Abstract
The chitinase-like protein YKL-40, encoded by the CHI3L1 gene, is a biomarker and functional effector of chronic inflammatory and allergic diseases. In the lung it is associated with asthma severity and reduced lung function. The cellular sources of YKL-40 in human airways and the mechanisms regulating YKL-40 expression are poorly understood. We previously showed that mechanical stress similar to that experienced during bronchoconstriction triggers epithelial cell signaling through epidermal growth factor receptor (EGFR), fibrotic mediator release, and goblet cell hyperplasia consistent with airway remodeling in asthma. We now show that well differentiated normal human bronchial epithelial cells express CHI3L1 and secrete YKL-40 under base-line culture conditions. Mechanical stress (30-cm H2O transcellular compressive stress) applied for 3 h induces CHI3L1 expression by ∼4-fold compared with time matched controls, resulting in increased secretion of YKL-40 by 3.6-fold 24 h after onset of the 3-h stimulus. Inhibition of EGFR or MEK1/2 (ERK kinase) significantly but incompletely attenuates mechanical stress-induced up-regulation of CHI3L1 expression in normal human bronchial epithelial cells. Direct activation of EGFR utilizing EGF-family ligands induces CHI3L1 expression. Our results reveal that human airway epithelial cells are a source of YKL-40 and demonstrate that mechanical stress potently induces CHI3L1 expression leading to increased secretion of YKL-40 protein in an EGFR and MEK1/2-dependent pathway. In the asthmatic airway mechanical stress may contribute to enhanced YKL-40 levels.
Keywords: Epithelium, Gene Expression, Lung, Protein Secretion, Signal Transduction, EGFR, YKL-40, Asthma, Bronchial Epithelium, Mechanotransduction
Introduction
Asthma is characterized by chronic inflammation dominated by Th2 cytokines and hyperresponsiveness of the airways. Airway epithelial cells experience altered physical forces during airway constriction (1), but the role that these physical forces plays in airway inflammation in asthma is not well understood. Previously, our laboratory showed that compressive mechanical stress similar in magnitude to that caused by bronchoconstriction initiates a program of signaling and gene expression supportive of airway remodeling (2–5). These studies identified a novel mechanotransduction mechanism acting through constitutively shed epidermal growth factor receptor (EGFR)2 ligands accumulating in a dynamically shrinking interstitial space (3, 5), with EGFR activation also observed during airway constriction in situ (3). How this mechanical signaling response alters airway function and inflammation and the secreted factors participating in such responses remain to be determined. In the current study we expand on this prior work to investigate the role of compressive mechanical stress in regulating the expression of the key inflammatory gene CHI3L1 and its protein product YKL-40.
YKL-40 is a chitinase like glycoprotein expressed and secreted by a variety of cell types including articular chondrocytes, synoviocytes, osteoblasts, macrophages, neutrophils, and epithelial cells (6–10). Since it was identified as a secreted product of articular chondrocytes and synovial cells in 1993 (11), a growing body of evidence has accumulated showing that high serum levels of YKL-40 are associated with various pathological conditions including chronic inflammation (7, 8, 12, 13), hepatic fibrosis (14, 15), poor prognosis cancers (16), and allergic diseases (17–19).
In the lung YKL-40 is both a biomarker of various disease states and an important functional mediator of asthma-like disease. High serum levels of YKL-40 are associated with asthma (20, 21), chronic obstructive pulmonary disease (22), and lung cancer (23). In asthma, increased serum levels of YKL-40 and expression of CHI3L1 in the lung are correlated with disease severity, airway remodeling, and decreased pulmonary function (24). Ober et al. (21) reported that CHI3L1 is a susceptibility gene for asthma, bronchial hyperresponsiveness, and decreased lung function. Several single nucleotide polymorphisms have been identified for CHI3L1, and these variants are correlated with serum levels of YKL-40 and the incidence of asthma (21, 25). Recently, Lee et al. (9) showed in a mouse model of ovalbumin sensitization and challenge that BRP-39, the mouse homologue for YKL-40, is prominently up-regulated in the airways, that BRP39-deficient mice have blunted Th2 responses and airway hyperresponsiveness, and that reconstitution of YKL-40 expression in pulmonary epithelium of BRP-39-deficient mice restores these prototypical responses to ovalbumin. These studies indicate a critical role for lung epithelial BRP39/YKL-40 expression in airway inflammation and reactivity. However, the role of human airway epithelial cells in YKL-40 production and the intracellular signaling pathways regulating YKL-40 production in human airway epithelium are not yet known.
We show here for the first time that well differentiated human bronchial epithelial cells express CHI3L1 and secrete YKL-40 under air-liquid interface culture conditions. Application of compressive mechanical stress mimicking bronchoconstriction significantly induces CHI3L1 expression and YKL-40 secretion in a magnitude of mechanical stress-dependent manner. We show that mechanical induction of CHI3L1 is driven in part by an EGFR/MEK1/2 dependent mechanism, and EGF-family ligands induce CHI3L1 expression in NHBE cells. Our findings establish a link between the airway mechanical environment and a critical regulator of airway inflammation and provide the first evidence that airway epithelial expression of CHI3L1 is regulated through an EGFR-dependent pathway.
MATERIALS AND METHODS
Culture of Bronchial Epithelial Cells
Primary human normal bronchial epithelial (NHBE) cells were expanded and maintained in a humidified 95% air, 5% CO2 incubator as described previously (26). Briefly, Passage-2 NHBE cells were plated on a 12- Transwell plate (Corning, Inc., Corning, NY) coated with 50 ng/ml type 1 rat tail collagen (BD Biosciences) at a density of 2 × 104 cells/cm2. NHBE cells were maintained in submerged culture conditions until cells reached confluence, at which point an air/liquid interface was established by removing media from the apical surface of cells. NHBE cells were fed with a 1:1 mixture of bronchial epithelial basal medium and high glucose (4.5 g/liter) Dulbecco's modified Eagle's medium supplemented as described previously (4). Primary cells from three different donors were used.
Permeability Assay; Dextran-FITC Flux
To assess the integrity of airway epithelial cells after exposure to the mechanical stress, a dextran-FITC flux assay was performed (27). Briefly, well differentiated NHBE cells were exposed to 30-cm H2O-compressive mechanical stress for 3 h, 1 μg/ml of Dextran-FITC (40 KDa; Invitrogen) was added to the apical surface of cells, and basal media were collected at the indicated time point in the figure. As a positive control for disrupting integrity of epithelial cells, cells were incubated with media containing various concentrations of EDTA (0.5, 1, and 2 mm) basally for 3 h followed by removal of the EDTA containing medium before the apical addition of dextran-FITC. 100 μl of1.5 ml basal medium was used to measure the accumulation of fluorescence signal across the epithelium using a Fluoroskan Ascent Fluorometer (Thermo Scientific, Waltham, MA) using an excitation wavelength of 485 nm and an emission wavelength of 520 nm.
Exposure of NHBE cells to Compressive Mechanical Stress or EGF-family Ligands
For all experiments, cells maintained in the air/liquid interface condition for 13–15 days were used. To expose cells to mechanical stress, non-toxic silicon plugs were press fit into the top of each Transwell with an access port for pressure application, which creates a sealed pressure chamber over the apical surface of the NHBE cells (1, 2). Each plug was connected to a 5% CO2 (balance room air) pressure cylinder via a humidified chamber maintained at 37 °C with gas pressure regulated as noted. After cells were connected to plugs, they were stabilized for at least 2 h before exposure to transcellular pressure. The pressure in the apical chamber was increased by 30 cm of H2O for the indicated duration, whereas the basal surface and medium remained at atmospheric pressure. The compressive stress (30 cm of H2O) is comparable with that present in the airway epithelium during bronchoconstriction and orders of magnitude higher than the stress experienced by the airway epithelium during normal breathing (1, 28). Application of transcellular mechanical stress on the epithelial cells has been routinely used in our laboratory, and physiologic relevance of this system has been documented previously (1, 3).
To investigate the direct activation of EGFR using various EGF ligands on the induction of CHI3L1, recombinant human EGF, Heparin binding EGF-like growth factor and transforming growth factor- α (TGF-α) were applied to cells at indicated concentrations. Recombinant ligands were purchased from R&D systems (R&D Systems, Minneapolis, MN). For detecting CHI3L1 expression, NHBE cells were exposed to compressive stress (30 cm of H2O) or various EGF ligands for 3 h. For detecting phosphorylation of ERK, cells were exposed to compressive stress for up to 30 min. Control cells were treated identically to compressive stress samples, including placement of the wells in the experimental apparatus, but were not exposed to the pressure gradient.
Inhibitor Studies
To investigate the signaling pathway regulating CHI3L1 expression, pharmacologic inhibitors were preincubated in the basal media for 30 min before application of mechanical stress (EGFR inhibitor AG1478 (IC50 = 3 nm), PKC inhibitor bisindolylmaleimide 1 (IC50 = 8–20 nm), MEK1/2 inhibitor U0126 (IC50 = 72 nm for MEK1, 58 nm for MEK2), Rac inhibitor NSC23766 (IC50 = 50 μm, Rho inhibitor), C3 transferase (IC50 = 0.5 μg/ml), Rho-associated kinase inhibitor Y27632 (IC50 = 0.14 μm), focal adhesion kinase (FAK) inhibitor I-14 (IC50 = 1 μm)). DMSO was used for preparing an initial stock solution of AG1478, bisindolylmaleimide 1, and U0126; the other inhibitors were dissolved in water to prepare a stock solution. Stock solutions were dissolved to final concentrations in minimal media at the concentration described in each figure. DMSO or H2O was used for a vehicle control for inhibitors. The final concentration of DMSO in the basal media was less than 0.01% in all conditions.
Actinomycin D Treatment
To determine whether mechanical stress induced CHI3L1 expression through the synthesis of new mRNA, 5 μg/ml of actinomycin D was preincubated with cells for 30 min before mechanical stress.
Western Blot Analysis
To detect phosphorylation of signaling proteins, Western blot analysis was performed using 20∼ 50 μg of protein lysates per lane. The protein concentration of cell lysates was quantified by a BCA assay (Pierce). Each sample was boiled in 2× SDS-PAGE sample buffer (125 mm Tris-Cl (pH 6.8), 25% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.04% bromphenol blue) for 10 min, loaded on 7.5% or 10% SDS-polyacrylamide electrophoresis gels and transferred to a polyvinylidene difluoride membrane (PVDF) (Schleicher & Schuell). After blocking with 5% skim milk, membranes were incubated with the specific primary antibody followed by horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies. All antibodies to detect phosphorylated protein were purchased from Cell Signaling Technology (Danvers, MA). After detecting phosphorylated form of each protein, PVDF membranes were stripped in a prewarmed solution as previously described (29). The deprobed membrane was blocked with 5% skim milk, reprobed with an antibody for the total expression of each protein, and used as a loading control for phosphorylated protein. Final development was performed by incubating with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce), and protein bands were visualized by the ChemiGenius Bioimaging system (Syngene, Frederick, MD). The relative amount of phospho-protein was analyzed using GeneSnap software (Syngene) and normalized to the total amount of each protein.
To detect YKL-40 secreted into the basal media in response to mechanical stress, basal media samples were collected at the indicated periods of time after mechanical stress or in the absence of stress as a control. 20% trichloroacetic acid was used to concentrate equal amounts of protein from the basal media (30). Pellets were washed with ice-cold acetone 3 times and suspended in 30 μl of 2× SDS-PAGE buffer, boiled, and loaded on the 10% SDS-PAGE as descried above. Antibody against human YKL-40 and recombinant human YKL-40 for a positive control were purchased from R&D systems.
Real Time PCR Analysis
RNA was purified from cell lysates with RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Two μg of total RNA was used for synthesizing cDNA using MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA). 20 ng of cDNA was used for the real time PCR reaction in the mixture of primers and the 2× SYBR Green PCR master mix. The forward and reverse primers for CHI3L1 (GenBankTM code NM_001276) and GAPDH endogenous control were generated by Primer Express 3.0 software (Applied Biosystems) (Table 1). All primer sets used in this paper were validated against GAPDH using various amounts of cDNA to ensure similar levels of PCR efficiency. Fold changes were calculated by the comparative ΔΔCt method (31).
TABLE 1.
| Gene | Primer sequence | Reference | |
|---|---|---|---|
| CHI3L1 | Forward | 5′-TTCTGTGGCCAGGATCTG-3′ | Primer express 3 |
| Reverse | 5′-TTGCAGCGAGTGCATCCTT-3′ | ||
| GAPDH | Forward | 5′-TGGGCTACACTGAGCACCAG-3′ | Primer express 3 (49) |
| Reverse | 5′-GGGTGTCGCTGTTGAAGTCA-3′ | ||
Statistical Analysis
Data are presented as the mean ± S.D. Results were evaluated by Student's t tests (p values < 0.05 considered significant) with Bonferroni posttest correction for multiple comparisons.
RESULTS
Bronchial Epithelial Cells Are the Source of YKL-40 Production
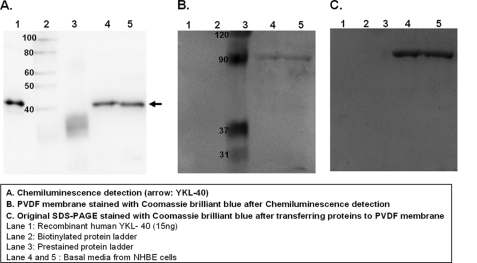
To determine whether NHBE cells synthesize the YKL-40 protein product encoded by CHI3L1, total secreted proteins in basal culture media from air-liquid interface cultures of NHBE cells were concentrated by trichloroacetic acid precipitation and separated using SDS-PAGE(10% separating gel). 15 ng of recombinant human YKL-40 was loaded as a positive control. Western blot analysis was performed using an antibody against human YKL-40 (R&D Systems). As shown in Fig. 1A, secretion of YKL-40 protein from well differentiated NHBE cells was detectable under base-line culture conditions. Coomassie Brilliant Blue R250 staining of the PVDF membrane (Fig. 1B) after chemiluminescence detection of YKL-40 confirmed the loading of protein onto the transfer membrane and low abundance of overall protein staining at the molecular weight of YKL-40. Similarly, Coomassie Brilliant Blue R250 staining of the original SDS-PAGE (Fig. 1C) gel after protein transfer confirmed efficient transfer at the molecular weight of YKL-40 but with residual high molecular weight protein remaining in the gel. This remaining protein band on the gel after transfer was used as a loading control for normalization of YKL-40 in further analysis after mechanical stress.
FIGURE 1.
YKL-40 protein is secreted from bronchial epithelial cells. A, YKL-40 protein secreted from NHBE cells was detected by Western blot analysis. Recombinant human YKL-40 was used as a positive control. B, Coomassie Blue R250 staining of PVDF membrane after chemiluminescence detection is shown. C, shown is Coomassie Blue R250 staining of original SDS-PAGE stained after transferring proteins to the PVDF membrane.
Mechanical Stress Induces CHI3L1 Expression in Well Differentiated NHBE Cells
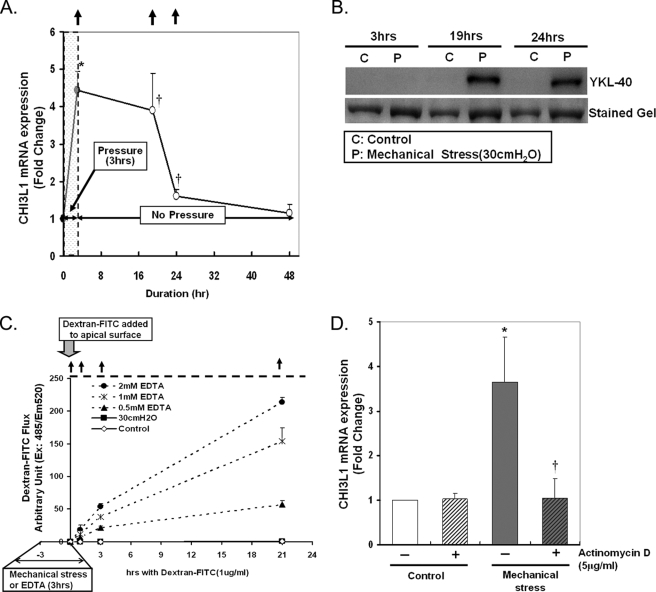
In preliminary experiments, well differentiated NHBE cells grown in air/liquid interface conditions were exposed to 30 cm of H2O transcellular compressive stress for 1, 2, 3, or 8 h. Peak induction of CHI3L1 expression was identified at 3 h (data not shown). As shown in Fig. 2A, these preliminary findings were confirmed as illustrated by our finding that 3 h of mechanical stress increased CHI3L1 expression by ∼4-fold compared with the time matched control. Induction of CHI3L1 expression by mechanical stress was sustained up to 24 h (1.6-fold) after 3 h of mechanical stress. Induction of YKL-40 protein secretion in response to 3 h of mechanical stress was apparent at 19 h of post-mechanical stress, but no increase in secretion was evident at 3 h (Fig. 2B) or 8 h (data not shown), suggesting that the accumulation of YKL-40 in the media in response to mechanical stress was driven by increased transcription of CHI3L1 followed by translation and secretion of YKL-40 rather than simply by enhanced exocytosis of intracellular YKL-40.
FIGURE 2.
Mechanical stress induces CHI3L1 mRNA expression. A, well differentiated NHBE cells were exposed to 30 cm of H2O transcellular compressive stress for 3 h, and pressure was released for the rest of period. CHI3L1 mRNA expression was measured by real-time PCR. Fold increase was calculated by ΔΔCt method. CHI3L1 mRNA expression was significantly up-regulated by 3 h of compressive stress by 4.43-fold and sustained up to 24 h by 1.6-fold (*, p < 0.005 versus control; †, p < 0.05 versus control). Data are represented as the mean ± S.E. from independent experiments (n = 7 (for 3 h) or 4 (for other time points)). The arrows indicate the points of basal media collection used for detection of YKL-40 secretion. B, YKL-40 secretion was detected by Western blotting on the basal media collected at different time points (arrows in A) after 3 h of compressive stress. Induction of YKL-40 secretion response to mechanical stress follows the up-regulation of CHI3L1 mRNA expression. C, a dextran-FITC flux assay was performed to assess epithelial cell integrity after exposure to 30 cm of H2O of transcellular compressive stress or various concentrations of EDTA (0, 0.5, 1, or 2 mm) for 3 h. D, to understand the mechanism of mRNA induction in response to mechanical stress, 5 μg/ml actinomycin D was pretreated to the basal media for 30 min before exposure to mechanical stress for 3 h. CHI3L1 mRNA expression was measured by a real-time PCR as described previously. (*, p < 0.05 versus control; †, p < 0.01 versus mechanical stress alone). Data are represented as the mean ± S.D. from independent experiments (n = 3).
An alternative explanation is that YKL-40 accumulated in the basal media due to breakdown in epithelial barrier function under mechanical loading, leading to apical to basal flux of YKL-40. To test this possibility, fluorescent-labeled Dextran was added to the apical cell surface at the conclusion of mechanical stress, and its flux into the basal media was measured by fluorescence detection. As illustrated in Fig. 2C, exposure to 30 cm of H2O of mechanical stress for 3 h does not disrupt the epithelial integrity assessed by dextran-FITC flux. As a positive control for disruption of epithelial integrity, various concentrations of EDTA were added to the basal media, and EDTA-dependent dextran-FITC flux was detected (Fig. 2C).
To test whether new transcription was involved in the CHI3L1 expression response to the mechanical stress, the transcription inhibitor actinomycin D (5 μg/ml) was preincubated with cells for 30 min before exposure to the mechanical stress. As illustrated in Fig. 2D, pretreatment with actinomycin D completely attenuated mechanical stress-induced CHI3L1 expression in NHBE cells. These results demonstrate that mechanical stress induces CHI3L1 expression and that the response is dependent on the synthesis of new RNA.
Compressive Mechanical Stress Is a Physiologically Strong Stimulator of CHI3L1 Expression and YKL-40 Secretion from NHBE Cells
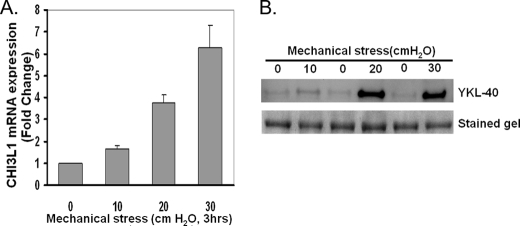
To test the dependence of CHI3L1 induction on the magnitude of mechanical stress applied, we compared the responses to three increasing levels of mechanical stress (10, 20, or 30 cm H2O). CHI3L1 expression (Fig. 3A) was induced in a magnitude-dependent manner with a maximal response at 30 cm of H2O. YKL-40 protein secretion was relatively unchanged by 10 cm of H2O of mechanical stress but prominently increased by 20 and 30 cm of H2O of mechanical stress. These responses to mechanical stress in the induction of CHI3L1 and YKL-40 secretion occur within the physiological range expected in the epithelium of constricted airways (32).
FIGURE 3.
Mechanical stress induces YKL-40 production in a magnitude of compressive mechanical stress-dependent manner. A, CHI3L1 mRNA was measured after exposure to increasing magnitude of compressive mechanical stress for 3 h. CHI3L1 expression is up-regulated in a magnitude-dependent manner. Data are represented as the mean ± S.D. from independent experiments (n = 2). B, YKL-40 secretion was detected at 24 h after 3 h of compressive mechanical stress. Induction of YKL-40 secretion was detected by either 20 or 30 cm of H2O of transcellular compressive stress applied.
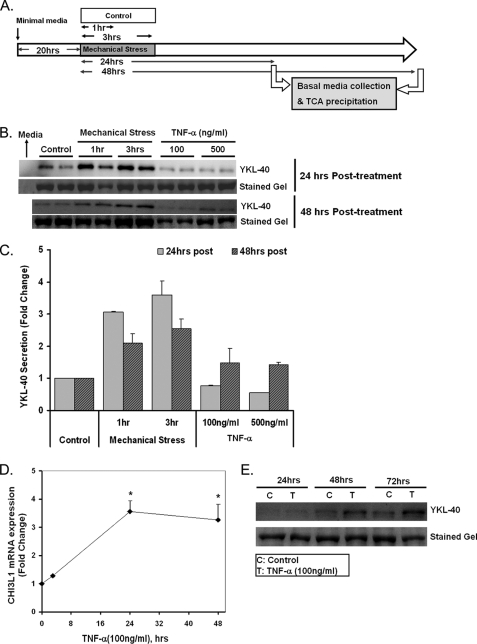
To investigate the kinetics of YKL-40 secretion from NHBE cells, basal media samples were collected at 8, 24, or 48 h after initiation of mechanical stress of either 1 or 3 h of duration (Fig. 4A). Both 1 and 3 h of mechanical stress induced YKL-40 secretion from NHBE cells measured at 24 h by 3.07- and 3.59-fold, respectively (Fig. 4, B and C). This result demonstrates that as little as 1 h duration of mechanical stress provokes YKL-40 production in NHBE cells. For comparison, we tested the role of tumor necrosis factor-α (TNF-α) in stimulation of YKL-40 production. TNF-α has been shown to induce YKL-40 production in a variety of cell types including chondrocytes, colonic epithelial cells, and alveolar macrophages (8, 22, 33). In contrast to mechanical stress, TNF-α elicited a delayed and more muted increase in YKL-40 secretion (Fig. 4, B and C) of ∼1.5-fold at 48 h. Culture media alone did not produce an immunoreactive band, confirming that YKL-40 secretion was produced specifically by NHBE cells. The YKL-40 response to transient mechanical stress appeared to peak at 24 h as YKL-40 levels were reduced by 48 h (Fig. 4, B and C). The delayed induction of YKL-40 secretion response to 100 ng/ml of TNF-α was sustained up to 72 h (Fig. 4, C and E) and paralleled by slower increases in CHI3L1 gene expression relative to mechanical stress, with peak induction of CHI3L1 of 3.56-fold at 24 h (Fig. 4D).
FIGURE 4.
Secretion of YKL-40 is induced by mechanical stress. A, shown is an experimental plan for investigating kinetics of YKL-40 secretion by mechanical stress. Basal media samples were collected at 24 or 48 h after 1 or 3 h of mechanical stress. TCA, trichloroacetic acid. B, secretion of YKL-40 was increased 3.59-fold by mechanical stress at 24 h of post-3 h of pressure; 100 or 500 ng/ml TNF-α modestly increased YKL-40 secretion at 48 h. C, shown is densitometry analysis of YKL-40 secretion at 24 or 48 h post-treatment. D, CHI3L1 expression was induced by TNF-α treatment (100 ng/ml) for 24 and 48 h by 3.56- and 3.27-fold, respectively (*, p < 0.001 versus control). Data are represented as the mean ± S.D. from independent experiments (n = 3). E, YKL-40 secretion was induced by TNF-α treatment (100 ng/ml) and sustained up to 72 h.
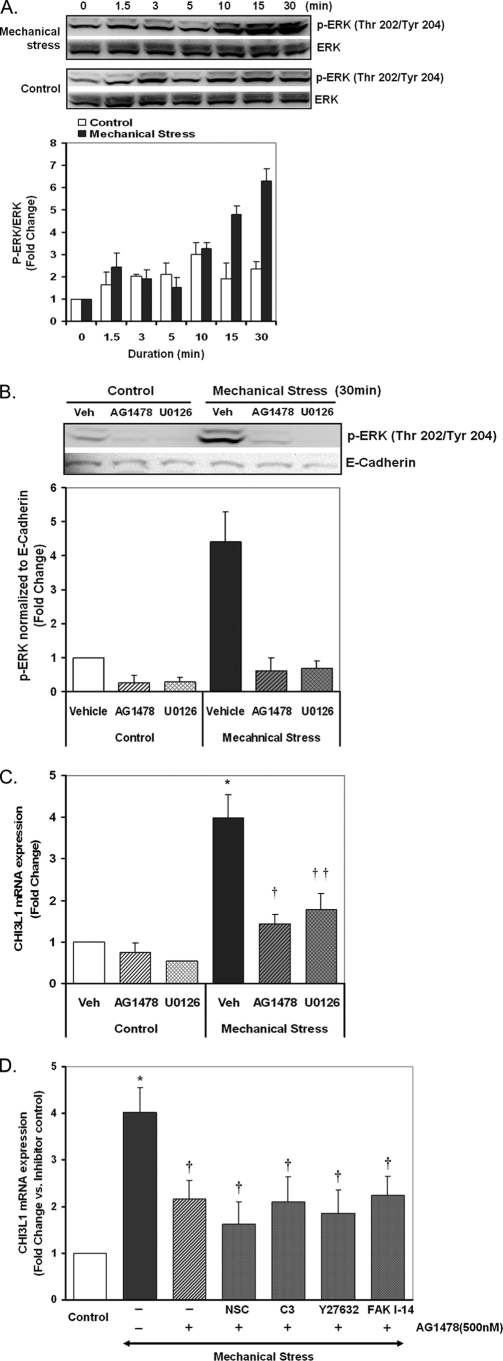
CHI3L1 Induction in Response to Mechanical Stress Is Mediated via an EGFR/MEK1/2 Pathway
Compressive mechanical stress is known to rapidly activate the EGFR, leading to enhanced phosphorylation of ERK1/2 (3). We confirmed the ERK1/2 response in NHBE cells (Fig. 5A), then designed experiments to test the role this pathway plays in enhanced expression of CHI3L1. We confirmed the efficacy of inhibitors of EGFR (AG1478, 500 nm) and ERK kinase ((also known as MEK1/2) U0126, 10 μm) by directly measuring the ERK phosphorylation response to mechanical stress (Fig. 5B). As illustrated in Fig. 5C, pretreatment with AG1478 (500 nm) and U0126 (10 μm) significantly but incompletely attenuated compressive stress-induced CHI3L1 expression as measured by real-time PCR. Mechanical stress-induced CH3L1 expression was 1.44- and 1.78-fold in the presence of AG1478 and U0126, respectively, whereas CHI3L1 expression was enhanced 3.99-fold by mechanical stress with vehicle control. These results demonstrate that the CHI3L1 response to mechanical stress is driven in part by an EGFR/ERK pathway but suggests that other pathways also contribute to this response to mechanical stress.
FIGURE 5.
Inhibition of EGFR or MEK1/2 attenuates mechanical stress-induced CHI3L1 expression. A, ERK phosphorylation response to mechanical stress was measured by Western blot analysis. B, ERK phosphorylation in the presence or absence of AG1478 (500 nm) and U0126 (10 μm) was measured by Western blotting and quantified by densitometry analysis. E-cadherin was used as a loading control. C, CHI3L1 expression in the presence or absence of AG1478 (500 nm) and U0126 (10 μm) was measured by a real-time PCR. Data are represented as the mean ± S.D. from independent experiments (n = 4) (*, p < 0.005 vehicle (Veh) + pressure versus control; †, p < 0.005 AG1478 + pressure versus vehicle + pressure; ††, p < 0.05 U0126 + pressure versus vehicle + pressure). D, CHI3L1 expression in the presence or absence of AG1478 (500 nm) alone or in combination with each inhibitor (NSC23766 (NSC, 100 μm), C3 transferase (C3, 2 μg/ml), Y27632 (10 μm), or FAK inhibitor-14 (FAK I-14, 50 μm)) was measured by real-time PCR. Data are represented as the mean ± S.E. from independent experiments (n = 3) (*, p < 0.005 vehicle + pressure versus control; †, p < 0.05 versus vehicle + pressure).
Among the many pathways previously linked to mechanical signaling, the small Rho family GTPases and FAK have been widely implicated in mechanotransduction (34, 35). Rac, Rho, and the downstream Rho-associated kinase are the components of the Rho GTPases most commonly implicated in mechanical signaling. Thus, we used C3 transferase to inhibit Rho, Y27632 to inhibit Rho-associated kinase, and NSC23766 to inhibit Rac. None of these compounds alone significantly attenuated the CHI3L1 expression response to mechanical stress (data not shown). When added in combination with the EGFR inhibitor AG1478, none of these compounds significantly enhanced the effect of AG1478 alone (Fig. 5D); the mechanical response was inhibited by 62–79%, with no statistically significant difference between treatment conditions containing AG1478 and other inhibitors or AG1478 alone. FAK inhibitor 14, which targets the tyrosine 397 activation domain of FAK, by itself modestly attenuated the CHI3L1 response to mechanical loading (p < 0.05, data not shown), but it also failed to enhance the inhibitory effect of AG1478 (Fig. 5D). We conclude that Rac, Rho, Rho-associated kinase, and FAK signaling do not contribute significantly to the EGFR-independent expression of CHI3L1 in response to mechanical loading.
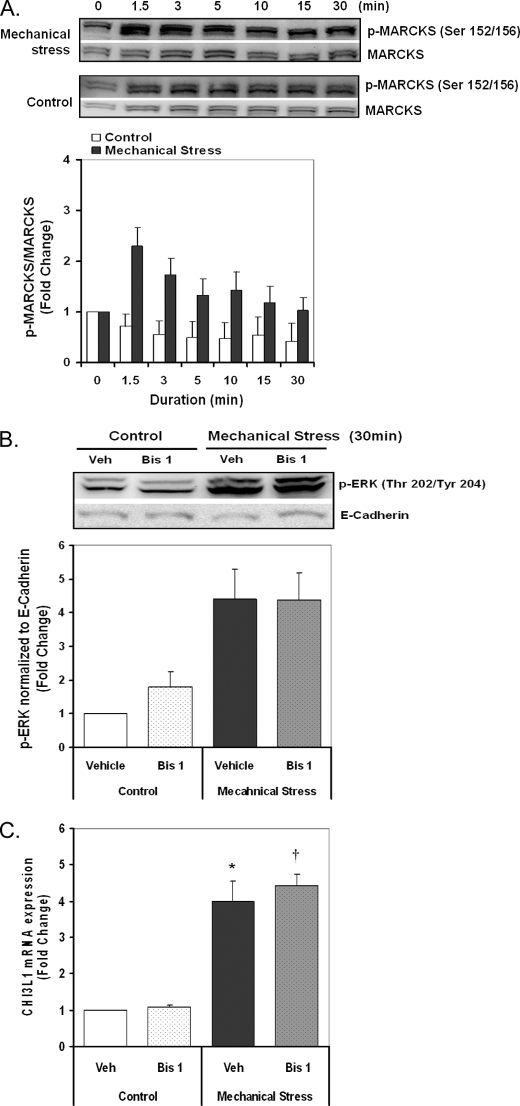
Protein kinase C (PKC) is another candidate to mediate mechanoactivation of gene expression through both ERK-dependent and ERK-independent mechanisms (36). To assess the activation of PKC by mechanical stress, phosphorylation of myristoylated alanine-rich protein kinase C substrate (MARCKS), which is a well known substrate of PKC, was measured by Western blot analysis (29, 37). MARCKS was rapidly phosphorylated by compressive stress with a maximum response at 1.5 min and dephosphorylated at later time points. Inhibition of PKC by bisindolylmaleimide 1 (1 μm) completely attenuated the MARCKS phosphorylation response to mechanical stress (not shown) but did not alter the ERK phosphorylation response (Fig. 6B). However, pretreatment with bisindolylmaleimide 1 did not alter the CHI3L1 response to mechanical stress (Fig. 6C), demonstrating that the induction of CHI3L1 is independent of mechanoactivation of PKC.
FIGURE 6.
PKC is activated by mechanical stress but does not play a role in CHI3L1 induction. A, phosphorylation of MARCKS was detected to assess the PKC activation response to mechanical stress over the various time points and quantified by densitometry analysis. Total MARCKS protein was detected after stripping of phospho-MARCKS antibody as a loading control. B, ERK phosphorylation response to mechanical stress was measured by Western blotting and quantified by densitometry analysis in the presence or absence of the PKC inhibitor bisindolylmaleimide 1 (Bis 1; 1 μm). E-Cadherin was used as a loading control. C, CHI3L1 expression was measured by real-time PCR in the presence or absence of the PKC inhibitor bisindolylmaleimide 1 (1 μm). Data are represented as the mean ± S.D. from independent experiments (n = 3) (*, p < 0.005 vehicle + pressure versus control; †, p < 0.05 bisindolylmaleimide 1 + pressure versus vehicle + pressure).
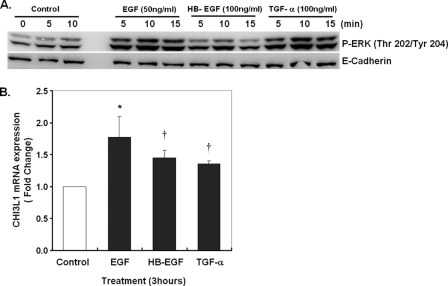
EGF-family Ligands Induce CHI3L1 Expression
To directly test the role of the EGFR pathway in CHI3L1 expression, we stimulated NHBE cells with various EGF-family ligands. Direct ligand stimulation with EGF (50 ng/ml), HB-EGF (100 ng/ml), or TGF-α (100 ng/ml) applied to differentiated NHBE cells led to ERK phosphorylation peaking at 10 min (Fig. 7A). As illustrated in Fig. 7B, EGF, heparin binding-EGF, and TGF-α all significantly, but moderately induce CHI3L1 expression in well differentiated NHBE cells. The strongest response to EGF was 45% that mediated by mechanical stress. Together these results confirm that the EGFR pathway is upstream of CHI3L1 expression in NHBE cells and emphasize that mechanical stress provides a potent stimulus for CHI3L1 induction.
FIGURE 7.
Direct ligand activation of EGFR enhances CHI3L1 expression. NHBE cells were exposed to EGF (50 ng/ml), heparin binding (HB)-EGF (100 ng/ml), or TGF-α (100 ng/ml) for 5–15 min for analysis of ERK phosphorylation or 3 h for analysis of CHI3L1 expression. A, phosphorylation of ERK in response to EGF-family members detected by Western blot. E-cadherin was used as a loading control. B, CHI3L1 expression was measured by real-time PCR (*, p < 0.01 versus Control; †, p < 0.05 versus control). Data are represented as the mean ± S.D. from independent experiments (n = 3).
DISCUSSION
Our experiments show that NHBE cells express CHI3L1 and secrete its protein product, YKL-40, to the basal media under air/liquid interface culture conditions. Compressive mechanical stress (30 cm of H2O) comparable in magnitude to that associated with bronchoconstriction enhanced CHI3L1 expression, and this response was dependent on new RNA synthesis. Up-regulation of CHI3L1 in response to mechanical stress resulted in enhanced secretion of YKL-40 protein product. Induction of CHI3L1 expression and YKL-40 secretion was dependent on the magnitude of transcellular mechanical stress applied over the physiological range experienced by the epithelium of constricted airways (32). Based on our previous demonstration that compressive mechanical stress activates EGFR (3), we tested whether this intracellular signaling pathway regulates CHI3L1 expression in bronchial epithelial cells. We found that mechanical stress induced CHI3L1 expression in an EGFR/MEK1/2-dependent manner in bronchial epithelial cells. Further supporting this conclusion, direct activation of EGFR by EGF-family ligands significantly induced CHI3L1 expression.
The regulation of CHI3L1 expression has been previously studied in a variety of cell types (8, 22, 33, 38, 39), but our understanding of the molecular mechanisms influencing its expression remains limited. The pro-inflammatory cytokine TNF-α has been shown to induce CHI3LI in chondrocytes, colonic epithelial cells, alveolar macrophages, and various cancer cells (8, 22, 33, 38), and we observed enhanced CHI3L1 expression and YKL-40 secretion by TNF-α with similar kinetics to those observed in chondrocytes (33) and macrophages (22). However, compared with the response to mechanical stress, the response to TNF-α was slower and weaker, indicating the relative potency of mechanical stress in CHI3L1 induction and YKL-40 secretion.
In mouse airways the expression of BRP-39, the homologue for human YKL-40, is enhanced by overexpression of IL-13 in lung epithelium. However, no data are currently available to assess whether this represents a direct effect of IL-13-mediated signaling on airway epithelial cells or a secondary effect of IL-13 overexpression. Intriguingly, several lines of evidence indicate that IL-13 results in shedding of EGF-family ligands and EGFR transactivation in airway epithelial cells grown in monolayer culture (40), although most bronchial epithelial responses to IL-13 stimulation appear distinct from those requiring EGFR activation (41, 42). Based on our observation that direct activation of the EGFR augments CHI3L1 expression, IL-13-mediated EGFR transactivation could play a role in the expression of CHI3L1 in inflamed airways.
Although EGFR clearly contributes to mechanoactivation of CHI3L1 in our cell culture system, it does not account for all of the response to compressive stress. Thus, mechanical stress appears to activate additional EGFR-independent pathways, which further enhance CHI3L1 expression. We evaluated PKC activation as one candidate for EGFR-independent mechanosignaling, and although we documented significant activation of this pathway, we found no evidence that it contributes to CHI3L1 expression, although it may contribute to other responses to mechanical stress (5). We also tested the contributions of Rac, Rho, Rho-associated kinase, and FAK and found that none could account for the EGFR-independent CHI3L1 response to mechanical loading. Together these results emphasize the important role for EGFR activation in response to mechanical stress in this system, which accounts for ∼60–70% of the CHI3L1 response to mechanical loading, and suggest that the additional pathways activated by mechanical stress are outside the widely known small Rho family GTPases and FAK. Additional studies to elucidate the molecular mechanisms linking mechanical stress to CHI3L1 expression will be needed to further our understanding of the regulatory control of this gene.
Although YKL-40 is best known as a biomarker of inflammation and lung disease (21, 22), its functional roles are slowly coming into focus. In a variety of cell types and contexts application of exogenous YKL-40 has been implicated in angiogenesis (43), cell proliferation (44, 45), apoptosis (9), macrophage differentiation (10), and inhibition of collagen degradation (46, 47). A closely related family member has been described that possesses potent eosinophil chemotactic activity (48). The finding that BRP39−/− mice are protected from ovalbumin-induced airway hyperresponsiveness and that adding back human YKL-40 restores the response is the clearest evidence of an important functional role for YKL-40 in airway disease (9). In particular, YKL-40 was shown to play an essential role in antigen sensitization and IgE induction, dendritic cell accumulation and activation, and alternative activation of macrophages as well as protection from inflammatory cell apoptosis. In alveolar macrophages YKL-40 promotes the release of inflammatory cytokines (IL-8 and MCP-1) and matrix metalloproteases-9 (22). Together these findings suggest that overproduction of YKL-40 from the airway epithelium could be an important cue for prolonged inflammation. Based on our observations that mechanical stress activates CHI3L1 expression in vitro and EGFR in constricted airways in situ (3), we propose that CHI3L1/YKL-40 represents a novel link between the mechanical stress caused by airway constriction and the airway inflammation and airway remodeling characteristic of chronic asthma.
In summary, our findings demonstrate for the first time that NHBE cells are a source of YKL-40 secretion and that compressive mechanical stress comparable with that experienced during bronchoconstriction is a physiologically relevant stimulator of CHI3L1 expression and YKL-40 synthesis. The response to compressive mechanical stress is mediated in part through an EGFR/MEK1/2 pathway in NHBE cells, representing the first link between this pivotal signaling pathway and CHI3L1 expression. Based on the emerging evidence of important functional roles for YKL-40, further study of the roles of EGFR activation and YKL-40 expression in regulation of airway biology and disease appears strongly warranted.
This work was supported, in whole or in part, by National Institutes of Health Grants HL082856 and HL88028.
- EGFR
- EGF receptor
- NHBE cells
- normal human bronchial epithelial cells
- FAK
- focal adhesion kinase
- MARCKS
- myristoylated alanine-rich protein kinase C substrate.
REFERENCES
- 1.Ressler B., Lee R. T., Randell S. H., Drazen J. M., Kamm R. D. (2000) Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L1264–L1272 [DOI] [PubMed] [Google Scholar]
- 2.Tschumperlin D. J., Shively J. D., Swartz M. A., Silverman E. S., Haley K. J., Raab G., Drazen J. M. (2002) Am. J. Physiol. Lung Cell. Mol. Physiol. 282, L904–L911 [DOI] [PubMed] [Google Scholar]
- 3.Tschumperlin D. J., Dai G., Maly I. V., Kikuchi T., Laiho L. H., McVittie A. K., Haley K. J., Lilly C. M., So P. T., Lauffenburger D. A., Kamm R. D., Drazen J. M. (2004) Nature 429, 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J. A., Tschumperlin D. J. (2009) Am. J. Respir. Cell Mol. Biol. 41, 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojic N., Chung E., Kho A. T., Park J. A., Huang A., So P. T., Tschumperlin D. J. (2010) FASEB J. 24, 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansen J. S., Olee T., Price P. A., Hashimoto S., Ochs R. L., Lotz M. (2001) Arthritis Rheum. 44, 826–837 [DOI] [PubMed] [Google Scholar]
- 7.Rathcke C. N., Vestergaard H. (2006) Inflamm. Res. 55, 221–227 [DOI] [PubMed] [Google Scholar]
- 8.Mizoguchi E. (2006) Gastroenterology 130, 398–411 [DOI] [PubMed] [Google Scholar]
- 9.Lee C. G., Hartl D., Lee G. R., Koller B., Matsuura H., Da Silva C. A., Sohn M. H., Cohn L., Homer R. J., Kozhich A. A., Humbles A., Kearley J., Coyle A., Chupp G., Reed J., Flavell R. A., Elias J. A. (2009) J. Exp. Med. 206, 1149–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehli M., Niller H. H., Ammon C., Langmann S., Schwarzfischer L., Andreesen R., Krause S. W. (2003) J. Biol. Chem. 278, 44058–44067 [DOI] [PubMed] [Google Scholar]
- 11.Hakala B. E., White C., Recklies A. D. (1993) J. Biol. Chem. 268, 25803–25810 [PubMed] [Google Scholar]
- 12.Huang K., Wu L. D. (2009) J. Int. Med. Res. 37, 18–24 [DOI] [PubMed] [Google Scholar]
- 13.Koutroubakis I. E., Petinaki E., Dimoulios P., Vardas E., Roussomoustakaki M., Maniatis A. N., Kouroumalis E. A. (2003) Int. J. colorectal Dis. 18, 254–259 [DOI] [PubMed] [Google Scholar]
- 14.Pungpapong S., Nunes D. P., Krishna M., Nakhleh R., Chambers K., Ghabril M., Dickson R. C., Hughes C. B., Steers J., Nguyen J. H., Keaveny A. P. (2008) Liver Transpl. 14, 1294–1302 [DOI] [PubMed] [Google Scholar]
- 15.Johansen J. S., Christoffersen P., Møller S., Price P. A., Henriksen J. H., Garbarsch C., Bendtsen F. (2000) J. Hepatol. 32, 911–920 [DOI] [PubMed] [Google Scholar]
- 16.Cintin C., Johansen J. S., Christensen I. J., Price P. A., Sørensen S., Nielsen H. J. (1999) Br. J. Cancer 79, 1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shuhui L., Mok Y. K., Wong W. S. (2009) Int. Arch. Allergy Immunol. 149, 369–377 [DOI] [PubMed] [Google Scholar]
- 18.Elias J. A., Homer R. J., Hamid Q., Lee C. G. (2005) J. Allergy Clin. Immunol. 116, 497–500 [DOI] [PubMed] [Google Scholar]
- 19.Sohn M. H., Lee J. H., Kim K. W., Kim S. W., Lee S. H., Kim K. E., Kim K. H., Lee C. G., Elias J. A., Lee M. G. (2009) Am. J. Respir. Crit. Care Med. 179, 449–456 [DOI] [PubMed] [Google Scholar]
- 20.Hartl D., Lee C. G., Da Silva C. A., Chupp G. L., Elias J. A. (2009) Curr. Opin. Allergy Clin. Immunol. 9, 60–66 [DOI] [PubMed] [Google Scholar]
- 21.Ober C., Tan Z., Sun Y., Possick J. D., Pan L., Nicolae R., Radford S., Parry R. R., Heinzmann A., Deichmann K. A., Lester L. A., Gern J. E., Lemanske R. F., Jr., Nicolae D. L., Elias J. A., Chupp G. L. (2008) N. Engl. J. Med. 358, 1682–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Létuvé S., Kozhich A., Arouche N., Grandsaigne M., Reed J., Dombret M. C., Kiener P. A., Aubier M., Coyle A. J., Pretolani M. (2008) J. Immunol. 181, 5167–5173 [DOI] [PubMed] [Google Scholar]
- 23.Johansen J. S., Drivsholm L., Price P. A., Christensen I. J. (2004) Lung Cancer 46, 333–340 [DOI] [PubMed] [Google Scholar]
- 24.Chupp G. L., Lee C. G., Jarjour N., Shim Y. M., Holm C. T., He S., Dziura J. D., Reed J., Coyle A. J., Kiener P., Cullen M., Grandsaigne M., Dombret M. C., Aubier M., Pretolani M., Elias J. A. (2007) N. Engl. J. Med. 357, 2016–2027 [DOI] [PubMed] [Google Scholar]
- 25.Rathcke C. N., Holmkvist J., Husmoen L. L., Hansen T., Pedersen O., Vestergaard H., Linneberg A. (2009) PloS One 4, e6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J. A., He F., Martin L. D., Li Y., Chorley B. N., Adler K. B. (2005) Am. J. Pathol. 167, 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sallee J. L., Burridge K. (2009) J. Biol. Chem. 284, 14997–15006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiggs B. R., Hrousis C. A., Drazen J. M., Kamm R. D. (1997) J. Appl. Physiol. 83, 1814–1821 [DOI] [PubMed] [Google Scholar]
- 29.Park J. A., Crews A. L., Lampe W. R., Fang S., Park J., Adler K. B. (2007) Am. J. Pathol. 171, 1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim M., Smart R. C. (2003) J. Biol. Chem. 278, 19674–19681 [DOI] [PubMed] [Google Scholar]
- 31.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32.Ding D. J., Martin J. G., Macklem P. T. (1987) J. Appl. Physiol. 62, 1324–1330 [DOI] [PubMed] [Google Scholar]
- 33.Recklies A. D., Ling H., White C., Bernier S. M. (2005) J. Biol. Chem. 280, 41213–41221 [DOI] [PubMed] [Google Scholar]
- 34.Chen C. S., Tan J., Tien J. (2004) Annu. Rev. Biomed. Eng. 6, 275–302 [DOI] [PubMed] [Google Scholar]
- 35.Clark E. A., Brugge J. S. (1995) Science 268, 233–239 [DOI] [PubMed] [Google Scholar]
- 36.Kim S. W., Hong J. S., Ryu S. H., Chung W. C., Yoon J. H., Koo J. S. (2007) Mol. Cell. Biol. 27, 6933–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herget T., Oehrlein S. A., Pappin D. J., Rozengurt E., Parker P. J. (1995) Eur. J. Biochem. 233, 448–457 [DOI] [PubMed] [Google Scholar]
- 38.Bhat K. P., Pelloski C. E., Zhang Y., Kim S. H., deLaCruz C., Rehli M., Aldape K. D. (2008) FEBS Lett. 582, 3193–3200 [DOI] [PubMed] [Google Scholar]
- 39.Junker N., Johansen J. S., Hansen L. T., Lund E. L., Kristjansen P. E. (2005) Cancer Sci. 96, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allahverdian S., Harada N., Singhera G. K., Knight D. A., Dorscheid D. R. (2008) Am. J. Respir. Cell Mol. Biol. 38, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Booth B. W., Adler K. B., Bonner J. C., Tournier F., Martin L. D. (2001) Am. J. Respir. Cell Mol. Biol. 25, 739–743 [DOI] [PubMed] [Google Scholar]
- 42.Zhen G., Park S. W., Nguyenvu L. T., Rodriguez M. W., Barbeau R., Paquet A. C., Erle D. J. (2007) Am. J. Respir. Cell Mol. Biol. 36, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao R., Hamel K., Petersen L., Cao Q. J., Arenas R. B., Bigelow C., Bentley B., Yan W. (2009) Oncogene 28, 4456–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Recklies A. D., White C., Ling H. (2002) Biochem. J. 365, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Ceuninck F., Gaufillier S., Bonnaud A., Sabatini M., Lesur C., Pastoureau P. (2001) Biochem. Biophys. Res. Commun. 285, 926–931 [DOI] [PubMed] [Google Scholar]
- 46.Bigg H. F., Wait R., Rowan A. D., Cawston T. E. (2006) J. Biol. Chem. 281, 21082–21095 [DOI] [PubMed] [Google Scholar]
- 47.Iwata T., Kuwajima M., Sukeno A., Ishimaru N., Hayashi Y., Wabitsch M., Mizusawa N., Itakura M., Yoshimoto K. (2009) Biochem. Biophys. Res. Commun. 388, 511–516 [DOI] [PubMed] [Google Scholar]
- 48.Owhashi M., Arita H., Hayai N. (2000) J. Biol. Chem. 275, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 49.Chu E. K., Cheng J., Foley J. S., Mecham B. H., Owen C. A., Haley K. J., Mariani T. J., Kohane I. S., Tschumperlin D. J., Drazen J. M. (2006) Am. J. Respir. Cell Mol. Biol. 35, 628–638 [DOI] [PMC free article] [PubMed] [Google Scholar]