Abstract
Although early studies of inhibitor of apoptosis proteins (IAPs) suggested that cIAP1 directly binds and inhibits caspases similarly to X-linked IAP (XIAP), a recent one found that micromolar concentrations of cIAP1 only weakly inhibit caspase-3, -7, or -9. Here, we show that cIAP1 specifically and cooperatively blocks the cytochrome c-dependent apoptosome in vitro. Hence, cIAP1 prevented the activation of procaspase-3 but had no effect on the processing of procaspase-9 or the activity of prior activated caspase-3. Like cIAP1, XIAP had no effect on procaspase-9 processing and was a more potent inhibitor of procaspase-3 activation than of already activated caspase-3 activity. Inhibition of procaspase-3 activation depended on BIR2 and BIR3 of cIAP1 and was independent of BIR1, RING, CARD, and UBA domains. Smac prevented cIAP1 from inhibiting procaspase-3 activation and reversed the inhibition by prior addition of cIAP1. A procaspase-9 mutant (D315A) that cannot produce the p12 subunit was resistant to inhibition by cIAP1. Therefore, the N-terminal Ala-Thr-Pro-Phe motif of the p12 subunit of the caspase-9 apoptosome facilitates apoptosome blockade. Consequently, cIAP1 cooperatively interacts with oligomerized processed caspase-9 in the apoptosome and blocks procaspase-3 activation.
Keywords: Apoptosis, Caspase, Cell Death, Cytochrome c, Protease, Smac/DIABLO, XIAP, Apoptosome, cIAP1, Inhibitor of Apoptosis Protein
Introduction
Inhibitor of apoptosis proteins (IAPs)2 are a family of eight human proteins that have one or three baculovirus IAP repeat (BIR) domains. Although IAPs have been thought of as primarily inhibitors of apoptosis, it is now known that they play important roles in mitotic chromosome segregation, cellular morphogenesis, copper homeostasis, and intracellular signal transduction (1, 2). IAPs with three BIR domains (XIAP, cIAP1, cIAP2, and neuronal apoptosis inhibitor protein (NAIP)) can directly bind caspases via BIR2 and BIR3 and down-regulate caspase activity (3–7). Caspases are highly specific cysteinyl proteases that implement the cell death program by processing hundreds of different intracellular proteins after one or a few aspartyl residues (8). XIAP, which is the best characterized member of the family, inhibits the hydrolytic activity of caspase-3, -7, and -9 (9–11). Similarly to XIAP, NAIP directly binds and inhibits caspase-3, -7, and -9 (6, 7). Recent work on cIAP1 and cIAP2 indicates that they can directly bind processed caspase-3 or -7 (12) but have little effect on the activity of caspase-3, -7, or -9 (13).
Importantly, cIAPs are E3 ubiquitin ligases, which catalyze both trans- and autoubiquitination, as are four other IAPs (XIAP, melanoma IAP (also called Livin), ILP2 (also called testis-specific IAP), and Apollon (also called BRUCE)) (14–21). Interestingly, cIAPs can mediate either nondegradative Lys-63 polyubiquitination or degradative Lys-48 polyubiquitination of a protein target. For example, Lys-63 polyubiquitination of RIP1 mediates TNFα-evoked NFκB activation, whereas Lys-48 auto- or transubiquitination mediates proteasomal degradation of cIAPs or of a cIAP-binding protein such as a caspase or Smac/DIABLO (14, 16, 22–26). Hence, cIAPs may transubiquitinate caspases, which can trigger proteasomal degradation and/or down-regulation of proteolytic activity. Conversely, Smac can selectively reduce the levels of cIAP1 and cIAP2 but not XIAP (24). Additionally, the Smac homodimer binds BIR domains via the N-terminal IAP-binding motif (IBM), which frees the caspase from the IAP.
Here, we show that cIAP1 potently inhibits the processing and activation of procaspase-3 by the cytochrome c-dependent Apaf-1·caspase-9 apoptosome. The BIR domains of cIAP1 were sufficient for the inhibition of procaspase-3 activation, which was independent of the RING and CARD domains of cIAP1. Smac potently prevented and rapidly reversed blockade of the apoptosome by cIAP1. These findings support a model in which cIAP1 binds the apoptosome and sterically hinders access of procaspase-3 substrate to the catalytic center of the Apaf-1·caspase-9 complex.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
911 cells were grown in Dulbecco's modified Eagle's medium/Ham's F-12 (50:50) containing 10% FBS (25). Cells were transfected by calcium phosphate as described (25). Cells were plated (4 × 106/100-mm diameter tissue culture dish) and incubated for 6 h in a humidified atmosphere of 5% CO2 and 95% air. After 16 h, the transfection medium was removed and replaced with fresh growth medium. The cells were cultured for another 48 h before preparing lysates as described below. A full-length clone, Apaf-1XL (27), was subcloned from pcDNA3 into pcDNA3.1 by restriction digestion with BamHI and XhoI to produce Apaf-1 (apoptotic protease-activating factor-1) with a C-terminal V5 epitope tag and hexahistidine tags. Empty vector transfections were done with the pcDNA3.1 vector lacking a cDNA insert.
Preparation of S100 and S64 Fractions
Cell lysates were prepared from 20 confluent 911 cultures (100-mm diameter) 3 days after seeding essentially as described (28). Cultures were washed twice with ice-cold PBS, and cells were harvested by scraping and centrifugation. After resuspension in an equal volume of buffer containing 250 mm sucrose, 20 mm Tris-HCl (pH 7.5), 10 mm KCl, 1.5 mm MgCl2 (1 m Ultra; Fluka), 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm Pefabloc (Pierce), and 10 μg/ml each leupeptin and aprotinin, the cells were disrupted with a Dounce homogenizer (100 strokes with a tight-fitting pestle). Homogenates were centrifuged at 100,000 × g for 1 h at 4 °C to produce the S100 fraction from nontransfected 911 cells. S64 was prepared from Apaf-1-transfected cells using the following modifications of the S100 method. 1) The PBS-rinsed cells were suspended with 0.5 ml of modified sucrose lysis buffer, which contained no EDTA, 0.2 mm EGTA, and 1 mm MgCl2. 2) The cells were disrupted by five passes through an 18-gauge needle followed by five passes through a 26-gauge syringe needle. 3) The lysate was centrifuged at 16,000 × g for 30 min at 4 °C in a microcentrifuge, followed by a 45-min centrifugation at 30,000 rpm (64,000 × g × h). Protein concentrations of S64 and S100 fractions were determined by the Bradford method with bovine serum albumin as a standard and the Bio-Rad reagent. S64 has 2–3 times higher apoptosome activity than S100, which indicates that Apaf-1 is rate-limiting for apoptosome activity (data not shown).
Apoptosome Assay of Caspase-3-like Activity and Western Blot Analysis
S100 or S64 (1–1.5 μg/μl) was incubated for the indicated interval (15–60 min) in buffer containing 25 mm Tris-HCl (pH 7.5), 50 mm KCl, 0.2 mm EGTA (pH 7.5), 1 mm DTT, and 5 μm PS-341 (bortezomib), a proteasome inhibitor, in the presence and absence of the indicated concentration of bovine cytochrome c (Sigma) and 1 mm Na2ATP (Sigma) unless indicated otherwise. Following incubation with or without cytochrome c and ATP, a 4-μl sample was diluted with 36 μl of caspase assay buffer, which contained 25 mm Tris-HCl (pH 7.5), 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.2 mm EGTA, and 0.25 mm Ac-DEVD-AMC (a fluorogenic tetrapeptide substrate) (25). Following a 30-min incubation at 37 °C, the caspase reaction was stopped by dilution with ice-cold buffer containing 10 mm Tris-HCl (pH 7.5) and 1 mm EDTA. The fluorescence of freed AMC was measured at 440 nm (excitation at 380 nm) at room temperature by dilution of a 0.1-ml sample of each reaction with 1.9 ml of 10 mm Tris-HCl (pH 7.5) and 1 mm EDTA. Fluorescence units were converted to moles of AMC produced by constructing a standard curve, which was linear over the range of AMC measured (10–400 pmol). IC50 values, EC50 values, Hill coefficients, and statistical parameters were determined by nonlinear regression with GraphPad Prism 5. Values were not corrected for minor differences in the purity of the IAPs.
S100 or S64 proteins were size-fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and immunostained with antibody to caspase-9 (mouse monoclonal MAB8301, R&D Systems) or caspase-3 (rabbit monoclonal anti-cleaved caspase-3, 8G10, Cell Signaling Technology). Immunostaining was detected with the LumiGLO chemiluminescent substrate of horseradish peroxidase (Kirkegaard & Perry Laboratories). Exposed x-ray film (B-Plus blue, Medlink Imaging) was scanned with an Epson Precision 4870 photo scanner.
Caspase-9 Cloning and in Vitro Expression
Caspase-9 cDNA was excised from pCMV6. Plasmid caspase-9XL4 (Origene SC119362) was digested with HindIII and NotI and ligated into pcDNA3.1-V5-His (Invitrogen). D315A and D330A mutants of caspase-9 were generated by QuikChange site-directed mutagenesis (Stratagene) with the following primers: D315A, 5′-CTGGCAGTAACCCCGAGCCAGCTGCCACCCCGTTCCAG (forward) and 5′-CCTTCCTGGAACGGGGTGGCAGCTGGCTCGGGGTTACTGC (reverse); and D330A, 5′-GAGGACCTTCGACCAGCTGGCCGCCATATCTAGTTTGCCC (forward) and 5′-GTGTGGGCAAACTAGATATGGCGGCCAGCTGGTCGAAGG (reverse). In vitro transcription and translation were done with a rabbit reticulocyte master mixture (Promega) as recommended by the manufacturer.
Gateway Cloning of IAPs into Destination Vectors
Plasmids containing IAP cDNA sequences were purchased from American Type Culture Collection. cIAP1 (IMAGE ID 4831062), the α-isoform of Livin (IMAGE ID 4859588), Survivin (IMAGE ID 2961114), and plasmid pcDNA3.1-XIAP-GeneStorm (Invitrogen) were used templates for cloning IAPs into pENTR-D-Topo (Invitrogen). The following gene-specific primers were used to PCR-amplify IAP inserts, which were cloned into pENTR-D-Topo according to the manufacturer's protocol: cIAP1, 5′-CACCATGCACAAAACTGCCTCCCAAAG (forward) and 5′-TCATTAAGAGAGAAATGTACGAACAG (reverse); Livin, 5′-CACCATGGGACCTAAAGACAGTGCC (forward) and 5′-TCACTAGGACAGGAAGGTGCGCACGCG (reverse); Survivin, 5′-CACCATGGGTGCCCCGACGTTGCCC (forward) and 5′-TCATCAATCCATGGCAGCCAGCTGCTC (reverse); and XIAP, 5′-CACCATGACTTTTAAGAGTTTTGAAG (forward) and 5′-TTATTAAGACATAAAAATTTTTTGCTTG (reverse). The PCR cycling conditions were as follows: 94 °C for 1 min; 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 2 min for 25 cycles; and 72 °C for 5 min. pENTR-IAP plasmids were recombined with either pDEST15 encoding an N-terminal GST tag or pBAD-DEST49 encoding an N-terminal His-patch thioredoxin (HPT) and a C-terminal V5-His6 tag using LR Clonase II (Invitrogen) according to the manufacturer's protocol.
Cloning of cIAP1 Truncation Variants
cIAP1-(339–618) and cIAP1-(453–618) were PCR-amplified using pENTR-cIAP1 as a template with the cIAP1 339Met forward primer (5′-CACATGAAAGGCCAAGAGTTTGTTG) and cIAP1 453Met forward primer (5′-CACCATGGCATCAGATGATTTGTC) and the previously mentioned cIAP1 reverse primer. PCR products were ligated into pENTR-D-Topo, followed by recombination with pDEST15 or pBAD-DEST49. pDEST15- or pBAD-DEST49-cIAP1-(1–450) and -cIAP1-(1–565), respectively, were produced converting the codon for amino acids 451 and 566 to stop codons by QuikChange site-directed mutagenesis. The following primers were used: cIAP1 451 stop, 5′-GAGGAGAAGGAAAAACAAGCTTAATAAATGGCATCAGATG (forward) and 5′-CATCTGATGCCATTTATTAAGCTTGTTTTTCCTTCTCCTC (reverse); and cIAP1 566 stop, 5′-GAACAATTGAGGAGGTTGCAATAATAACGAACTTGTAAAGTG (forward) and cIAP1 566 stop, 5′-CACTTTACAAGTTCGTTATTATTGCAACCTCCTCAATTGTTC(reverse).
Purification of GST- and HPT-IAPs
Escherichia coli strain BL21-AI transformed with pDEST15-XIAP (or wild-type cIAP1, a truncation mutant of cIAP1, Livin, or Survivin) was grown overnight in LB broth containing 0.1 mg/ml ampicillin at 37 °C. Two 100-ml cultures of LB broth containing 0.2 mg/ml carbenicillin and 50 μm ZnSO4 were inoculated with 2 ml of the overnight culture and grown to an absorbance of ∼0.6 at 600 nm. l-Arabinose was added to 0.2% (w/v) to induce GST-XIAP expression at 37 °C. Bacteria were harvested after 8 h, frozen, and stored at −80 °C. Bacteria were suspended with 8 ml of lysis buffer, which contained 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm DTT, 0.2 mm Na2EGTA (pH 7.5), 0.8 mm benzamidine, and 0.25 mm Pefabloc SC. Additionally, the lysis buffer contained the following protease inhibitors 10 μg/ml leupeptin, 5 μg/ml pepstatin, and 2 μg/ml aprotinin. Recombinant lysozyme (2 μl, 30 kilounits/μl; Novagen) and Benzonase (4.8 μl, 25 units/μl; Novagen) were added, and the bacteria were rotated slowly for 25 min at room temperature. The bacteria were lysed by sonication, and lysate was centrifuged for 30 min at 100,000 × g. The supernatant was incubated at 4 °C with 10 mg of GSH-agarose for 3 h. The agarose was pelleted in a microcentrifuge for 5 min and washed twice with TED buffer (50 mm Tris-HCl (pH 7.5), 0.2 mm Na2EGTA (pH 7.5), and 1 mm DTT) containing 0.5 m NaCl and twice with TED buffer. GST-XIAP was eluted with 0.2-ml samples of buffer 25 mm Tris-HCl (pH 8.2), 10 mm GSH, 0.1 mm Na2EGTA, and 1 mm DTT. Protein concentration was determined with the Bradford reagent (Bio-Rad) and BSA as a standard. Proteins were frozen in liquid nitrogen and stored at −80 °C. Each of the GST-IAPs was ∼65–90% pure based on densitometric analysis of Coomassie Blue-stained SDS gels.
Recombinant human cIAP1 (amino acids 144–356) with a C-terminal 15-amino acid AviTag was obtained from Sino Biological Inc. His10-cIAP1 and Smac56-His6 were purchased from R&D Systems.
RESULTS
cIAP1 Prevents Procaspase-3 Activation by the Cytochrome c-dependent Caspase-9 Apoptosome
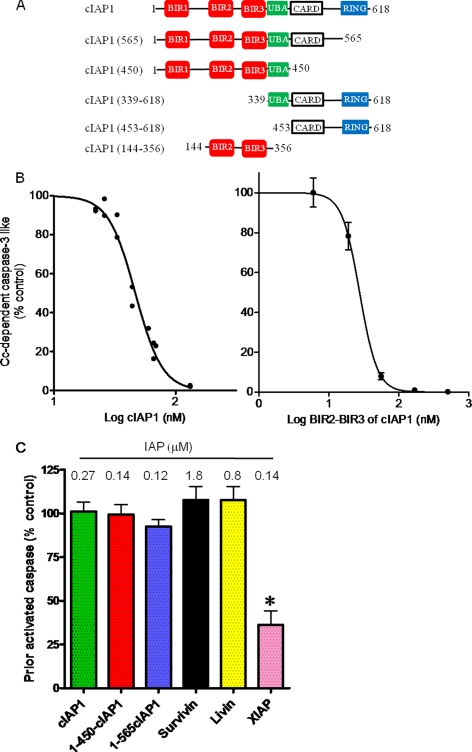
Apoptosome activation in vitro was entirely dependent on the addition of cytochrome c and a nucleotide such as ATP (data not shown). Caspase-3 activation was assayed with the fluorogenic substrate Ac-DEVD-AMC by diluting a sample of the incubation with cytochrome c 10-fold into caspase assay buffer. Activation of caspase-3 was confirmed by Western blot analysis, which showed that cytochrome c evoked accumulation of the large subunit (17 kDa), which is generated by processing of the 32-kDa procaspase-3 (see below). Caspase activity increased linearly following a 15–90-min incubation of the S100 fraction with cytochrome c (data not shown). The apoptosome reaction was initiated by the addition of the S100 fraction to buffer containing cytochrome c, ATP, and the indicated IAP. IAPs with an N-terminal GST tag were expressed and purified from E. coli. Only XIAP and cIAP1 potently blocked caspase activation by the cytochrome c-dependent apoptosome (Fig. 1 and Table 1). The IC50 values of XIAP and cIAP1 were 17 and 46 nm, respectively (Table 1). Micromolar concentrations of GST itself (≤7 μm) or of GST-Survivin (≤6 μm) had no effect on cytochrome c-dependent apoptosome activation. It appears that XIAP and cIAP1 may be sufficiently potent to regulate apoptosome activation intracellularly. Livin was ∼17 times less potent than cIAP1, which suggests that Livin (α-isoform) is unlikely to down-regulate apoptosome activation intracellularly (Table 1).
FIGURE 1.
cIAP1 potently blocks cytochrome c-dependent apoptosome activation of procaspase-3-like activity but does not affect already activated caspase-3-like activity. A, schematic of wild-type cIAP1 and truncation mutants. BIR domains and CARD motifs mediate protein-protein interactions. The UBA motif enables binding of Lys-63-linked polyubiquitin chains. RING binds ubiquitin-conjugating enzymes. B, inhibition of the cytochrome c-dependent apoptosome by cIAP1 and the BIR2-BIR3 segment of cIAP1. Cytochrome c (Cc)-dependent apoptosome activation of caspase-3 was assayed using an S100 preparation and the indicated concentrations of GST-cIAP1 and the BIR2-BIR3 segment of cIAP1. After a 30-min incubation of S100 with cytochrome c and ATP as described under “Experimental Procedures,” the apoptosome reaction was stopped by dilution with caspase assay buffer. Caspase-3 activity was determined with Ac-DEVD-AMC as substrate. C, effect of IAPs on already activated caspase-3-like activity. The caspase-3 activity of the S100 fraction was activated by the addition of cytochrome c and ATP in the absence of an added IAP. Caspase activation was stopped by diluting a sample of the apoptosome reaction by 10-fold into caspase assay buffer that contained the indicated micromolar concentration of each GST-IAP. Values are means ± S.E. (n = 3). Control activity was 12.2 ± 1.3 nmol of AMC produced per mg/min. *, statistically significant effect of XIAP (p < 0.01) by analysis of variance (GraphPad Prism 5).
TABLE 1.
Kinetic parameters of the inhibition of cytochrome c-dependent caspase-3 activation by IAPs
Apoptosome activation of S100 was assayed as described under “Experimental Procedures.” Kinetic and statistical parameters were determined by nonlinear regression curve fitting with Prism 5.
| IAP | IC50 | 95% confidence limit of IC50 | Hill coefficient | No. of experiments |
|---|---|---|---|---|
| nm | nm | |||
| GST-XIAP | 17 | 15–19 | 3.80 ± 0.51 | 3 |
| GST-cIAP1 | 46 | 44–49 | 3.91 ± 0.32 | 3 |
| BIR2-BIR3 cIAP1 | 28 | 25–31 | 3.45 ± 0.36 | 3 |
| GST-Livin | 771 | 549–1081 | 0.92 ± 0.13 | 3 |
| His10-cIAP1 | 180 | 168–193 | 2.68 ± 0.19 | 3 |
| HPT-cIAP1 | 104 | 95–115 | 4.73 ± 0.88 | 5 |
| HPT-cIAP1-(1–450) | 75 | 65–86 | 3.97 ± 0.89 | 4 |
Although early and current studies of IAPs have used GST-tagged IAPs, some have suggested that the GST tag can affect of the potency of the IAP by >100-fold (13, 29). Table 1 shows that the IC50 values of HPT-cIAP1 and the truncation mutant HPT-cIAP1-(1–450) were 104 and 75 nm, respectively. Additionally, BIR2 and BIR3 of cIAP1 had an IC50 of 28 nm. Hence, the GST tag has little, if any, effect on the potency of GST-cIAP1. Additionally, commercially prepared cIAP1 with an N-terminal decahistidine tag was found to be less potent (IC50 = 180 nm) than either GST- or HPT-tagged cIAP1 (Table 1). Differences in the purification of cIAP1 probably contribute to differences in the potency of the various preparations.
Note that cIAP1 inhibited cytochrome c- and apoptosome-dependent processing of procaspase-3 in a cooperative manner. The Hill coefficients of the various preparations of cIAP1 ranged from 2.68 to 4.73 (Table 1). The positive cooperativity of the inhibition suggests that the inhibition is caused by binding of more than one cIAP1 to the oligomerized apoptosome, i.e. the Apaf-1·caspase-9 holoenzyme. By contrast to cIAP1, blockade of the apoptosome by Livin was not cooperative (Table 1). Livin has a single BIR domain, whereas cIAP1 has three. The BIR2-BIR3 segment of cIAP1 may be required for cooperativity because truncated cIAP1 that contained only the BIR2-BIR3 segment cooperatively blocked apoptosome activation of procaspase-3 (Fig. 1B and Table 1).
Importantly, XIAP and cIAP1 were much less potent inhibitors of active caspase-3 than of the activation of procaspase-3 by the apoptosome. Thus, 0.27 μm cIAP1 had no effect on the caspase activity of processed caspase-3, and 0.14 μm XIAP inhibited the activity of processed caspase-3 by ∼75% (Fig. 1C). Survivin and Livin at 1.8 and 0.8 μm had no effect on the activity of already activated caspase-3 (Fig. 1C).
BIR2 and BIR3 of cIAP1 Are Sufficient to Block Apoptosome Activation
Truncation mutants of cIAP1 that lacked the RING domain (GST-cIAP1–565) or both the RING and the CARD domains (GST-cIAP1–450 or HPT-cIAP1–450) were essentially as potent as full-length GST-cIAP1 (Fig. 1, A and B, and Table 1) (data not shown). Moreover, a truncation mutant consisting of BIR2-BIR3 (amino acids 144–356) was as potent as wild-type cIAP1 (Fig. 1B and Table 1). By contrast, 1.4 μm GST-cIAP1-(339–618) or 0.8 μm GST-cIAP1-(453–618) had no effect on apoptosome activation of procaspase-3 (data not shown). Thus, BIR2, BIR3, and the short segment between them was sufficient to potently and cooperatively prevented the caspase-9 apoptosome from activating caspase-3-like activity.
cIAP1 Has No Effect on the Processing of Procaspase-9
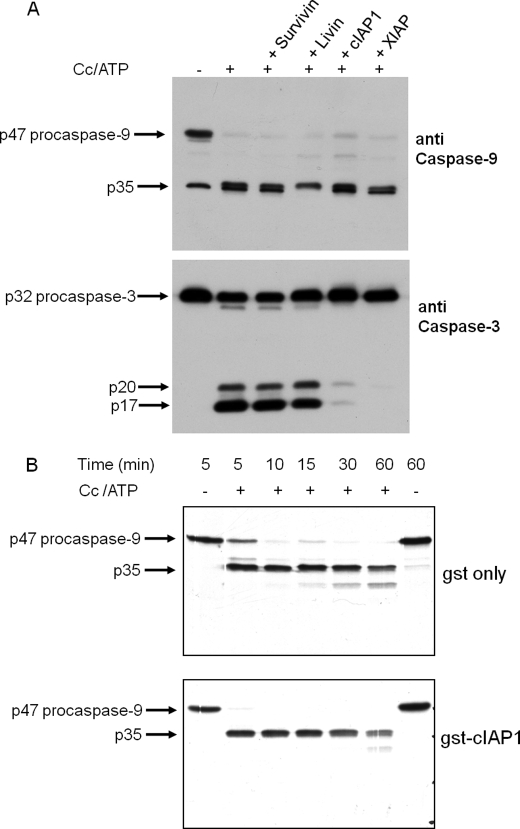
cIAP1 had no effect on the processing of procaspase-9 to the 35-kDa large subunit, which was evoked by cytochrome c plus ATP (Fig. 2A). Similarly, XIAP had no effect on the production of the large subunit of caspase-9 (Fig. 2A). By contrast to the lack of effect of cIAP1 and XIAP on the processing of procaspase-9, each of these IAPs essentially abolished the processing of procaspase-3 to the 20- and 17-kDa forms of the large subunit (Fig. 2A). The processing of procaspase-9 to p35 was confirmed by the addition of 35S-labeled procaspase-9 or an 35S-labeled procaspase-9 mutant (D315A or D330A) to the S64 fraction from Apaf-1-transfected cells, which has 2–3 times greater apoptosome activity (Fig. 3B). Although 35S-labeled procaspase-9 D315A was processed at Asp-330 in response to cytochrome c plus ATP, 35S-labeled wild-type procaspase-9 was predominantly processed at Asp-315 (Figs. 2B and 3B). Under conditions that essentially abolished procaspase-3 activation, GST-cIAP1 had no effect on the processing of 35S-labeled wild-type procaspase-9 (Fig. 2B). Additionally, GST-cIAP1 had no effect on the processing of either the D315A or D330A mutant of 35S-labeled procaspase-9 (data not shown).
FIGURE 2.
cIAP1 and XIAP prevent the processing of procaspase-3 by the cytochrome c-dependent apoptosome but have no effect on the processing of procaspase-9. A, Western blot analysis of the effects of IAPs on the processing of procaspase-9 and -3. The apoptosome reaction was carried out with S64 from Apaf-1-transfected cells at 1.5 μg/μl for 30 min at 37 °C in the presence or absence of bovine cytochrome c (Cc; 20 μg/ml) plus 1 mm ATP. A sample of the apoptosome reaction (24 μg of protein) was used for SDS-PAGE (12% gel), and Western analysis of caspase-9 and -3 was done as described under “Experimental Procedures.” After immunostaining caspase-9 with the mouse monoclonal antibody, the membrane was reprobed with rabbit monoclonal antibody to cleaved caspase-3. The apoptosome reaction was carried out in the absence of cytochrome c (−) or in the presence of cytochrome c plus the indicated IAP. The concentrations of GST-Survivin, GST-Livin, GST-cIAP1, and GST-XIAP were 1.8, 1.5, 0.27, and 0.13 μm, respectively. B, autoradiogram of the time course of 35S-labeled procaspase-9 processing in the presence of GST (gst only) or GST-cIAP1 (gst-cIAP1). The 20-μl apoptosome reaction included 1.0 μg/μl S64 from Apaf-1-transfected cells and 1 μl of 35S-labeled procaspase-9 and was incubated for the indicated intervals at 37 °C in the presence or absence of bovine cytochrome c (100 μg/ml) plus 1 mm ATP. The reactions contained 500 nm purified GST or 150 nm GST-cIAP1 as indicated. After the indicated intervals, 10 μl of the reaction was fractionated by SDS-PAGE (15% gel) and autoradiographed to visualize 35S-labeled p47 procaspase-9 and the p35 large subunit.
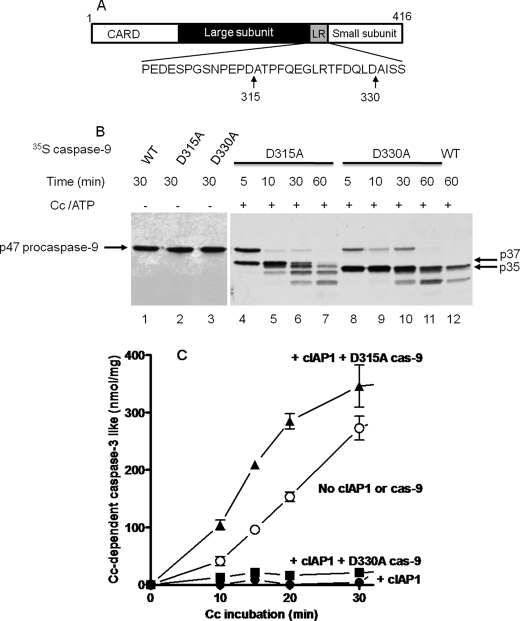
FIGURE 3.
The procaspase-9 D315A mutant, but not the D330A mutant, reverses the inhibition of the cytochrome c-dependent apoptosome by cIAP1. A, schematic of human procaspase-9 and the positions of the autocatalytic cleavage site (Asp-315) and the caspase-3 cleavage site (Asp-330) within the linker region (LR) between the large and small subunits of mature caspase-9. B, autoradiogram showing the processing of 35S-labeled p47 procaspase-9 mutants (D315A and D330A) by the apoptosome. The experiment was done as described in the Fig. 2B legend except the 35S-labeled procaspase-9 used was wild-type or the indicated point mutant. C, the procaspase-9 D315A mutant, but not the D330A mutant, prevents cIAP1 inhibition of the apoptosome. The cytochrome c (Cc)-dependent apoptosome activation of procaspase-3 was assayed as described for B in the presence (▲, ■, and ●) or (○) absence of 500 nm GST-cIAP1. The indicated D315A or D330A mutant of procaspase-9 (cas-9; 4 μl of TnT reaction/20 μl of apoptosome reaction) was added to the apoptosome reaction. Samples (4 μl) of the apoptosome reaction were removed after the indicated intervals, diluted 10 times with caspase assay buffer, and assayed for caspase-3-like activity with Ac-DEVD-AMC as substrate. Values are means ± S.E. (n = 3) for AMC produced (nmol/mg of S64 protein).
Importantly, the addition of the procaspase-9 D315A mutant to the S64 fraction completely overcame the inhibition of the apoptosome by GST-cIAP1 and moderately increased caspase activation compared with the “no cIAP1 or caspase-9” control (Fig. 3C). By contrast, the procaspase-9 D330A mutant failed to overcome apoptosome inhibition by GST-cIAP1 (Fig. 3C). The D315A mutant likely is transprocessed by activated caspase-3 at Asp-330, which produces a p10 subunit that begins with Ala-Ile-Ser-Ser, whereas autoprocessing at Asp-315 produces the p12 subunit, which begins with Ala-Thr-Pro-Phe. The proline at the third position is known to be critical for high affinity binding of BIR3 (30). These results indicate that the N-terminal Ala-Thr-Pro-Phe motif of the p12 subunit of the caspase-9 apoptosome facilitates apoptosome blockade.
Smac Antagonizes cIAP1-inhibited Apoptosome Activation of Procaspase-3
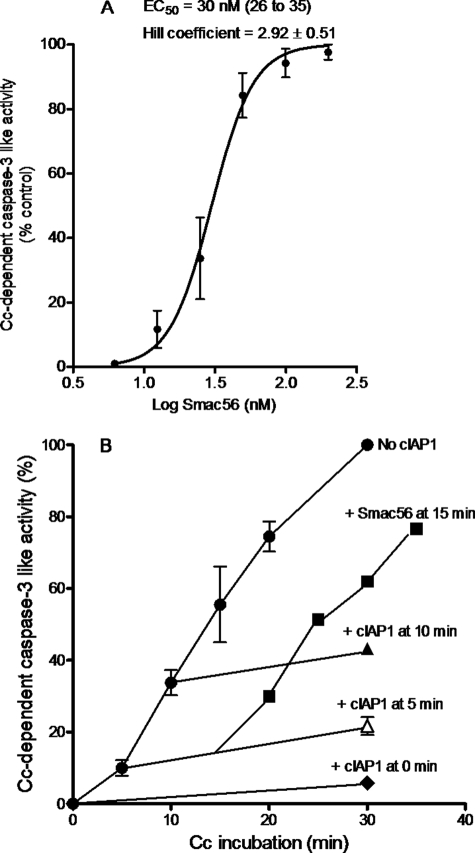
Mature Smac56 (amino acids 56–239) potently prevented cIAP1 from inhibiting apoptosome activation of procaspase-3 (Fig. 4A). The EC50 of bacterially expressed mature Smac with a hexahistidine tag was 30 nm (Fig. 4A). The EC50 of Smac-GST expressed in 911 cells was similar to that of recombinant Smac from bacteria (data not shown). Mature Smac prevented the inhibition of the caspase-9 apoptosome by each of the cIAP1 constructs indicated in Table 1 (data not shown). Thus, each of the purified cIAP1 proteins specifically inhibited the apoptosome by a mechanism that was potently prevented by mature Smac.
FIGURE 4.
Smac prevents and reverses cIAP1-evoked inhibition of procaspase-3 activation. A, mature Smac56 (amino acids 56–239) prevents cIAP1 from inhibiting the cytochrome c-dependent apoptosome. Values are means ± S.E. (n = three or four experiments). Cytochrome c (Cc)-dependent apoptosome activation was assayed with 1 μg/μl S100 fraction in the presence of 40 nm GST-cIAP1 and the indicated concentration of Smac56-His6. B, mature Smac56 reverses the cIAP1 inhibition of the apoptosome. Values are means ± S.D. (n = 3–7) for six different experiments with S64 (1.5 μg/μl) from Apaf-1-transfected cells. Smac56-His6 was added as indicated to 200 nm at 15 min (■) to the apoptosome reaction that received 250 nm GST-cIAP1 at 5 min (△). Other apoptosome reactions received no GST-cIAP1 (●) or 250 nm GST-cIAP1 at 0 min (♦) or 10 min (▲). Samples (4 μl) of the apoptosome reaction were removed after the indicated intervals, diluted 10 times with caspase assay buffer, and assayed for caspase-3-like activity with Ac-DEVD-AMC as substrate.
Smac is an exceptionally stable dimeric protein that antagonizes XIAP with a stoichiometry of two Smac protomers/XIAP (31, 32). Hence, the potency of the Smac dimer is similar to that of cIAP1 (30 compared with 46 nm) (Fig. 4A and Table 1). The addition of Smac to the cIAP1-inhibited apoptosome rapidly and fully reversed apoptosome activation (Fig. 4B). This result indicates that the mechanism of cIAP1 was fully reversible whether Smac was added 5 or 15 min after the start of the incubation of S100 with cytochrome c, ATP, and cIAP1 (Fig. 4B).
DISCUSSION
Here, we have shown that cIAP1 potently inhibited the processing and activation of procaspase-3 by the cytochrome c-dependent Apaf-1·caspase-9 apoptosome complex. This action of cIAP1 exhibited positive cooperativity and specificity because cIAP1 was 2.5 times less potent than XIAP and 17 times more potent than the α-isoform of Livin (Table 1). BIR2 and BIR3 were sufficient to prevent procaspase-3 activation (Fig. 1B and Table 1). The IC50 of cIAP1 or BIR2 and BIR3 of cIAP1 was 2–3 times that of XIAP (Table 1). Smac antagonized the inhibitory action of cIAP1 with an EC50 of 30 nm (Fig. 4A). The Smac dimer is known to directly bind and displace XIAP from caspases (32). The two IBMs of dimeric Smac bind to BIR2 and BIR3 of XIAP, which also mediate the interaction with caspases (33). It is likely that Smac binding to the BIR domains of cIAP1 is similar to that of XIAP because the IBM-interacting residues of BIR2 and BIR3 of XIAP are highly conserved in cIAP1 (13). Thus, the IC50 of cIAP1 (46 nm) or of BIR2 and BIR3 of cIAP1 (28 nm) is expected to be similar to the EC50 of dimeric Smac (30 nm), as observed (Fig. 4A and Table 1).
Interestingly, like XIAP, cIAP1 had no effect on the processing of procaspase-9 between the large and small subunits (Fig. 2A). Procaspase-9 is known to autoprocess following assembly of the active apoptosome, although autoprocessing is not required for activation (34–36). Thus, cytochrome c evoked the formation of the active apoptosome in the presence of cIAP1 or XIAP because neither IAP affected the autoprocessing of procaspase-9 (Fig. 2A). However, in the presence of cIAP1, the apoptosome was unable to process procaspase-3, as reported previously for XIAP with recombinant Apaf-1 and caspase-9 (35). In fact, under conditions that essentially abolished procaspase-3 activation, cIAP1 had no effect on the Apaf-1·caspase-9 complex, which was isolated by velocity sedimentation in a glycerol gradient (data not shown). The addition of Smac to the cIAP1-inhibited apoptosome rapidly and fully reversed the inhibition of procaspase-3 activation (Fig. 4B). This reversal supports the finding that cIAP1 did not disrupt apoptosome assembly.
BIR3 of XIAP interacts with the N-terminal IBM (i.e. Ala-Thr-Pro-Phe) of the p12 subunit of processed caspase-9 (35, 37). Moreover, XIAP has no effect on procaspase-3 activation by the apoptosome unless procaspase-9 has been processed to p35/p12 subunits (37). Hence, XIAP and cIAP1 bind to the oligomerized processed caspase-9 apoptosome and prevent procaspase-3 activation (Fig. 2A and Table 1).
A recent report showed that differences in the amino acid sequences of BIR2 and BIR3 of cIAP1 compared with XIAP account for the lack of a direct inhibition of caspase-3 and -7 by cIAP1 (30). Furthermore, the peptide-binding specificity of BIR3 of cIAP1 is essentially the same as that of XIAP (30). Livin, which has a single BIR domain that is a type III BIR domain like BIR3 of cIAP1 and XIAP (30), was substantially less potent than cIAP1 and failed to exhibit cooperative inhibition of the apoptosome (Table 1). The Livin result is consistent with the two-site avidity enhancement model with a secondary interaction provided by BIR2 of cIAP1 and XIAP (30). Interestingly, cIAP1 fails to inhibit monomeric purified caspase-9 activity toward a peptidyl-AMC substrate (13), yet cIAP1 is a specific cooperative inhibitor of the oligomerized Apaf-1·caspase-9 apoptosome assayed with a physiological substrate, procaspase-3 (Figs. 1B and 2 and Table 1). Because Apaf-1 heptamerizes in the apoptosome, the cooperativity of the inhibition by cIAP1 may result from the interaction of multiple cIAP1 monomers with oligomerized caspase-9 in the apoptosome. Unfortunately, the stoichiometry of caspase-9 in the apoptosome is unknown (38, 39).
It appears that BIR domains can inhibit caspase-9 by more than one mechanism. For example, XIAP inhibits monomeric caspase-9 activity assayed with a peptidyl-AMC substrate, but cIAP1 does not (13). Mutation of four residues of cIAP1 that are close to the C terminus of BIR3 to the corresponding amino acids of XIAP enables cIAP1 to inhibit caspase-9 (13). Thus, cIAP1 appears to block the apoptosome by preventing the processing and activation of effector caspases such as procaspase-3, as shown here (Fig. 2). Although it is not excluded that added cIAP1 depends on an endogenous protein in the cell lysate to block the apoptosome, this seems unlikely. BIR2 and BIR3 of cIAP1 can associate with caspase-9 (13). Deletion of the other protein interaction domains of cIAP1 (BIR1, CARD, RING, and UBA) had no effect on the potency to block the caspase-9 apoptosome. Thus, the BIR2-BIR3 segment of cIAP1 was as potent as cIAP1 (Table 1). Additionally, mature Smac with a C-terminal GST tag was used to deplete IAPs such as XIAP, cIAPs, and Apollon from a cell lysate and found to have no effect on the inhibition of the apoptosome by BIR2 and BIR3 of cIAP1 (data not shown). Therefore, it appears that cIAP1 inhibition is independent of XIAP and Apollon.
A model that can account for the action of cIAP1 is that it binds to the IBM of the processed p12 subunit of the Apaf-1·caspase-9 apoptosome and sterically hinders it from processing procaspase-3, as was proposed for XIAP (35). Hence, it was suggested that the close proximity of the N terminus of the p12 subunit and the catalytic center residues of caspase-9 may hinder the entry of substrate (35). The crystal structure of Smac complexed with BIR3 of XIAP is the basis of the model (40), which assumes that cIAP1 binds the caspase-9 holoenzyme, but not procaspase-3, as was reported recently (12). Moreover, the model explains why cIAP1 fails to inhibit the autoprocessing of procaspase-9 in the assembled apoptosome complex, namely because procaspase-9 lacks the free amino group of the first residue of the IBM, which hydrogen bonds to Glu-314 of BIR3 of XIAP (40) or the corresponding Asp of cIAP1. Accordingly, XIAP and cIAP1 failed to bind and prevent autoprocessing of procaspase-9 because exposure of the p12 small subunit IBM requires autoprocessing, which tethers the IAP to the active center. Hence, XIAP and cIAP1 inhibited only the caspase-9 apoptosome processed at Asp-315 but not that processed at Asp-330 (Fig. 3C).
The cIAP1 and cIAP2 proteins are widely expressed in normal human tissues and in human cancers (41–44). The cellular level of cIAPs varies widely, and higher cIAP1 protein levels are associated with resistance to anticancer agents (41). The intracellular concentrations of cIAPs are unknown, and even if they were known, the significance would be complicated by compartmentalization and complexation with other cellular proteins (43, 44). Interestingly, cells from XIAP-deficient mice have been shown to overexpress cIAP1 and cIAP2 proteins (45). This result is consistent with the redundancy of the functional blockade of procaspase-3 processing and activation by the apoptosome shown here for cIAP1 and XIAP (Fig. 2A). A further effort is needed to evaluate the role of cIAP1 and cIAP2 in vivo in the inhibition of effector caspase activation by the apoptosome.
In summary, our results show that cIAP1 specifically and cooperatively blocks the oligomerized processed caspase-9 apoptosome by a mechanism that Smac antagonizes. Additional studies are needed to clarify the extent to which the anti-apoptosome function of cIAP1 down-regulates programmed death of mammalian cells, in which the levels of the cIAPs are known to be a critical determinant of cell fate (46, 47).
This work was supported, in whole or in part, by National Institutes of Health Grant GM60383.
- IAP
- inhibitor of apoptosis protein
- BIR
- baculovirus IAP repeat
- XIAP
- X-linked IAP
- cIAP
- cellular IAP
- Smac
- second mitochondrial derived activator of caspases
- IBM
- IAP-binding motif
- Ac-DEVD-AMC
- acetyl-Asp-Glu-Val-Asp 7-amino-4-methylcoumarin
- HPT
- His-patch thioredoxin.
REFERENCES
- 1.Srinivasula S. M., Ashwell J. D. (2008) Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed J. C., Doctor K. S., Godzik A. (2004) Sci. STKE 2004, re9. [DOI] [PubMed] [Google Scholar]
- 3.Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. (1997) Nature 388, 300–304 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi R., Deveraux Q., Tamm I., Welsh K., Assa-Munt N., Salvesen G. S., Reed J. C. (1998) J. Biol. Chem. 273, 7787–7790 [DOI] [PubMed] [Google Scholar]
- 5.Roy N., Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. (1997) EMBO J. 16, 6914–6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier J. K., Lahoua Z., Gendron N. H., Fetni R., Johnston A., Davoodi J., Rasper D., Roy S., Slack R. S., Nicholson D. W., MacKenzie A. E. (2002) J. Neurosci. 22, 2035–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davoodi J., Lin L., Kelly J., Liston P., MacKenzie A. E. (2004) J. Biol. Chem. 279, 40622–40628 [DOI] [PubMed] [Google Scholar]
- 8.Lüthi A. U., Martin S. J. (2007) Cell Death Differ. 14, 641–650 [DOI] [PubMed] [Google Scholar]
- 9.Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. (2001) Cell 104, 791–800 [DOI] [PubMed] [Google Scholar]
- 10.Chai J., Shiozaki E., Srinivasula S. M., Wu Q., Alnemri E. S., Shi Y., Dataa P. (2001) Cell 104, 769–780 [DOI] [PubMed] [Google Scholar]
- 11.Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y. (2003) Mol. Cell 11, 519–527 [DOI] [PubMed] [Google Scholar]
- 12.Choi Y. E., Butterworth M., Malladi S., Duckett C. S., Cohen G. M., Bratton S. B. (2009) J. Biol. Chem. 284, 12772–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckelman B. P., Salvesen G. S. (2006) J. Biol. Chem. 281, 3254–3260 [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. (2000) Science 288, 874–877 [DOI] [PubMed] [Google Scholar]
- 15.Huang H., Joazeiro C. A., Bonfoco E., Kamada S., Leverson J. D., Hunter T. (2000) J. Biol. Chem. 275, 26661–26664 [DOI] [PubMed] [Google Scholar]
- 16.Hu S., Yang X. (2003) J. Biol. Chem. 278, 10055–10060 [DOI] [PubMed] [Google Scholar]
- 17.Ma L., Huang Y., Song Z., Feng S., Tian X., Du W., Qiu X., Heese K., Wu M. (2006) Cell Death Differ. 13, 2079–2088 [DOI] [PubMed] [Google Scholar]
- 18.Hauser H. P., Bardroff M., Pyrowolakis G., Jentsch S. (1998) J. Cell Biol. 141, 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao Y., Sekine K., Kawabata A., Nakamura H., Ishioka T., Ohata H., Katayama R., Hashimoto C., Zhang X., Noda T., Tsuruo T., Naito M. (2004) Nat. Cell Biol. 6, 849–860 [DOI] [PubMed] [Google Scholar]
- 20.Vucic D., Stennicke H. R., Pisabarro M. T., Salvesen G. S., Dixit V. M. (2000) Curr. Biol. 10, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 21.Kasof G. M., Gomes B. C. (2001) J. Biol. Chem. 276, 3238–3246 [DOI] [PubMed] [Google Scholar]
- 22.Bertrand M. J., Milutinovic S., Dickson K. M., Ho W. C., Boudreault A., Durkin J., Gillard J. W., Jaquith J. B., Morris S. J., Barker P. A. (2008) Mol. Cell 30, 689–700 [DOI] [PubMed] [Google Scholar]
- 23.Varfolomeev E., Goncharov T., Fedorova A. V., Dynek J. N., Zobel K., Deshayes K., Fairbrother W. J., Vucic D. (2008) J. Biol. Chem. 283, 24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q. H., Du C. (2004) J. Biol. Chem. 279, 16963–16970 [DOI] [PubMed] [Google Scholar]
- 25.Chen L., Smith L., Wang Z., Smith J. B. (2003) Mol. Pharmacol. 64, 334–345 [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y., Nakabayashi Y., Takahashi R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benedict M. A., Hu Y., Inohara N., Núñez G. (2000) J. Biol. Chem. 275, 8461–8468 [DOI] [PubMed] [Google Scholar]
- 28.Liu X., Kim C. N., Yang J., Jemmerson R., Wang X. (1996) Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 29.Eckelman B. P., Salvesen G. S., Scott F. L. (2006) EMBO Rep. 7, 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckelman B. P., Drag M., Snipas S. J., Salvesen G. S. (2008) Cell Death Differ. 15, 920–928 [DOI] [PubMed] [Google Scholar]
- 31.Gonçalves R. B., Sanches D., Souza T. L., Silva J. L., Oliveira A. C. (2008) Biochemistry 47, 3832–3841 [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., Rich R. L., Myszka D. G., Wu H. (2003) J. Biol. Chem. 278, 49517–49522 [DOI] [PubMed] [Google Scholar]
- 33.Pop C., Salvesen G. S. (2009) J. Biol. Chem. 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez J., Lazebnik Y. (1999) Genes Dev. 13, 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., Alnemri E. S. (2001) Nature 410, 112–116 [DOI] [PubMed] [Google Scholar]
- 36.Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. (1998) Mol. Cell 1, 949–957 [DOI] [PubMed] [Google Scholar]
- 37.Bratton S. B., Lewis J., Butterworth M., Duckett C. S., Cohen G. M. (2002) Cell Death Differ. 9, 881–892 [DOI] [PubMed] [Google Scholar]
- 38.Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X., Acehan D., Ménétret J. F., Booth C. R., Ludtke S. J., Riedl S. J., Shi Y., Wang X., Akey C. W. (2005) Structure 13, 1725–1735 [DOI] [PubMed] [Google Scholar]
- 40.Wu G., Chai J., Suber T. L., Wu J. W., Du C., Wang X., Shi Y. (2000) Nature 408, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 41.Tamm I., Kornblau S. M., Segall H., Krajewski S., Welsh K., Kitada S., Scudiero D. A., Tudor G., Qui Y. H., Monks A., Andreeff M., Reed J. C. (2000) Clin. Cancer Res. 6, 1796–1803 [PubMed] [Google Scholar]
- 42.Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. (2002) Cancer Res. 62, 4860–4866 [PubMed] [Google Scholar]
- 43.Vischioni B., van der Valk P., Span S. W., Kruyt F. A., Rodriguez J. A., Giaccone G. (2006) Human Pathol. 37, 78–86 [DOI] [PubMed] [Google Scholar]
- 44.Qi S., Mogi S., Tsuda H., Tanaka Y., Kozaki K., Imoto I., Inazawa J., Hasegawa S., Omura K. (2008) Int. J. Oral Maxillofac. Surg. 37, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 45.Harlin H., Reffey S. B., Duckett C. S., Lindsten T., Thompson C. B. (2001) Mol. Cell. Biol. 21, 3604–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 47.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]