypomethylated status in hMSCs is reasonable considering the potential of these cells to differentiate. However, the core-promoter in terminally differentiated cells of a remote cell-lineage such as white blood cells was free from methylation and thus in a reversible state for gene expression (Fig. 2A). At present, we have no data to explain why some types of cells use DNA methylation to inhibit the expression of ChM-I and others do not. Cells of chondrogenic and osteogenic lineages are closely related, and may share a considerable proportion of transcriptional machinery. Therefore DNA methylation might be required to inhibit the expression of genes specific to chondro- or osteogenic lineages. The reporter assay gave almost identical results in hMSCs as in hPCs (Fig. 5, A and B), indicating that transcriptional machinery n hMSC to be ready to induce the expression of ChM-I, although there is no endogenous expression of the gene. This result strongly suggested that epigenetic machinery regulated the lineage-specific gene expression in stem cells.
The epigenetic status of each cell had been considered static, but recent a study demonstrated a dynamic nature to these modifications (25). We and others gave examples in which modification of the histone code induced a change of DNA methylation (3, 26). Notably, the modification of H3K9 strongly correlated with DNA methylation; dimethylated H3K9 correlated with hypomethylation and deacetylated H3K9 correlated with hypermethylation (9, 27). The recent discovery that the forced expression of transcription factors can reverse the epigenetic status of differentiated to that of ES cells is an extreme example of the dynamic nature of epigenetic status (28, 29). We showed that the expression of ChM-I was down-regulated by histone modifications in stem cells and some types of differentiated cells. The epigenetic status is induced and maintained by a number of intrinsic histone modifiers (30). In this study, we found that YY1 and p300 are main modifiers of histone associated with the core-promoter region of the ChM-I gene. YY1 is a DNA-binding zinc finger transcription factor, which has dual functions as an activator and a repressor (14). YY1 inhibits the transcription of target genes by competing for DNA-binding sites with activators, binding directly to activators, or recruiting co-repressors (14). One of these co-repressors is HDAC2, which was first identified as a binding partner of YY1 (13, 14). Previously, we demonstrated that HDAC2 bound to a histone tail associated with the core-promoter region of ChM-I in ChM-I-negative cells (3). In the present study, we showed that forced expression of YY1 deacetylated H3 associated with the core-promoter region in ChM-I (Fig. 3E), whereas HDAC2 was dissociated by the inhibition of YY1, causing acetylation of H3 (Fig. 6, B, D, and F). These results indicated that YY1 repressed the expression of ChM-I by recruiting HDAC2 to induce deacetylation of H3. Another repressive mechanism is the direct binding of p300, a binding partner of YY1 (14). p300 acts as an activator for target gene expression through intrinsic HAT activity (11, 12). Inhibition of p300 by siRNA resulted in the deacetylation of H3 and the repression of ChM-I expression (Fig. 5C), indicating the role of p300 as HAT for ChM-I expression. p300 also acts as a transcriptional co-activator for Sp3, not Sp1 (19), which is the main transcription factor of ChM-I (2). The exogenous expression of p300 enhanced the promoter activity in the reporter assay, suggesting the role of p300 as a co-activator. Inhibition of the promoter activity by the YYI expression vector in the reporter assay indicated that YY1 acted as a direct repressor for p300 in addition to acting as a recruiter of HDAC (Fig. 6, A, C, and E). These results suggested that YY1 and p300 are involved in the regulation of ChM-I expression through the modification of histones and also the regulation of each others function.
It remains unclear how the repression of YY1 is relieved in chondrogenic cells. One possible mechanism is a post-translational modification of the YY1 protein. YY1 is glycosylated by O-linked N-acetylglucosaminylation, and glycosylated YY1 fails to bind to DNA (31). O-linked glucosamine is expressed in cartilage tissue (32). Such tissue-specific modifications may determine the expression of tissue-specific genes. It is also likely that tissue-specific chromatin-remodeling factors other than YY1 and p300 are involved in the regulation. Analyzing these issues may help to elucidate how the direction of differentiation is determined in stem cells.
Supplementary Material
Acknowledgments
We thank Dr. M. Nakanishi for providing MS-275 and helpful suggestions, Dr. K. Miyazono for the p300 expression vector, and Dr. E. Seto for the YY1 expression vector.
This work was supported by Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science, from the Ministry of Education, Culture, Sports, Science, and Technology, and from the Ministry of Health, Labor, and Welfare.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ChM
- chondromodulin
- MSC
- mesenchymal stem cell
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- OND
- oligonucleotides.
REFERENCES
- 1.Hiraki Y., Tanaka H., Inoue H., Kondo J., Kamizono A., Suzuki F. (1991) Biochem. Biophys. Res. Commun. 175, 971–977 [DOI] [PubMed] [Google Scholar]
- 2.Aoyama T., Okamoto T., Nagayama S., Nishijo K., Ishibe T., Yasura K., Nakayama T., Nakamura T., Toguchida J. (2004) J. Biol. Chem. 279, 28789–28797 [DOI] [PubMed] [Google Scholar]
- 3.Aoyama T., Okamoto T., Kohno Y., Fukiage K., Otsuka S., Furu M., Ito K., Jin Y., Nagayama S., Nakayama T., Nakamura T., Toguchida J. (2008) Biochem. Biophys. Res. Commun. 365, 124–130 [DOI] [PubMed] [Google Scholar]
- 4.Noer A., Sorensen A. L., Boquest A. C., Collas P. (2006) Mol. Biol. Cell. 17, 3543–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan A. I. (1991) J. Orthop. Res. 9, 641–650 [DOI] [PubMed] [Google Scholar]
- 6.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 7.Grunstein M. (1997) Nature 389, 349–352 [DOI] [PubMed] [Google Scholar]
- 8.Zhao W., Soejima H., Higashimoto K., Nakagawachi T., Urano T., Kudo S., Matsukura S., Matsuo S., Joh K., Mukai T. (2005) J. Biochem. 137, 431–440 [DOI] [PubMed] [Google Scholar]
- 9.Kondo Y., Shen L., Issa J. P. (2003) Mol. Cell. Biol. 23, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan Q., Yoshida T., McDonald O. G., Owens G. K. (2007) Stem Cells 25, 2–9 [DOI] [PubMed] [Google Scholar]
- 11.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 12.Bannister A. J., Kouzarides T. (1996) Nature 384, 641–643 [DOI] [PubMed] [Google Scholar]
- 13.Yang W. M., Yao Y. L., Sun J. M., Davie J. R., Seto E. (1997) J. Biol. Chem. 272, 28001–28007 [DOI] [PubMed] [Google Scholar]
- 14.Gordon S., Akopyan G., Garban H., Bonavida B. (2006) Oncogene 25, 1125–1142 [DOI] [PubMed] [Google Scholar]
- 15.Aoyama T., Liang B., Okamoto T., Matsusaki T., Nishijo K., Ishibe T., Yasura K., Nagayama S., Nakayama T., Nakamura T., Toguchida J. (2005) J. Bone. Miner. Res. 20, 377–389 [DOI] [PubMed] [Google Scholar]
- 16.Shibata K. R., Aoyama T., Shima Y., Fukiage K., Otsuka S., Furu M., Kohno Y., Ito K., Fujibayashi S., Neo M., Nakayama T., Nakamura T., Toguchida J. (2007) Stem Cells. 25, 2371–2382 [DOI] [PubMed] [Google Scholar]
- 17.Suske G. (1999) Gene 238, 291–300 [DOI] [PubMed] [Google Scholar]
- 18.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 19.Lee J. S., Galvin K. M., See R. H., Eckner R., Livingston D., Moran E., Shi Y. (1995) Genes Dev. 9, 1188–1198 [DOI] [PubMed] [Google Scholar]
- 20.Kishikawa S., Murata T., Kimura H., Shiota K., Yokoyama K. K. (2002) Eur. J. Biochem. 269, 2961–2970 [DOI] [PubMed] [Google Scholar]
- 21.Spivakov M., Fisher A. G. (2007) Nat. Rev. Genet. 8, 263–271 [DOI] [PubMed] [Google Scholar]
- 22.Bernstein B. E., Meissner A., Lander E. S. (2007) Cell 128, 669–681 [DOI] [PubMed] [Google Scholar]
- 23.Esteller M. (2007) Nat. Rev. Genet. 8, 286–298 [DOI] [PubMed] [Google Scholar]
- 24.Reik W., Dean W., Walter J. (2001) Science 293, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 25.Klose R. J., Zhang Y. (2007) Nat. Rev. Mol. Cell Biol. 8, 307–318 [DOI] [PubMed] [Google Scholar]
- 26.Nakao M. (2001) Gene 278, 25–31 [DOI] [PubMed] [Google Scholar]
- 27.Rougeulle C., Chaumeil J., Sarma K., Allis C. D., Reinberg D., Avner P., Heard E. (2004) Mol. Cell. Biol. 24, 5475–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 29.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 30.Pasini D., Bracken A. P., Agger K., Christensen J., Hansen K., Cloos P. A., Helin K. (2008) Cold Spring Harb. Symp. Quant. Biol. 73, 253–263 [DOI] [PubMed] [Google Scholar]
- 31.Hiromura M., Choi C. H., Sabourin N. A., Jones H., Bachvarov D., Usheva A. (2003) J. Biol. Chem. 278, 14046–14052 [DOI] [PubMed] [Google Scholar]
- 32.Thonar E. J., Lohmander L. S., Kimura J. H., Fellini S. A., Yanagishita M., Hascall V. C. (1983) J. Biol. Chem. 258, 11564–11570 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.