Abstract
Posttranslational modifications, e.g. proteolysis, glycosylation, and citrullination regulate chemokine function, affecting leukocyte migration during inflammatory responses. Here, modification of CXCL5/epithelial cell-derived neutrophil-activating protein-78 (ENA-78) by proteases or peptidylarginine deiminases (PAD) was evaluated. Slow CXCL5(1–78) processing by the myeloid cell marker aminopeptidase N/CD13 into CXCL5(2–78) hardly affected its in vitro activity, but slowed down the activation of CXCL5 by the neutrophil protease cathepsin G. PAD, an enzyme with a potentially important function in autoimmune diseases, site-specifically deiminated Arg9 in CXCL5 to citrulline, generating [Cit9]CXCL5(1–78). Compared with CXCL5(1–78), [Cit9]CXCL5(1–78) less efficiently induced intracellular calcium signaling, phosphorylation of extracellular signal-regulated kinase, internalization of CXCR2, and in vitro neutrophil chemotaxis. In contrast, conversion of CXCL5 into the previously reported natural isoform CXCL5(8–78) provided at least 3-fold enhanced biological activity in these tests. Citrullination, but not NH2-terminal truncation, reduced the capacity of CXCL5 to up-regulate the expression of the integrin α-chain CD11b on neutrophils. Truncation nor citrullination significantly affected the ability of CXCL5 to up-regulate CD11a expression or shedding of CD62L. In line with the in vitro results, CXCL5(8–78) and CXCL5(9–78) induced a more pronounced neutrophil influx in vivo compared with CXCL5(1–78). Administration of 300 pmol of either CXCL5(1–78) or [Cit9]CXCL5(1–78) failed to attract neutrophils to the peritoneal cavity. Citrullination of the more potent CXCL5(9–78) lowers its chemotactic potency in vivo and confirms the tempering effect of citrullination in vitro. The highly divergent effects of modifications of CXCL5 on neutrophil influx underline the potential importance of tissue-specific interactions between chemokines and PAD or proteases.
Keywords: Chemokines, Chemotaxis, Immunology, Inflammation, Neutrophil, Posttranslational Modification, Peptidylarginine Deiminase
Introduction
Upon infection or tissue injury, launching an appropriate inflammatory response is indispensable. Key players in the first line of host defense against invading microbial agents are neutrophils circulating in the peripheral blood (1–3). These leukocytes sense and start migrating toward the site of infection in response to exogenous and endogenous mediators (chemotactic factors). Upon arrival, neutrophils can phagocytose and degrade microbial agents using oxidative and non-oxidative mechanisms. By secreting enzymes and chemotactic factors such as chemokines, neutrophils boost the subsequent invasion of additional, well defined subsets of immune cells, thereby modulating and amplifying the immune response (4–7). Trafficking of neutrophils and other leukocytes is strongly regulated by chemokines produced at the site of inflammation. Immobilized on glycosaminoglycans, chemokines build up a chemotactic gradient and are presented on the endothelium of post-capillary venules (8). This results in the selective, chemokine receptor-dependent recruitment and activation of various leukocyte subsets. Chemokines are divided into 4 subsets, based on the number of cysteines and their location in the primary amino acid sequence. If the characteristic NH2-terminal cysteine residues are separated by a random amino acid, the chemokines are labeled CXC chemokines. The subset of ELR+CXC chemokines possesses a Glu-Leu-Arg (ELR) motif preceding the characteristic CXC sequence. These chemokines, in particular, are responsible for the recruitment and activation of neutrophils through binding to and activation of the G protein-coupled receptors (GPCR)4 CXCR1 or CXCR2. Although neutrophils play an essential role in the innate immune response, excessive neutrophil accumulation and activation has detrimental effects and may lead to the development of inflammatory diseases or autoimmunity (9). On the contrary, a reduced neutrophil number or defective neutrophil function can lead to recurrent infections, as seen in patients with leukocyte adhesion deficiency or neutropenia (10, 11). The correct coordination and timely resolution of the neutrophil influx is therefore critical in maintaining the balance between health and disease. Obviously, efficient up- and down-regulation of chemokine function are required for rapid induction and termination of an inflammatory response. Different regulatory mechanisms have been described. Classic mechanisms, such as regulation of transcriptional activity and mRNA stability, coordinate chemokine and chemokine receptor gene expression (12–16). Such expression is influenced by pro-inflammatory cytokines and bacterial or viral products in the microenvironment of the cell. Moreover, in vivo chemokine activity depends on proper secretion and on presentation by glycosaminoglycans (17–19). Furthermore, decoy receptors, such as D6, Duffy Antigen Receptor for chemokines (DARC), and Chemocentryx chemokine receptor (CCX-CKR), control chemokine function (20). These decoy receptors bind and constitutively internalize specific chemokine subsets, but are not coupled to conventional signaling pathways. Thus, they may affect the local availability of chemokines, functioning as a “molecular trap.” Under specific conditions, signal transduction pathways of some classic chemokine receptors can be uncoupled too (21). This way, decoy-like or “frozen” receptors are generated. In addition, chemokine receptor dimerization, chemokine gene duplication resulting in a new chemokine variant, and synergy between chemokines further refine chemokine function (22–25). Finally, posttranslational processing of secreted chemokines significantly contributes to chemokine function control (26). Chemokine activity can be both positively and negatively affected upon enzymatic modification, such as proteolytic processing. This can result in a more/less active chemoattractant by changing the affinity for the receptor (GPCR or decoy receptor) or the receptor-signaling potency (26, 27). Minimal NH2-terminal truncation of chemokines can generate a structural variant with antagonistic properties, as well. More profound proteolysis can result in total destruction and inactivation. Interestingly, inducible inflammatory chemokines are more sensitive to and more frequently affected by posttranslational modification than constitutive chemokines, which emphasizes its role in the regulation of an inflammatory response.
This study focuses on the regulation of the function of the ELR+CXC chemokine CXCL5/epithelial cell-derived neutrophil-activating protein-78 (ENA-78) by posttranslational modification. CXCL5 attracts neutrophils through interaction with CXCR2 (28, 29). We here describe the susceptibility of this ELR+CXC chemokine to modification by the aminopeptidase CD13 and peptidylarginine deiminase (PAD). CD13/aminopeptidase N is a myeloid cell marker (30) and has already been shown to process CD26-truncated CXCL11, removing NH2-terminal amino acids one by one and thereby reducing the anti-angiogenic activity of this molecule (31). PAD and CXCL5 are up-regulated in the synovial fluid of rheumatoid arthritis patients (32, 33) and PAD has recently been discovered to modify chemokines, such as CXCL8, CXCL10, CXCL11, and CXCL12, by converting one specific or multiple Arg residues into citrulline (Cit) (34–36). Furthermore, the effects of posttranslational modification by NH2-terminal processing and citrullination on the biological activity of CXCL5 are investigated by comparing the chemically synthesized isoforms CXCL5(2–78), CXCL5(8–78), CXCL5(9–78), [Cit9]CXCL5(1–78), and [Cit9]CXCL5(9–78) with the unmodified CXCL5(1–78) molecule in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Reagents and Cells
Human neutrophil cathepsin G was obtained from Calbiochem (San Diego, CA). Porcine kidney CD13/aminopeptidase N and PAD2 purified from rabbit skeletal muscle were from Sigma. Recombinant CXCL5 was from R&D Systems (Minneapolis, MN). CXCL11(3–73) was chemically synthesized as described (31). The competitive serine protease inhibitor benzamidine was purchased from Acros Organics (Geel, Belgium). Human embryonic kidney (HEK) 293 cells transfected with human CXCR2 were provided by Dr. J. M. Wang (National Cancer Institute, Frederick, MD) (37) and cultured in Dulbecco's modified Eagle's medium (DMEM; Lonza, Basel, Switzerland) supplemented with 10% (v/v) fetal bovine serum (FBS) and 800 μg/ml of geneticin (Invitrogen, Merelbeke, Belgium).
Enzymatic Modification of CXCL5 in Vitro
To evaluate the potential enzymatic modification, chemokines were incubated with cathepsin G, PAD2, or CD13 at 37 °C for the indicated time intervals at enzyme-substrate (E/S) molar ratios of 1/100, 1/20 or 1/25, respectively. Cathepsin G and CD13 cleavage were accomplished in phosphate-buffered saline (PBS) (pH 7.4), whereas incubations with PAD were carried out in 40 mm Tris with 2 mm CaCl2 (pH 7.4). In addition, 10 mm benzamidine was added when incubating CXCL5 with CD13 to inhibit contaminating serine proteases present in the purchased CD13 (31). CXCL5 processing was stopped by adding 0.1% (v/v) trifluoroacetic acid (TFA). Processed preparations were either desalted on C4 ZipTips (Millipore, Billerica, MA) and analyzed by electrospray ion trap mass spectrometry (Esquire LC ion trap mass spectrometer; Bruker, Bremen, Germany) or blotted on polyvinylidene difluoride (PVDF) membranes in a ProSorb cartridge prior to Edman degradation on a 491 Procise cLC protein sequencer (Applied Biosystems, Foster City, CA).
Chemical Synthesis of CXCL5 and Its Posttranslationally Modified Variants
Intact CXCL5(1–78) and its truncated and deiminated isoforms, i.e. CXCL5(2–78), CXCL5(8–78), CXCL5(9–78), [Cit9]CXCL5(1–78), and [Cit9]CXCL5(9–78), were chemically synthesized based on fluorenyl methoxycarbonyl (Fmoc) chemistry using a 433A solid phase peptide synthesizer (Applied Biosystems), as described in Loos et al. (38). The anticipated piperidide formation (situated at Asp72–Gly73) was overcome by use of the preformed dipeptide Fmoc-Asp(OtBu)-(Dmb)Gly-OH (Novabiochem, EMD Chemicals, Gibbstown, NJ) (38). Intact synthetic peptides were separated from incompletely synthesized by-products by reversed phase chromatography (RP-HPLC) on a Source 5-RPC column (4.6 × 150 mm; GE Healthcare) applying an acetonitrile gradient in 0.1% (v/v) TFA. 0.7% of the column effluent was analyzed by online ion trap mass spectrometry (Esquire LC). Fractions containing the protein with the correct relative molecular mass (Mr) were folded overnight and subsequently repurified on a C8 Aquapore-RP 300 column (2.1 × 220 mm; PerkinElmer Life Sciences) combined with online mass spectrometry. Homogenous CXCL5 fractions were pooled, lyophilized, and dissolved in ultrapure water (MilliQ; Millipore). Multiple assays were applied to determine the protein concentrations, i.e. enzyme-linked immunosorbent assay (ELISA), bicinchoninic acid protein assay (Pierce), SDS-PAGE, and NH2-terminal sequencing based on Edman degradation.
Gel Filtration Chromatography
Gel filtration chromatography of CXCL5(1–78) and [Cit9]CXCL5(1–78) was performed on a Superdex 75 PC3.2/30 column (GE Healthcare). 1 μg CXCL5 (50 μl) was injected in 10 mm phosphate buffer (pH 7.4), containing 150 mm NaCl and 0.05% Tween 20 at a flow rate of 100 μl/min. Eluting proteins were detected by UV absorbance (λ = 214 nm).
Isolation of Granulocytes from Fresh Human Buffy Coats
Fresh human buffy coats were obtained from the Blood Transfusion Center of the Red Cross (Leuven, Belgium). The bulk of erythrocytes were removed by sedimentation in erythrocyte-aggregating hydroxyethyl starch (Plasmasteril, Fresenius AG, Bad Homburg, Germany) for 30 min. The supernatant was layered on Ficoll-sodium diatrizoate medium (Lymphoprep, Axis-Shield PoC AS, Oslo, Norway) to separate mononuclear cells from granulocytes by density gradient centrifugation (30 min, 400 × g). Residual erythrocytes in the granulocyte pellet were eliminated by hypotonic shock (30 s) in ultrapure water (MilliQ). Purified granulocytes were used for chemotaxis and Ca2+ signaling experiments.
Signal Transduction through CXCR2
The capacity of the CXCL5 isoforms to induce signaling through CXCR2 resulting in an increase of the intracellular Ca2+ concentration ([Ca2+]i) was tested on both freshly purified granulocytes and CXCR2-transfected HEK293 cells (39). Briefly, 107 cells/ml, suspended in RPMI 1640 (Lonza) + 2% (v/v) FBS (granulocytes) or DMEM + 10% (v/v) FBS (HEK293 cells), were loaded for 30 min at 37 °C with 2.5 μm of the ratiometric fluorescent dye Fura-2 (Molecular Probes, Invitrogen, Merelbeke, Belgium) in the presence of 0.01% (w/v) Pluronic F-127 (Sigma). 107 cells were kept aside for autofluorescence measurements. After washing, the cells were resuspended at 106 cells/ml in calcium buffer, i.e. Hanks' balanced salt solution (HBSS) with 1 mm Ca2+, and 0.1% (v/v) FBS, buffered with 10 mm HEPES/NaOH to pH 7.4. At 37 °C, the cells were stimulated with various concentrations of CXCL5. Meanwhile, upon alternating excitation at 340 and 380 nm, the fluorescence intensity ratio (R) of Fura-2 at 510 nm was continuously measured in a LS50B luminescence spectrophotometer (PerkinElmer). The values for Rmax and Rmin were determined by stimulating the cells with 50 μm digitonin and 10 mm EGTA in 20 mm Tris (pH 8.5), respectively. Incorporation of these values in the equation of Grynkiewicz et al. (40) allowed converting fluorescence intensities into [Ca2+]i. The Mann-Whitney U test was used to detect statistically significant different increases in [Ca2+]i.
Phosphorylation of extracellular signal-regulated kinase-1/2 (ERK1/2) upon chemokine treatment was measured as previously described (41). Briefly, 106 HEK293 cells transfected with CXCR2 (2 ml) were seeded in 6-well plates (9 cm2; Techno Plastic Products AG, Trasadingen, Switzerland) in DMEM enriched with 10% (v/v) FBS. After 24 h the growth medium was removed and the cells were cultured overnight in starvation medium (serum-free DMEM). Subsequently, the starvation medium was replaced by DMEM containing 0.5% (w/v) BSA (15 min, 37 °C). The cells were stimulated for 5 min at 37 °C with a range of CXCL5 concentrations. Then, signaling was stopped and the cells were lysed. The bicinchoninic acid protein assay was used to determine the total protein concentration in the supernatant of the lysate. The amount of ERK1/2 in the supernatant (in picograms) was estimated using an ELISA specifically measuring both phosphorylated ERK1 (Thr202/Tyr204) and phosphorylated ERK2 (Thr185/Tyr187) (R&D Systems). For comparison of signaling by various CXCL5 isoforms with the corresponding concentrations of intact CXCL5(1–78), the Wilcoxon matched-pairs signed ranks test was used. The Wilcoxon single-sample signed ranks test was performed to compare ERK1/2 phosphorylation in CXCL5-treated cells with ERK1/2 phosphorylation in medium-treated cells (100% pERK).
Internalization of CXCR2 was analyzed by flow cytometry. HEK293 cells transfected with human CXCR2 were grown in DMEM enriched with 10% (v/v) FBS and 800 μg/ml of geneticin. After trypsinization, the cells were allowed to recover for 2 h in DMEM containing 10% (v/v) FBS at room temperature. Cells were washed with serum-free medium and resuspended to 5 × 106 cells/ml in pre-heated (37 °C) RPMI1640 supplemented with 1% (w/v) bovine serum albumin (endotoxin-free; Sigma). Aliquots of 5 × 105 cells were challenged with various concentrations of the CXCL5 isoforms or with medium (baseline receptor expression level) for 10 min at 37 °C. After incubation, the cells were chilled on ice for 10 min and washed once in ice-cold assay buffer (PBS + 2% (v/v) FBS). All further procedures were performed at 4 °C. The cells were incubated for 30 min with purified mouse anti-human CD182 (CXCR2) (50 μg/ml; BD Biosciences, San Jose, CA) or purified mouse IgG1 isotype control (50 μg/ml; eBioscience, San Diego, CA). Afterward, the cells were washed twice with ice-cold assay buffer, followed by incubation of the cells for another 30 min with phycoerythrin-labeled goat anti-mouse IgG (1.3 μg/ml; BD Biosciences), diluted in ice-cold assay buffer. Once again the cells were washed with assay buffer and finally fixed in assay buffer + 2% (w/v) formaldehyde. In a similar experiment performed at 4 °C we demonstrated that preincubation of the cells with chemokine does not interfere with the interaction between CXCR2 and the anti-human CXCR2 antibody (data not shown). The level of phycoerythrin bound to the cells was evaluated by flow cytometry (FACScalibur; BD Biosciences). The relative surface expression of CXCR2 was calculated as a percentage as: [(MFICXCL5 − MFIisotype control)/(MFImedium − MFIisotype control)] × 100. MFICXCL5 and MFImedium represent the mean fluorescence intensities of cells incubated with CXCL5 or medium, respectively, and stained with anti-human CXCR2 antibody, whereas MFIisotype control stands for the mean fluorescence intensity of medium-stimulated cells stained with isotype control antibody (baseline staining). Statistical analysis was performed using the Wilcoxon matched-pairs signed ranks test for comparison of the CXCL5 isoforms or the Wilcoxon single-sample signed ranks test for comparison with the medium-treated control (100% receptor binding).
Neutrophil Migration in Vitro and in Vivo
The chemotactic activity of the different CXCL5 isoforms for neutrophil granulocytes in vitro was compared using the Multiscreen plate-based chemotaxis assay (Millipore), as previously described (41). Both chemokine test samples and purified granulocytes (2.5 × 106 cells/ml) were diluted in Hanks' balanced salt solution enriched with 0.1% (w/v) BSA (endotoxin-free; Sigma). Neutrophils were allowed to migrate across the 5-μm hydrophilic polycarbonate filter in response to the concentration gradient for 45 min at 37 °C. After incubation, migrated cells in the lower wells of the Multiscreen plate were quantified using the luminescence ATP detection assay (PerkinElmer). By adding luciferase and d-luciferin, the production of light is elicited, proportional to the ATP concentration and, consequently, to the amount of migrated cells. The emitted light was measured in a luminescence reader (FL600 microtiter plate fluorescence reader; Bio-Tek Instruments, Winooski, VT). The chemotactic index was calculated by dividing the luminescence intensity of the test sample through the luminescence intensity of the control buffer. Statistical analysis was performed using the Mann-Whitney U test.
In vivo blood vessel extravasation of circulating neutrophil granulocytes into the peritoneal cavity was examined by intraperitoneal injection of endotoxin-free CXCL5 (in 0.9% (w/v) NaCl) or 0.9% (w/v) NaCl alone in female NMRI mice (6–10 weeks, raised pathogen-free; Elevage Janvier, Le Genest Saint Isle, France). Endotoxin concentrations were evaluated with the Limulus amoebocyte lysate test (Cambrex, East Rutherford, NJ). After 2 h, mice were sacrificed and the peritoneal cavity was washed with 5 ml of saline enriched with 2% (v/v) FBS and 20 units/ml of heparin. Total leukocyte counts in the peritoneal lavage were determined. 105 cells/ml were used for the preparation of 2 cytospins/mouse. After Hemacolor (Merck, Darmstadt, Germany) staining, percentages of neutrophils in the peritoneal lavage were determined in duplicate by differential microscopic counting of at least 100 leukocytes/cytospin. Neutrophil percentages or total neutrophil counts in the peritoneal lavage were statistically compared between CXCL5-treated mice and control mice (intraperitoneal injection of saline) using the Mann-Whitney U test. The experimental protocol (involving the use of laboratory animals) was reviewed by the Animal Ethical Committee of the K.U. Leuven and Belgian and European guidelines concerning the handling of laboratory animals were followed.
Regulation of the Expression of Adhesion Molecules by Neutrophil Granulocytes
Whole blood from healthy volunteers was collected in lithium-heparin vacutainer collection tubes (BD Biosciences) and diluted to 106 leukocytes/ml in warm PBS (37 °C) (42). Leukocytes were stimulated with the indicated concentrations of CXCL5 or 10−7 m formyl-MLF (fMLF; as a positive control) diluted in warm PBS or with warm PBS alone (negative control), for 2 or 10 min at 37 °C. To stop signaling, the cells were placed on ice and 1 ml of ice-cold PBS was added. After centrifugation, PBS was replaced by ice-cold assay buffer and the cells were placed on ice for 30 min to block the Fc-receptors. Afterward, the cells were incubated on ice for 30 min with the appropriate antibodies, diluted in assay buffer (allophycocyanin-conjugated anti-human CD62L from eBioscience; phycoerythrin-Cy5-conjugated anti-human CD11b, phycoerythrin-conjugated anti-human CD16, and fluorescein isothiocyanate-conjugated anti-human CD11a from BD Biosciences). The cells were washed 3 times with assay buffer and fixed with assay buffer containing 2% (w/v) formaldehyde. Red blood cells were removed using FACS lysing solution (BD Biosciences). The cells were washed and fixed definitively prior to analysis on a BD FACSCalibur cytometer. Based on forward/side scatter and CD16 expression, neutrophils were selected and analyzed for CD62L, CD11a, and CD11b expression. The Wilcoxon single-sample signed ranks test was performed to compare the adhesion molecule expression statistically with baseline adhesion molecule expression (100%).
RESULTS
Generation of Posttranslationally Modified CXCL5 Isoforms by CXCL5-processing Enzymes
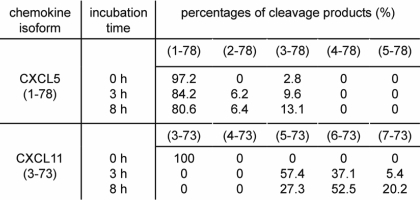
In view of the efficient cleavage of the CXC chemokine CXCL11(3–73) by CD13 (31), we examined the susceptibility of CXCL5 to processing by CD13 (Table 1). CD13 is a metalloprotease that specifically removes NH2-terminal amino acids one by one (43–45). Upon incubation of recombinant CXCL5(1–78) with CD13 (E/S molar ratio 1/25; final enzyme concentration 100 pmol/ml), the NH2-terminal Ala residue and the subsequent Gly residue were partially removed after 3 h, as shown by mass spectrometry analysis (Table 1). As could be expected based on the specificity of CD13, the third amino acid, i.e. a Pro residue, was resistant to cleavage. In parallel, synthetic CXCL11(3–73) was incubated with CD13 as a positive control. Taken together, we can conclude that CXCL5 was processed more slowly by CD13 compared with CXCL11(3–73). Furthermore, less amino acid residues were removed.
TABLE 1.
Processing of CXCL5 by CD13/aminopeptides N
Recombinant CXCL5(1–78) or synthetic CXCL11(3–73) (as a positive control) were incubated with CD13 at an E/S molar ratio of 1/25. After either 3 or 8 h, the reaction was stopped with TFA and percentages of the cleavage products were identified by mass spectrometry.
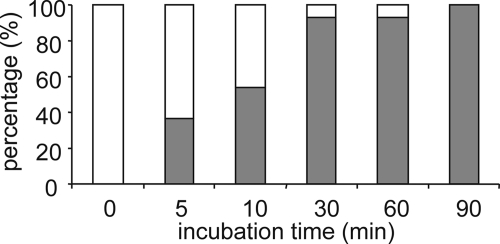
In addition to proteolytic truncation, posttranslational modification of chemokines through deimination by PADs of specific Arg residues located in the NH2-terminal region was reported for natural CXCL8 and CXCL10 (34, 35). In contrast, PAD-induced deimination in CXCL12 was observed on all Arg present in this chemokine (36). To investigate whether CXCL5 can be citrullinated, CXCL5 was incubated with rabbit PAD2 for different time intervals at a 1/20 E/S molar ratio (Fig. 1). Changes in the NH2-terminal amino acid sequence and the relative molecular mass of CXCL5 (conversion of Arg to Cit increases the molecular mass by one mass unit) were uncovered making use of Edman degradation-based amino acid sequencing, combined with ion trap mass spectrometry. CXCL5 holds 2 Arg residues, situated at positions 9 and 12. The most NH2-terminal Arg of CXCL5, Arg9, was rapidly converted into a Cit residue. After 5 min approximately one-third of the Arg9 residues were deiminated. Upon 30 min of incubation Arg9 was almost completely modified. In contrast, after 30 min of incubation, no Cit residues were detected at position 12, i.e. the position of the second Arg residue of CXCL5.
FIGURE 1.
Citrullination of CXCL5 by PAD. Recombinant CXCL5(1–78) was incubated with rabbit PAD2 at an E/S molar ratio of 1/20. At several time points the reaction was stopped with TFA, and the reaction mixture was analyzed by ion trap mass spectrometry combined with Edman degradation-based NH2-terminal sequencing to define the conversion rate. The defined percentages of CXCL5(1–78) (open bar) and [Cit9]CXCL5(1–78) (filled bar) are shown at the indicated time points.
Effect of Citrullination and CD13 Processing on Cathepsin G Cleavage
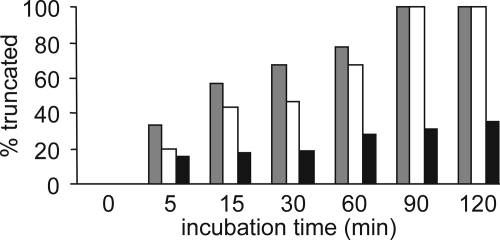
NH2-terminally truncated isoforms of CXCL5, such as CXCL5(8–78) and CXCL5(9–78), have been identified from natural sources (46). Characterization of the biological activity of CXCL5(9–78) revealed enhanced neutrophil-activating and chemotactic potency in vitro (47). Cleavage of CXCL5 by cathepsin G, a serine protease present in the primary granules of polymorphonuclear granulocytes (5, 7), has been shown to result in the generation of CXCL5(9–78) (47). Because citrullination hinders the cleavage of CXCL8 by plasmin and thrombin (35), the effect of citrullination on the susceptibility of CXCL5 to cathepsin G processing was investigated. Therefore, the ability of cathepsin G to cleave both CXCL5(1–78) and [Cit9]CXCL5(1–78) was tested (Fig. 2). Although for both molecules 100% removal of the NH2-terminal octapeptide was observed after 90 min at a 1/100 E/S molar ratio, conversion of [Cit9]CXCL5(1–78) was moderately more efficient than conversion of CXCL5(1–78). Overall, the effect of citrullination of CXCL5 on the susceptibility to activation by cathepsin G was small compared with the earlier reported effects of citrullination on the efficiency of CXCL8 truncation by plasmin and thrombin (35). In contrast, the removal of the NH2-terminal amino acid by CD13, resulting in generation of CXCL5(2–78), clearly lowered the susceptibility of CXCL5 to cleavage by cathepsin G. After 120 min only 35% of CXCL5(2–78) was converted to CXCL5(9–78) (Fig. 2).
FIGURE 2.
Effect of citrullination and cleavage by CD13 on the susceptibility of CXCL5 to NH2-terminal cleavage by cathepsin G. CXCL5(1–78) is a substrate for the serine protease cathepsin G, generating the more potent CXCL5(9–78) (47). Percentages of generated CXCL5(9–78) in function of time are depicted for CXCL5(1–78) (open bar), [Cit9]CXCL5(1–78) (grey bar), and CXCL5(2–78) (black bar).
Chemical Synthesis of Posttranslationally Modified CXCL5 Variants
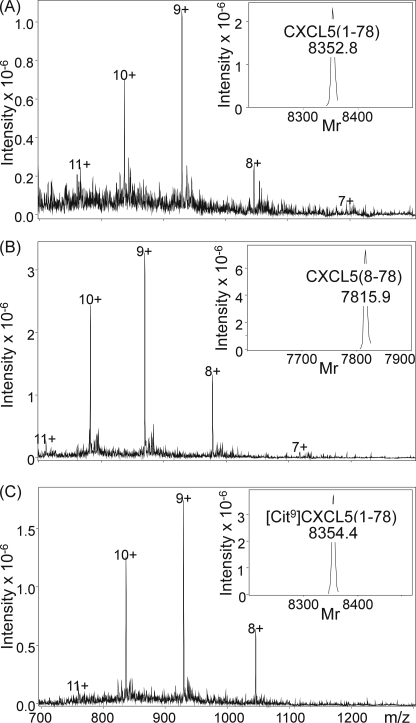
To obtain sufficient amounts of homogenously pure CXCL5 isoforms, full-length CXCL5(1–78), the CD13-truncated isoform CXCL5(2–78), [Cit9]CXCL5(1–78), and [Cit9]CXCL5(9–78) and the naturally occurring CXCL5(8–78) and CXCL5(9–78) were chemically synthesized by solid phase peptide synthesis using Fmoc chemistry. Subsequently, the synthetic proteins were deprotected and purified by RP-HPLC. Cysteine bridges were formed by incubating the proteins in a mixture of oxidized and reduced glutathione and finally, the chemokines were repurified chromatographically. The molecular mass of the chemically synthesized proteins was verified by ion trap mass spectrometry (Fig. 3) and found to correspond to their theoretical relative molecular mass, i.e. 8351.9 for CXCL5(1–78), 8280.9 for CXCL5(2–78), 7814.3 for CXCL5(8–78), 8352.9 for [Cit9]CXCL5(1–78), 7702.1 for CXCL5(9–78), and 7703.1 for [Cit9]CXCL5(9–78). Edman degradation-based sequencing was used to confirm the NH2-terminal sequence of the synthesized molecules.
FIGURE 3.
Chemical synthesis of CXCL5 and modified isoforms. Synthesized CXCL5 isoforms were purified by RP-HPLC combined with on-line mass spectrometry. Fractions containing homogenous, folded CXCL5 were pooled, lyophilized, and dissolved in ultrapure water. Finally, the quality of the synthetic CXCL5(1–78), CXCL5(8–78), or [Cit9]CXCL5(1–78) was confirmed by mass spectrometry analysis, as depicted in panels A–C, respectively. The raw mass spectra were averaged over the chromatographic peaks and show the multiply charged ions. Based on these ions, Bruker deconvolution software allowed for calculation of the Mr of the uncharged molecule, which is shown as an inset and corresponds to the theoretical Mr.
As a control, synthetic CXCL5(1–78) and recombinant CXCL5(1–78) were compared in the Ca2+ signaling assay to guarantee identical specific activity and no difference was noticed (data not shown). On gel filtration chromatography citrullinated and non-citrullinated CXCL5 eluted at identical retention volumes. Thus, citrullination did not affect multimerization of CXCL5 (data not shown).
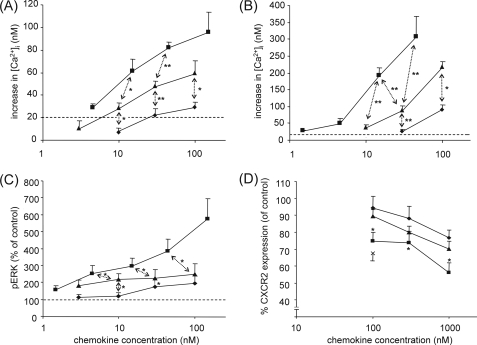
NH2-terminal Truncation Enhances, whereas Citrullination Diminishes the Signaling and Chemotactic Properties of CXCL5
To compare the signaling properties of the different posttranslationally modified isoforms of CXCL5 through its cognate receptor, CXCR2, both Ca2+ signaling and phosphorylation of the mitogen-activated protein (MAP) kinases ERK1/2 were evaluated. In Ca2+ signaling assays using CXCR2-transfected HEK293 cells, CXCL5(1–78) induced an increase in the intracellular Ca2+ concentration starting from 10 nm onward, whereas the minimal effective concentration of CXCL5(8–78) was at least 3 times lower and that of [Cit9]CXCL5(1–78) was at least 3 times higher (Fig. 4A). Based on the dose-response curves, it can be concluded that citrullination of Arg9 led to a 3–10-fold decrease of the Ca2+ signaling potency, whereas cleavage of 7 NH2-terminal amino acids clearly potentiated the CXCL5-induced cytoplasmic Ca2+ influx. A similar pattern was seen under more physiological conditions, i.e. when using freshly purified granulocytes (Fig. 4B). Quantification of the amount of phosphorylated ERK1/2 upon stimulation of HEK293-CXCR2 cells with the CXCL5 isoforms for 5 min confirmed these data (Fig. 4C). Indeed, the same rank order was observed for this assay, namely CXCL5(1–78) being less potent than CXCL5(8–78), but more efficacious than [Cit9]CXCL5(1–78). At high concentrations (100 nm), the difference between intact and truncated CXCL5 was even more pronounced.
FIGURE 4.
Signaling capacity of CXCL5 isoforms through CXCR2. CXCR2-transfected HEK293 cells (panel A) and granulocytes (panel B) were loaded with the ratiometric Ca2+-binding molecule Fura-2, and [Ca2+]i was measured upon stimulation of the cells with the indicated concentrations of CXCL5(1–78) (▴), CXCL5(8–78) (■), or [Cit9]CXCL5(1–78) (♦). Values represent the mean (±S.E.) increase of the [Ca2+]i (n ≥ 3). The dashed line indicates the detection limit (20 nm). Significant differences between CXCL5(1–78) and other CXCL5 isoforms were detected using the Mann-Whitney U test (*, p < 0.05; **, p < 0.01). The amount of phosphorylated ERK1/2 in CXCR2-transfected HEK293 cells after 5 min of stimulation with the indicated concentrations of CXCL5(1–78) (▴), CXCL5(8–78) (■), and [Cit9]CXCL5(1–78) (♦) is depicted in panel C. The values shown are the percentages of ERK1/2 phosphorylation compared with medium-treated control cells (± S.E.) (n = 6). For comparison with the corresponding concentrations of intact CXCL5(1–78), the Wilcoxon matched-pairs signed ranks test was used (*, p < 0.05). Internalization of CXCR2 was tested on CXCR2-transfected HEK293 cells. Panel D shows the relative CXCR2 surface expression (compared with medium-treated control cells) following stimulation with the indicated concentrations of CXCL5(1–78) (▴), CXCL5(8–78) (■), and [Cit9]CXCL5(1–78) (♦) (n ≥ 6). 100 nm IL-8(1–77) (×) was enclosed as a positive control. Statistical comparison of the CXCL5 isoforms was done by use of the Wilcoxon matched-pairs signed ranks test (*, p < 0.05), and error bars indicate the standard error of the mean (S.E.).
In addition, the ability of the different isoforms to induce internalization of CXCR2 was examined (Fig. 4D). The internalization process is an important regulator for the level of CXCR2 expression on the cell surface and thereby controls the responsiveness of the cell to CXCR2 agonists. CXCR2-transfected HEK293 cells were incubated with the indicated concentrations of CXCL5 for 1 h at 37 °C, followed by fluorescent staining with anti-human CXCR2 antibodies. The amount of CXCR2 expressed on the cells was quantified by FACS analysis. The effect of the posttranslational modifications on this process was analogous to the effect on Ca2+ signaling and on phosphorylation of ERK1/2. Significant internalization of CXCR2 was observed upon stimulation of the HEK293 cells with 100 nm CXCL5(1–78) (p = 0.02), 100 nm CXCL5(8–78) (p = 0.03), and 1000 nm [Cit9]CXCL5(1–78) (p = 0.008). Based on the entire dose-response curves, CXCL5(8–78) was again a stronger inducer of CXCR2 internalization compared with CXCL5(1–78). No statistically significant difference between CXCL5(1–78) and [Cit9]CXCL5(1–78) was detected, although their minimal effective concentrations were clearly different. Thus, the removal of 7 NH2-terminal amino acids enhances in vitro the CXCR2-signaling properties of CXCL5, whereas citrullination restricts CXCR2 activation and signaling.
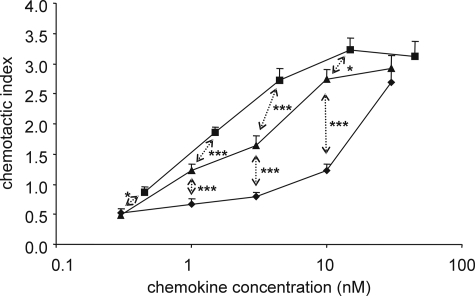
Subsequently, the more complex process of chemotaxis, which embodies multiple interconnected signaling pathways, was evaluated in response to the different CXCL5 isoforms. The chemotactic activity was compared in vitro on freshly purified neutrophil granulocytes using the Multiscreen chemotaxis assay (Fig. 5). 3 nm CXCL5(1–78) was necessary to induce a significant neutrophil chemotactic response compared with buffer (p < 0.0001). Contrary, the minimal effective concentrations of CXCL5(8–78) and [Cit9]CXCL5(1–78) were 1.5 (p < 0.0001) and 30 nm (p = 0.005), respectively. Again, CXCL5(1–78) was a stronger agonist than [Cit9]CXCL5(1–78). The potentiating effect of the NH2-terminal truncation on granulocyte chemotaxis, in the case of CXCL5(8–78), correlates with the enhanced activity detected in the signaling experiments. However, this truncation only resulted in a 2-fold increase of the chemotactic potency. Furthermore, equal chemotactic activity was detected when comparing CXCL5(1–78) with CXCL5(2–78), i.e. the CD13-processed isoform of CXCL5 (data not shown).
FIGURE 5.
In vitro chemotactic activity of CXCL5 isoforms for neutrophils. The Multiscreen chemotaxis assay was used to evaluate the chemotactic activity of CXCL5(1–78) (▴), CXCL5(8–78) (■), and [Cit9]CXCL5(1–78) (♦) in vitro (3 independent experiments). The chemotactic index is calculated by dividing the luminescence intensity of the test sample by the luminescence intensity of the control buffer. Statistical analysis of the CXCL5 isoforms with regard to each other and to the control buffer was performed using the Mann-Whitney U test (*, p < 0.05; ***, p < 0.001 for significant differences between CXCL5(1–78) and the other CXCL5 isoforms). Error bars indicate the S.E.
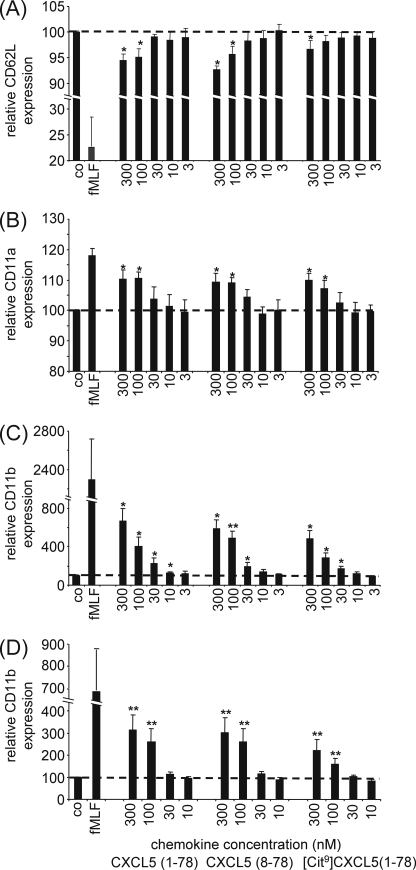
The Regulation of Adhesion Molecule Expression Is Moderately Affected by Posttranslational Modification of CXCL5
Chemokines, either produced by the underlying tissue and transported to the luminal surface of the endothelial vessel or synthesized by the endothelial cell itself, are presented to circulating leukocytes by binding to glycosaminoglycans on the luminal surface of the endothelial cell layer. Leukocyte migration in response to these chemokines involves a rapid transition from leukocyte rolling through weak, selectin-mediated interactions to firm adhesion via strong integrin-based attachment. In this context, the down-regulation of L-selectin (Fig. 6A) and the up-regulation of β2-integrins CD11a/CD18 (Fig. 6B) and CD11b/CD18 (Fig. 6C) were examined upon activation of neutrophils with various CXCL5 isoforms for 10 min. Neutrophils stimulated with CXCL5(1–78), indeed expressed lower yields of L-selectin and higher yields of CD11a and CD11b. Surprisingly, no striking differences were observed when stimulating the cells with comparable doses of CXCL5(1–78), CXCL5(8–78), or [Cit9]CXCL5(1–78), although a 3-fold difference in the minimal effective concentration for CD11b up-regulation and L-selectin down-regulation was observed when comparing CXCL5(1–78) with its deiminated variant. In contrast, CD11b up-regulation after 2 min of stimulation with CXCL5 (Fig. 6D) was clearly decreased upon citrullination of CXCL5(1–78). Neutrophilic CD11b up-regulation was significantly higher when stimulated with 300 pmol of CXCL5(1–78) compared with 300 pmol of [Cit9]CXCL5(1–78) (p < 0.05). Taken together, it can be concluded that, in contrast to signal transduction and chemotaxis, the expression of adhesion molecules, such as L-selectin, CD11a and CD11b, is only moderately affected by posttranslational modification of CXCL5.
FIGURE 6.
Regulation of the expression pattern of adhesion molecules on neutrophils by CXCL5 isoforms. Freshly isolated leukocytes were stimulated with the indicated concentrations of CXCL5 isoforms, 10−7 m fMLF as a positive control or vehicle (PBS) as a negative control (co) for 10 (panels A–C) or 2 min (panel D) at 37 °C. The expression patterns of L-selectin (CD62L) (panel A), CD11a (panel B), and CD11b (panels C and D) were evaluated by staining the stimulated cells with the appropriate antibodies, followed by FACS analysis. The mean fluorescence intensity upon stimulation with CXCL5 was divided by the mean fluorescence intensity upon PBS treatment and multiplied by 100 to obtain the relative expression of CD62L, CD11a, or CD11b. Values represent the mean relative expression (formulated as percentages) (± S.E.) of 5 independent experiments (5 different donors). The Wilcoxon single-sample signed ranks test was performed to statistically compare the relative adhesion molecule expression to buffer control (*, p < 0.05; **, p < 0.01).
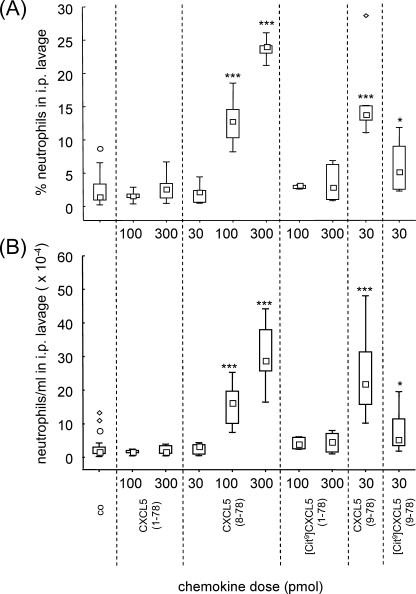
The Chemotactic Activity of CXCL5 in Vivo Is Strongly Enforced by Extended NH2-terminal Truncation and Reduced by Citrullination
Finally, the chemotactic activity of the CXCL5 isoforms was evaluated in vivo (Fig. 7). Mice were injected intraperitoneally with CXCL5 and after 2 h the leukocyte influx into the peritoneal cavity was evaluated microscopically. Interestingly, the influence of NH2-terminal truncation on the neutrophil chemotactic activity of CXCL5 was even more obvious in vivo than in vitro. Indeed, even a dose of 300 pmol of CXCL5(1–78) or [Cit9]CXCL5(1–78) failed to recruit neutrophils into the peritoneum, whereas injection of 100 pmol of CXCL5(8–78) already significantly elevated the percentage of neutrophil granulocytes in the peritoneal cavity from ∼2% to about 13%. Upon injection of 300 pmol of CXCL5(8–78) the relative amount of neutrophils further rose up to 24% (which corresponds to an absolute number of 29 × 104 neutrophils/ml in the peritoneum). The clearly increased chemotactic potency of truncated CXCL5 in vivo in combination with the fact that cathepsin G cleavage of CXCL5 is barely affected by citrullination, triggered the analysis of the effect of citrullination on the truncated isoform CXCL5(9–78). Interestingly, [Cit9]CXCL5(9–78) was able to induce a significant neutrophil influx into the peritoneum. However, the percentage of neutrophils recruited into the peritoneum was significantly lower upon injection of 30 pmol of [Cit9]CXCL5(9–78) in comparison to injection of 30 pmol of CXCL5(9–78) (p < 0.05). Thus, removal of 7 or 8 NH2-terminal amino acids of CXCL5 potentiates its in vivo activity, whereas citrullination of Arg9 reduces neutrophil recruitment to the peritoneal cavity.
FIGURE 7.
In vivo chemotactic activity of CXCL5 isoforms for neutrophils. In vivo neutrophil chemotaxis in mice in response to intraperitoneal injection of CXCL5(1–78), CXCL5(8–78), [Cit9]CXCL5(1–78), CXCL5(9–78), and [Cit9]CXCL5(9–78) was investigated. The total amount of leukocytes/ml in the intraperitoneal lavage was counted in Türk solution and the proportional amount of neutrophils in the peritoneal cavity was determined by differential counting of Hemacolor-stained cytospins. Panel A, the neutrophil percentages in the intraperitoneal lavage upon stimulation with buffer or CXCL5 are shown. Panel B, total neutrophil counts/ml are calculated and depicted in function of the CXCL5 isoform and concentration. Squares indicate the median neutrophil influx (formulated as percentages or total counts/ml); the bottom and the top of the rectangle denote the 25th and 75th percentile; whiskers represent the non-outlier range (coefficient 1.5) (CXCL5-treated mice: n = 3 to 9; control mice: n = 17). Outliers (○) and extremes (♢) are also plotted. Control mice (co), treated with vehicle, were included to quantify the spontaneous migration of neutrophils to the intraperitoneal cavity. The Mann-Whitney U test was used for statistical analysis (*, p < 0.05; ***, p < 0.001; compared with injection with vehicle).
DISCUSSION
The protein spectrum encoded by DNA is extensively diversified by posttranslational modifications such as proteolysis and glycosylation, which affect the structure and function of the molecule. In the case of chemokines, posttranslational modifications by proteolysis have been extensively studied and found to be extremely important for the regulation of chemokine activity. Thereby, proteases contribute to the further fine-tuning of the coordination of directional leukocyte migration (26). Many different chemokine-processing enzymes have been identified and diverging consequences for chemokine function have been described. NH2-terminal proteolysis can result in enhanced (e.g. CXCL8), reduced (e.g. CXCL10), or unaltered (e.g. CXCL6) chemokine activity or can even have a dual effect if the chemokine uses two receptors (e.g. CCR1 (decrease) and CCR5 (increase) for CCL5). Compared with NH2-terminal truncation, COOH-terminal proteolysis (e.g. CXCL10) has been detected less frequently and the impact on GPCR signaling is often less significant. Therefore, the NH2-terminal region, reported to be implicated in receptor binding and activation, is thought to be the principal target of proteolysis. Besides proteolytic cleavage, other types of posttranslational modification, such as glycosylation (e.g. CCL2) (48) and citrullination (e.g. CXCL8) (35) have been identified on chemokines isolated from natural sources. In general, for those chemokines evaluated, citrullination lowers the chemotactic potency (34, 35).
In this study, the capacity of the aminopeptidase CD13 and PAD to posttranslationally modify CXCL5 was evaluated. CD13 has been identified as a myeloid cell marker, expressed on both normal and transformed granulocytes and monocytes (30, 49). This aminopeptidase removes NH2-terminal amino acids one by one, preferably neutral amino acids such as Ala, Phe, and Leu (50). Pro, however, is rather resistant to CD13 cleavage. It has been shown that CD13 efficiently cleaves CXCL11(3–73), generated by CD26, thereby decreasing its angiostatic activity (31). Processing of CXCL5 by CD13, in contrast, is rather limited and blocked by the presence of a Pro residue at the third NH2-terminal position. A similar low conversion rate has been observed for CXCL8 (31). Comparison of CXCL5(1–78) and CXCL5(2–78) in vitro indicated that CD13 cleavage does not affect the neutrophil chemotactic potency of CXCL5. More extensive NH2-terminal truncation, in contrast, significantly enhances the chemokine activity of CXCL5. In particular, this has been shown for recombinant CXCL5(5–78), recombinant CXCL5(9–78), and a mixture of natural CXCL5(8–78) and CXCL5(9–78) (46, 47). In this study, pure CXCL5(8–78), a variant isolated from the conditioned medium of monocytes (46), was included in all assays. Our data confirm the strengthened in vitro activity for CXCL5(8–78). However, such NH2-terminal truncation does not alter the capacity of CXCL5 to modulate neutrophilic adhesion molecule expression. Interestingly, the data presented indicate that the significance of NH2-terminal truncation is even more pronounced in vivo, as CXCL5(8–78) is at least 3-fold more potent to induce a neutrophil influx into the intraperitoneal cavity.
A second enzyme that was tested for its ability to modify CXCL5, is PAD. PAD enzymes convert basic peptidylarginine residues to neutrally charged peptidylcitrulline residues by hydrolyzing the guanidino group (51, 52). The loss of a positive charge may destroy salt and hydrogen bonds, thus destabilizing the protein structure (53). To date, 5 different human PAD isozymes have been described, displaying different spatial tissue expression. PADs are primarily located intracellularly (52), but during inflammation, PAD2 and PAD4 have been detected extracellularly as well (33, 54, 55). Upon incubation of CXCL5 with natural rabbit PAD2, CXCL5 is citrullinated rapidly. CXCL5 holds 2 Arg residues, situated at positions 9 and 12. Arg9 is remarkably more prone to deimination than Arg12. This might be explained by the unstructured NH2-terminal region, which may favor Arg9 deimination and by the presence of a cysteine bridge adjacent to Arg12, which may restrict its accessibility to PAD (53). Similar substrate specificity, i.e. the preference for the primary NH2-terminal Arg of the chemokine, has been described for CXCL8 and CXCL10 (34, 35). Nevertheless, Arg5 of CXCL8 and Arg5 of CXCL10 are obviously converted more efficiently than Arg9 of CXCL5. This might be explained by the presence of a Glu residue behind Arg9 of CXCL5, which has been described to render the preceding Arg less susceptible to deimination (53).
Synthetic [Cit9]CXCL5(1–78) was used to evaluate the consequences of deimination of Arg9 by PAD. Based on the signal transduction and chemotaxis assays, it can be concluded that deimination impairs the CXCR2 signaling and the chemotactic activity of CXCL5 in vitro. Furthermore, the effect of citrullination on the capacity of CXCL5 to modulate adhesion molecule expression on the cell surface of neutrophils was evaluated. Soluble factors, such as interleukin-1 and lipopolysaccharide (LPS), have been shown to provoke the secretion of CXCL5 by cultured human endothelial cells (56, 57). Upon presentation by glycosaminoglycans, chemokines such as CXCL5 trigger the integrin-mediated arrest of circulating neutrophils. Neutrophil adhesion in the systemic circulation relies primarily on the β2-integrins CD11a/CD18 and CD11b/CD18 (10, 58, 59). Activation of neutrophils provokes up-regulation of the β2-integrin CD11b/CD18 (which is present in intracellular storage pools and rapidly translocated to the plasma membrane) together with shedding of L-selectin (CD62L) by either a disintegrin and metalloproteinase (ADAM)-17 or ADAM-8 (60–62). Leukocyte migration in response to chemokines involves a rapid transition from leukocyte rolling through weak, selectin-mediated interactions to firm adhesion via strong integrin-based attachment. The up-regulation of CD11a/b and the down-regulation of CD62L on neutrophils upon stimulation with CXCL5, promoting firm neutrophil adhesion, are moderately affected by posttranslational modification of the chemokine. The consequences of deimination for the chemotactic activity in vivo, as tested by intraperitoneal injection of CXCL5 in mice, could not be evaluated for intact CXCL5 as 300 pmol of neither CXCL5(1–78) nor [Cit9]CXCL5(1–78) induced a peritoneal neutrophil influx. The fact that, in contrast, 100 pmol of CXCL5(8–78) and 30 pmol of CXCL5(9–78) already induces a significant neutrophil influx underlines the importance of NH2-terminal truncation in vivo. Interestingly, citrullination of the truncated chemokine significantly reduced the in vivo chemotactic potency. Taken together, we can conclude that deimination impairs the inflammatory character of CXCL5 and that PAD-chemokine interaction may contribute to the resolution of inflammation.
At the site of inflammation, neutrophils secrete a broad spectrum of proteases, which are not only important for the degradation of the extracellular matrix, but can also orchestrate the inflammatory response by proteolytically modifying chemokines and cytokines (5, 7). Among these, cathepsin G, a serine protease present in the primary granules, has been identified as a candidate protease generating CXCL5(9–78) (47). As this isoform was expected, and later confirmed, to be more potent in vivo, we wondered whether [Cit9]CXCL5(1–78), CXCL5(2–78), and CXCL5(1–78) were differently susceptible to cathepsin G cleavage. Cleavage of CXCL5(1–78) is not impaired by deimination, on the contrary, [Cit9]CXCL5(1–78) is even slightly more susceptible to cleavage than CXCL5(1–78). This indicates that citrullination differentially protects CXCL5 and CXCL8 from proteolytic cleavage by cathepsin G and thrombin/plasmin, respectively. Surprisingly, CXCL5(2–78) is cleaved less efficiently by cathepsin G than CXCL5(1–78). Therefore, although CD13 cleavage is not expected to affect GPCR signaling by CXCL5, CD13 cleavage might play a more important role than expected, as it generates an isoform that is protected against cathepsin G cleavage.
In conclusion, this study substantiates the previously reported role for posttranslational modifications in the regulation of chemokine function. Although with opposite consequences, both NH2-terminal truncation and citrullination significantly affect the biological activity of CXCL5(1–78) and may both be implicated in the regulation of neutrophil extravasation during inflammation.
Acknowledgments
We thank Karolien Grauwen, Jean-Pierre Lenaerts and René Conings for technical assistance.
This work was supported in part by the Center of Excellence Grant EF/05/15 from K.U. Leuven, Concerted Research Actions Grant G.O.A./2007/15 from the Regional Government of Flanders, the Fund for Scientific Research of Flanders (F.W.O. Vlaanderen), the Interuniversity Attraction Poles Program-Belgian Science Policy, and the European Union 6FP EC contract INNOCHEM Grant LSHB-CT-2005-518167.
- GPCR
- G protein-coupled receptor
- [Ca2+]i
- intracellular calcium concentration
- PAD
- peptidylarginine deiminase
- fMLF
- formyl-MLF
- Fmoc
- fluorenyl methoxycarbonyl.
REFERENCES
- 1.Kennedy A. D., DeLeo F. R. (2009) Immunol. Res. 43, 25–61 [DOI] [PubMed] [Google Scholar]
- 2.Nauseef W. M. (2007) Immunol. Rev. 219, 88–102 [DOI] [PubMed] [Google Scholar]
- 3.Nathan C. (2006) Nat. Rev. Immunol. 6, 173–182 [DOI] [PubMed] [Google Scholar]
- 4.Eyles J. L., Roberts A. W., Metcalf D., Wicks I. P. (2006) Nat. Clin. Pract. Rheumatol. 2, 500–510 [DOI] [PubMed] [Google Scholar]
- 5.Pham C. T. (2008) Int. J. Biochem. Cell Biol. 40, 1317–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scapini P., Lapinet-Vera J. A., Gasperini S., Calzetti F., Bazzoni F., Cassatella M. A. (2000) Immunol. Rev. 177, 195–203 [DOI] [PubMed] [Google Scholar]
- 7.Pham C. T. (2006) Nat. Rev. Immunol. 6, 541–550 [DOI] [PubMed] [Google Scholar]
- 8.Rot A., von Andrian U. H. (2004) Annu. Rev. Immunol. 22, 891–928 [DOI] [PubMed] [Google Scholar]
- 9.Serhan C. N., Brain S. D., Buckley C. D., Gilroy D. W., Haslett C., O'Neill L. A., Perretti M., Rossi A. G., Wallace J. L. (2007) FASEB J. 21, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinauer M. C. (2007) Methods Mol. Biol. 412, 489–504 [DOI] [PubMed] [Google Scholar]
- 11.Newburger P. E. (2006) Hematology Am. Soc. Hematol. Educ. Program. 2006, 104–110 [DOI] [PubMed] [Google Scholar]
- 12.Fan J., Heller N. M., Gorospe M., Atasoy U., Stellato C. (2005) Eur. Respir. J. 26, 933–947 [DOI] [PubMed] [Google Scholar]
- 13.Hamilton T. A., Novotny M., Datta S., Mandal P., Hartupee J., Tebo J., Li X. (2007) J. Leukocyte Biol. 82, 213–219 [DOI] [PubMed] [Google Scholar]
- 14.Locati M., Riboldi E., Otero K., Martinez F. O., Riva F., Perrier P., Baviera S., Signorelli P., Bonecchi R., Allavena P., Sozzani S., Mantovani A. (2001) Immunobiology 204, 536–542 [DOI] [PubMed] [Google Scholar]
- 15.Loos T., Dekeyzer L., Struyf S., Schutyser E., Gijsbers K., Gouwy M., Fraeyman A., Put W., Ronsse I., Grillet B., Opdenakker G., Van Damme J., Proost P. (2006) Lab. Invest. 86, 902–916 [DOI] [PubMed] [Google Scholar]
- 16.Van Damme J., Wuyts A., Froyen G., Van Coillie E., Struyf S., Billiau A., Proost P., Wang J. M., Opdenakker G. (1997) J. Leukocyte Biol. 62, 563–569 [DOI] [PubMed] [Google Scholar]
- 17.Handel T. M., Johnson Z., Crown S. E., Lau E. K., Proudfoot A. E. (2005) Annu. Rev. Biochem. 74, 385–410 [DOI] [PubMed] [Google Scholar]
- 18.Colditz I. G., Schneider M. A., Pruenster M., Rot A. (2007) Thromb. Haemost. 97, 688–693 [PubMed] [Google Scholar]
- 19.Rot A. (1992) Immunol. Today 13, 291–294 [DOI] [PubMed] [Google Scholar]
- 20.Mantovani A., Bonecchi R., Locati M. (2006) Nat. Rev. Immunol. 6, 907–918 [DOI] [PubMed] [Google Scholar]
- 21.D'Amico G., Frascaroli G., Bianchi G., Transidico P., Doni A., Vecchi A., Sozzani S., Allavena P., Mantovani A. (2000) Nat. Immunol. 1, 387–391 [DOI] [PubMed] [Google Scholar]
- 22.Gouwy M., Struyf S., Proost P., Van Damme J. (2005) Cytokine Growth Factor Rev. 16, 561–580 [DOI] [PubMed] [Google Scholar]
- 23.Salanga C. L., O'Hayre M., Handel T. (2009) Cell Mol. Life Sci. 66, 1370–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struyf S., Burdick M. D., Proost P., Van Damme J., Strieter R. M. (2004) Circ. Res. 95, 855–857 [DOI] [PubMed] [Google Scholar]
- 25.Menten P., Struyf S., Schutyser E., Wuyts A., De Clercq E., Schols D., Proost P., Van Damme J. (1999) J. Clin. Invest. 104, R1–R5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortier A., Van Damme J., Proost P. (2008) Pharmacol. Ther. 120, 197–217 [DOI] [PubMed] [Google Scholar]
- 27.Savino B., Borroni E. M., Torres N. M., Proost P., Struyf S., Mortier A., Mantovani A., Locati M., Bonecchi R. (2009) J. Biol. Chem. 284, 26207–26215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahuja S. K., Murphy P. M. (1996) J. Biol. Chem. 271, 20545–20550 [DOI] [PubMed] [Google Scholar]
- 29.Walz A., Burgener R., Car B., Baggiolini M., Kunkel S. L., Strieter R. M. (1991) J. Exp. Med. 174, 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashmun R. A., Look A. T. (1990) Blood 75, 462–469 [PubMed] [Google Scholar]
- 31.Proost P., Mortier A., Loos T., Vandercappellen J., Gouwy M., Ronsse I., Schutyser E., Put W., Parmentier M., Struyf S., Van Damme J. (2007) Blood 110, 37–44 [DOI] [PubMed] [Google Scholar]
- 32.Koch A. E., Kunkel S. L., Harlow L. A., Mazarakis D. D., Haines G. K., Burdick M. D., Pope R. M., Walz A., Strieter R. M. (1994) J. Clin. Invest. 94, 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinloch A., Lundberg K., Wait R., Wegner N., Lim N. H., Zendman A. J., Saxne T., Malmström V., Venables P. J. (2008) Arthritis Rheum. 58, 2287–2295 [DOI] [PubMed] [Google Scholar]
- 34.Loos T., Mortier A., Gouwy M., Ronsse I., Put W., Lenaerts J. P., Van Damme J., Proost P. (2008) Blood 112, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 35.Proost P., Loos T., Mortier A., Schutyser E., Gouwy M., Noppen S., Dillen C., Ronsse I., Conings R., Struyf S., Opdenakker G., Maudgal P. C., Van Damme J. (2008) J. Exp. Med. 205, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struyf S., Noppen S., Loos T., Mortier A., Gouwy M., Verbeke H., Huskens D., Luangsay S., Parmentier M., Geboes K., Schols D., Van Damme J., Proost P. (2009) J. Immunol. 182, 666–674 [DOI] [PubMed] [Google Scholar]
- 37.Ben-Baruch A., Bengali K. M., Biragyn A., Johnston J. J., Wang J. M., Kim J., Chuntharapai A., Michiel D. F., Oppenheim J. J., Kelvin D. J. (1995) J. Biol. Chem. 270, 9121–9128 [DOI] [PubMed] [Google Scholar]
- 38.Loos T., Mortier A., Proost P. (2009) Methods Enzymol. 461, 3–29 [DOI] [PubMed] [Google Scholar]
- 39.Wuyts A., Van Osselaer N., Haelens A., Samson I., Herdewijn P., Ben-Baruch A., Oppenheim J. J., Proost P., Van Damme J. (1997) Biochemistry 36, 2716–2723 [DOI] [PubMed] [Google Scholar]
- 40.Grynkiewicz G., Poenie M., Tsien R. Y. (1985) J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 41.Gouwy M., Struyf S., Noppen S., Schutyser E., Springael J. Y., Parmentier M., Proost P., Van Damme J. (2008) Mol. Pharmacol. 74, 485–495 [DOI] [PubMed] [Google Scholar]
- 42.Loos T., Opdenakker G., Van Damme J., Proost P. (2009) Haematologica 94, 1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breljak D., Gabrilovac J., Boranic M. (2003) Haema 6, 453–461 [Google Scholar]
- 44.Riemann D., Kehlen A., Langner J. (1999) Immunol. Today 20, 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shipp M. A., Look A. T. (1993) Blood 82, 1052–1070 [PubMed] [Google Scholar]
- 46.Wuyts A., Govaerts C., Struyf S., Lenaerts J. P., Put W., Conings R., Proost P., Van Damme J. (1999) Eur. J. Biochem. 260, 421–429 [DOI] [PubMed] [Google Scholar]
- 47.Nufer O., Corbett M., Walz A. (1999) Biochemistry 38, 636–642 [DOI] [PubMed] [Google Scholar]
- 48.Jiang Y., Tabak L. A., Valente A. J., Graves D. T. (1991) Biochem. Biophys. Res. Commun. 178, 1400–1404 [DOI] [PubMed] [Google Scholar]
- 49.Look A. T., Ashmun R. A., Shapiro L. H., Peiper S. C. (1989) J. Clin. Invest. 83, 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner A. J. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) pp. 289–294, Elsevier Academic Press, London [Google Scholar]
- 51.György B., Tóth E., Tarcsa E., Falus A., Buzás E. I. (2006) Int. J. Biochem. Cell Biol. 38, 1662–1677 [DOI] [PubMed] [Google Scholar]
- 52.Vossenaar E. R., Zendman A. J., van Venrooij W. J., Pruijn G. J. (2003) Bioessays 25, 1106–1118 [DOI] [PubMed] [Google Scholar]
- 53.Tarcsa E., Marekov L. N., Mei G., Melino G., Lee S. C., Steinert P. M. (1996) J. Biol. Chem. 271, 30709–30716 [DOI] [PubMed] [Google Scholar]
- 54.Chang X., Yamada R., Suzuki A., Sawada T., Yoshino S., Tokuhiro S., Yamamoto K. (2005) Rheumatology 44, 40–50 [DOI] [PubMed] [Google Scholar]
- 55.Foulquier C., Sebbag M., Clavel C., Chapuy-Regaud S., Al Badine R., Méchin M. C., Vincent C., Nachat R., Yamada M., Takahara H., Simon M., Guerrin M., Serre G. (2007) Arthritis Rheum. 56, 3541–3553 [DOI] [PubMed] [Google Scholar]
- 56.Imaizumi T., Albertine K. H., Jicha D. L., McIntyre T. M., Prescott S. M., Zimmerman G. A. (1997) Am. J. Respir. Cell Mol. Biol. 17, 181–192 [DOI] [PubMed] [Google Scholar]
- 57.Lukacs N. W., Kunkel S. L., Allen R., Evanoff H. L., Shaklee C. L., Sherman J. S., Burdick M. D., Strieter R. M. (1995) Am. J. Physiol. Lung Cell. Mol. Physiol. 268, L856–L861 [DOI] [PubMed] [Google Scholar]
- 58.Strell C., Lang K., Niggemann B., Zaenker K. S., Entschladen F. (2007) Cell Mol. Life Sci. 64, 3306–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zarbock A., Ley K. (2009) Microcirculation 16, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Brazzell J., Herrera A., Walcheck B. (2006) Blood 108, 2275–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gómez-Gaviro M., Domínguez-Luis M., Canchado J., Calafat J., Janssen H., Lara-Pezzi E., Fourie A., Tugores A., Valenzuela-Fernández A., Mollinedo F., Sánchez-Madrid F., Díaz-González F. (2007) J. Immunol. 178, 8053–8063 [DOI] [PubMed] [Google Scholar]
- 62.Bainton D. F., Miller L. J., Kishimoto T. K., Springer T. A. (1987) J. Exp. Med. 166, 1641–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]