Abstract
We present an interdisciplinary approach that, by incorporating a range of experimental and computational techniques, allows the identification and characterization of functional/immunogenic domains. This approach has been applied to ArtJ, an arginine-binding protein whose orthologs in Chlamydiae trachomatis (CT ArtJ) and pneumoniae (CPn ArtJ) are shown to have different immunogenic properties despite a high sequence similarity (60% identity). We have solved the crystallographic structures of CT ArtJ and CPn ArtJ, which are found to display a type II transporter fold organized in two α-β domains with the arginine-binding region at their interface. Although ArtJ is considered to belong to the periplasm, we found that both domains contain regions exposed on the bacterial surface. Moreover, we show that recombinant ArtJ binds to epithelial cells in vitro, suggesting a role for ArtJ in host-cell adhesion during Chlamydia infection. Experimental epitope mapping and computational analysis of physicochemical determinants of antibody recognition revealed that immunogenic epitopes reside mainly in the terminal (D1) domain of both CPn and CT ArtJ, whereas the surface properties of the respective binding-prone regions appear sufficiently different to assume divergent immunogenic behavior. Neutralization assays revealed that sera raised against CPn ArtJ D1 partially reduce both CPn and CT infectivity in vitro, suggesting that functional antibodies directed against this domain may potentially impair chlamydial infectivity. These findings suggest that the approach presented here, combining functional and structure-based analyses of evolutionary-related antigens can be a valuable tool for the identification of cross-species immunogenic epitopes for vaccine development.
Keywords: Adhesion, Amino Acid Transport, Bacteria, Epitope Mapping, Protein Structure, Chlamydia, Computational Epitope Mapping, Immunogenicity, Protein Modeling, Structural Vaccinology
Introduction
One of the riddles in vaccine research is that subunit antigens showing high sequence homology among bacterial species may eventually display diverse antigenic and protective properties. For example, antigens belonging to very conserved protein families, such as those including glycolytic enzymes or heat shock proteins, are highly expressed and immunogenic in most bacterial pathogens but are found to be protective only for some of them (1). Therefore, it emerges that sequence conservation and structural similarities are necessary but not sufficient prerequisites to predict antigen immunologic properties. Experimental evidence that highly homologous proteins fail to elicit cross-protection against closely related heterologous strains may be ascribed to a variety of causes, including differential expression level and/or cellular localization. The in vivo capacity of an antigen to raise antibodies able to protect from bacterial infection, by either neutralizing bacterial entry or promoting their killing, could also depend on a complex combination of properties including structural complexity, dynamics, and epitope distribution. In this context, we performed a structural and functional analysis of the ArtJ orthologs expressed by Chlamydiae trachomatis (CT)5 and pneumoniae (CPn) that, although similar at the sequence level, show diverse immunogenic properties (2). ArtJ is annotated by analogy with the ART transport systems of Escherichia coli, which has five genes organized in two operons (3): artPIQM and artJ, which are responsible for the arginine transport. In CPn, however, the artPIQM genes are absent and, therefore, it appears that chlamydial ArtJ operates in a molecular context different from the E. coli model and must be peculiar to this species. Moreover, ArtJ is able to induce high antibody titers both in mouse models and human patients that experienced a C. trachomatis infection.6 However, although recombinant CPn ArtJ elicited antibodies able to neutralize Chlamydia infectivity the CT protein did not show this functional activity (2).
This evidence raised the question whether differences in structural features and related properties such as dynamics, specific intramolecular interactions, and electrostatics, between CT and CPn ArtJ may account, in addition to (or as a consequence of) sequence differences, for their different immunogenicity. In this study we investigated the antigenic properties of ArtJ in the two Chlamydia species, by exploiting new structural information. Epitope mapping and computational analysis of physicochemical determinants of antibody recognition revealed the presence of several antigenic regions in C. pneumoniae that were not immunogenic in C. trachomatis. Two domains were identified and used for functional characterization. A serum raised against a specific recombinant domain of C. pneumoniae ArtJ, but not against the homologous domain of C. trachomatis, was able to partially neutralize both C. pneumoniae and C. trachomatis infectivity in vitro. Based on these data, we postulate that structure-based protein engineering of conserved antigenic domains may be an important tool for designing better vaccines. Moreover, in this study we developed a method based on a number of molecular and computational tools that, by going beyond classic structural modeling, allow a rapid identification of functional/immunogenic domains.
EXPERIMENTAL PROCEDURES
Preparation of CT EBs and Chromosomal DNA
C. trachomatis serovar D strain D/UW-3/CX and Chlamydia pneumoniae FB/96 strain, a clinical isolate from a patient with pneumonia at the Sant'Orsola Polyclinic, Bologna, Italy, were grown in LLC-MK2 cell cultures (ATCC CCL7). CT and CPn elementary bodies (EBs) were harvested 72 h after cell culture infection and purified by density gradient centrifugation. Purified EBs were resuspended in sucrose-phosphate transport buffer and stored at −80 °C until use. When required, EB infectivity was heat inactivated by 3 h of incubation at 56 °C. Chromosomal DNA was prepared from gradient-purified EBs by lysing the cells overnight at 37 °C with 10 mm Tris-HCl, 150 mm NaCl, 3 mm EDTA, 0.6% SDS, proteinase K (100 μg/ml), sequential extraction with phenol and chloroform, alcohol precipitation, and resuspension in TE buffer, pH 8.
Cloning and Expression of Recombinant Proteins
CT and CPn artJ ORFs were PCR-amplified using the respective chromosomal DNA as template. PCR primers were designed to amplify genes without signal peptides. NdeI and XhoI cloning sites were inserted in the 5′ tails of the forward and reverse primers, respectively. PCR products were digested with NdeI and XhoI (New England Biolabs), purified from agarose gel (Qiaex Gel Extraction Kit, Qiagen), ligated with pET21b (Novagen) digested with the same enzymes, and transformed in chemically competent BL21-DE3 cells. Correct pET21-CT381His and pET21-CPn0482His constructs were selected using PCR screening, protein expression, and DNA sequencing of inserts for the plasmid. The constructs expressing D1 and D2 ArtJ domains were then derived from pET21-CT381His and pET21-CPn0482His using the polymerase incomplete primer extension method as previously described (4). PCR were set up containing 1 μm each of the forward and reverse primers, 1× Cloned Pfu DNA Polymerase Reaction Buffer, 2.5 units of Pfu Turbo DNA polymerase (Stratagene), 200 μm of each dNTP (Invitrogen), and 2 ng of plasmid DNA template. The reactions were treated as follows: initial denaturation for 2 min at 95 °C, then 25 cycles of 95 °C for 30 s, 55 °C for 45 s, and 68 °C for 14 min (except for the amplification of the D2 domains where only 2 min of extension time were used) followed by a cooldown to 4 °C. To obtain the mutant expressing D1 domain of CT ArtJ, plasmid pET21-CT381His was amplified using oligos 5′-GGAGGAAGGTGGATACGGGATTGGCGTTGC-3′ and 5′-TATCCACCTTCCTCCCCATAATAGGGAATCATAAGAA-3′. 1 μl of PCR product was then used to transform chemically competent HK100 cells (5) and correct transformants where selected by PCR screening and DNA sequencing. The clone coding for the D1 domain of CPn artJ was similarly obtained using PCR primers 5′-CGATGAGGGTGGCTGTGGTCTCGGCGT-3′ and 5′-CAGCCACCCTCATCGCCATAATAGGGAAGCAG-3′ and plasmid pET21-CPn0482His as DNA template for PCR. pET constructs created in HK100 cells were then prepared and used to transform BL21-DE3 cells to allow protein expression.
To generate clones expressing CT ArtJ D2 and CPn ArtJ D2, plasmids pET21-CT381His and pET21-CPn0482His were used as PCR templates to amplify the respective D2 domains. PCR products were then cloned in SpeedET/CcdB (4) using the polymerase incomplete primer extension cloning method and the above described PCR conditions. Briefly, a vector PCR (V-PCR) was performed using the SpeedET/CcdB vector as DNA template and oligos 5′-GCCCTGGAAGTACAGGTTTTCGTGATGATGATGATGATG-3′ and 5′-TAACGCGACTTAATTAAACGGTCTCCAGCTTGGCTGTTTTGGC-3′.Two insert PCRs were done to amplify D2 domains from pET21-CT381His and pET21-CPn0482His. The oligos for the amplification of CT ArtJ D2 were 5′-CTGTACTTCCAGGGCATAAAACACTTGGTTTTAGTGTTTAAAGGA-3′ and 5′-AATTAAGTCGCGTTATAAAACCCACTGATCTTCAGGGAG-3′. The oligos for CPn ArtJ D2 were 5′-CTGTACTTCCAGGGCGTTCAAGAGCTGATGGTGGTTTCT-3′ and 5′-AATTAAGTCGCGTTACAACACCCAACATTCAGGAGG-3′. 1 μl of V-PCR was mixed with 1 μl of each insert PCR, the mixtures were used to transform HK100 cells, and the correct clones were selected by PCR screening, protein expression, and DNA sequencing.
To express cloned proteins, BL21-DE3 clones containing pET constructs were grown in LB medium, 100 μg/ml of ampicillin at 25 °C until A600 = 0.5. Protein expression was then induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside and growing at the same temperature for and additional 4 h. To express proteins in HK100 cells containing SpeedET constructs, the clones were grown as above but kanamycin was used as antibiotic and 0.15% arabinose was used as inducer. Purification of His fusions were performed from the bacterial soluble fraction using nickel affinity chromatography as already described (6).
Protein Crystallization
Commercial screens were set up in 100-nl drop format using a “Honeybee” robot (Cartesian). Initial hits were optimized and transferred to a microliter scale in Limbro format. Final crystallization conditions were: sodium formate, 0.5 to 4 m; Tris, 100 mm, pH 6.8 to 8.7 (ratio of 1 volume of protein solution/1 volume of reservoir solution), for CT ArtJ and 0.2 m thiocyanate, 1.5–1.8; AmSO4 (ratio 2/1) for CPn ArtJ. Crystallization of CPn ArtJ required significant optimization and seeding from the initial “sea urchin”-shaped crystals.
Data Collection and Structure Resolution
Cryo conditions were directly obtained for CT ArtJ but 20% glycerol was added to the drop before crystal recovery for CPn ArtJ. Data were measured at the ESRF. The datasets were indexed and reduced using XDS (7). Initial molecular replacement attempts on CT ArtJ using the structures of some existing bilobal transporters failed. Thus, we used SAD to solve the structure of CT ArtJ. ShelX detected 16 selenium atoms in the unit cell and produced phases that were used by both “ArpWarp” (8) and “Resolve” (RESOLVE model-building) (9). This produced a density map that was refined further using Refmac5 and Coot. For CPn ArtJ the structure was solved first using the D1 domain of CT ArtJ (residues 38–117 and 218–259) as a model for CPn ArtJ in Molrep. Once this domain had been refined using Refmac5/Coot, the central domain (residues 124–206) was added to the model to complete the refinement. The structures have been deposited in the Protein Data Bank (PDB) with entry codes 3N26 (CPn ArtJ) and 3DEL (CT ArtJ).
Molecular Dynamics Simulation
All molecular dynamics (MD) simulations were performed with the software package GROMACS 3.3.1 using the GROMOS 45a3 force field with the SPC water model, and periodic boundary conditions. Temperature and pressure were kept at 300 K and 1 bar. The other MD parameters were set as described elsewhere (10). The simulations of CT ArtJ and CPn ArtJ included residues Glu-34 to Asn-257 and Arg-31 to Glu-255, respectively. The initial open apo conformation of CPn ArtJ, and the initial closed forms (both apo and bound) of CT ArtJ, were obtained by homology modeling using MODELLER (11–14) with the CT ArtJ and CPn ArtJ crystallographic structure described here as template, respectively. This generated six systems for simulation.
Protein structures taken every 25 ps in 100–200 ns of the five simulations performed for each system were clustered with the algorithm by Daura et al. (15, 16), using all C-α atoms (including that of the arginine ligand, when present) for calculation of the root mean square deviation and a cut-off of 0.2 nm. The resulting five largest structural clusters were used for the energy-based predictions of the location of epitopes.
Epitope Prediction by Energy Decomposition
The full description of the energy decomposition method can be found in Ref. 17. The description and benchmark of the extension of the energy decomposition method for use in large-scale epitope prediction efforts is part of Ref. 18. Briefly, for a protein of N residues, the NXN matrix of average non-bonded interactions between pairs of residues is built from the MD trajectory. The energy map is further simplified through Eigen value decomposition. Analysis of the N components of the eigenvector associated with the lowest Eigen value was shown to single out those residues (hot sites) behaving as strongly interacting and possible stabilizing centers (17). The energy decomposition analysis allows, in a complementary way, to single out residues that are not strongly coupled intra-molecularly to other residues, and whose interaction networks are not optimized. The latter may conveniently interact with a putative binding partner with minimal free-energy expense and with minimal perturbation of the three-dimensional fold-stability of the protein. The contact matrix-based topological analysis provides information on the spatial location and conformational properties of those residues (see supplemental Fig. S1). This allows highlighting specific regions of non-optimized interactions. It was shown that these regions cluster on the antigen surface and are most likely to be involved in antibody binding, translating in the actual physicochemical-based epitope prediction. Preliminary evaluation of the performance of this approach on a large test set of antigens with known epitopes showed high levels of accuracy, sensitivity, specificity, and positive predictive value (precision).
Epitope Prediction by Electrostatic Desolvation
The electrostatic properties of a protein surface were calculated solving the finite difference Poisson Boltzmann equation using the APBS program (19). A neutral probe with low dielectric constant was placed at various surface positions and the electrostatic energy difference of the system with a probe minus the protein alone was estimated for every probe placement. The probe perturbs the electric field and gives rise to an energetic penalty to replace the high dielectric aqueous region by a low dielectric probe. To assess the electrostatic desolvation penalty of a protein surface patch, the average desolvation energy of all probes within a distance cutoff of 10 Å to a given surface point was calculated and mapped onto the surface defined by the probe molecules (see supplemental Fig. S2). The surface regions with the lowest desolvation penalties are considered as putative conformational epitopes. The method was recently evaluated on a large set of known antibody-antigen complexes and has shown good selectivity and specificity to predict antigen epitope regions. Moreover, protein surfaces usually contain several possible epitope regions and the method is able to detect them as demonstrated for different lysozyme-antibody complexes (20). Both CT and CPn ArtJ x-ray structures, as well as their isolated domains (D1 and D2) and the simulation representatives extracted from MD simulations, were used for epitope predictions.
Preparation of Mouse Antisera
Mouse antisera were generated and treated as described (6). Briefly, groups of four 5–6-week-old CD1 female mice (Charles River) were immunized intra-peritoneally at days 1, 15, and 28 with 20 μg of purified recombinant protein in Freund's adjuvant. Preimmune and immune sera were prepared from blood samples collected on days 0 and 43, respectively, and pooled before use. Where specified, sera from mice immunized with 20 μg of E. coli contaminant proteins (IMAC-purified proteins from E. coli bacteria containing pET21b+ empty vector) were used as negative control. Western blot, ELISA, and flow cytometry of C. trachomatis EBs were performed as previously described (21). To reduce the amount of antibodies possibly elicited by contaminating E. coli antigens, the immune sera were incubated overnight at 4 °C with nitrocellulose strips adsorbed with an E. coli BL21 total protein extract.
In Vitro Neutralization Assay
In vitro neutralization assays were performed on LLC-MK2 (Rhesus monkey kidney) epithelial cell cultures. Serial dilutions of mouse immune and corresponding preimmune sera were prepared in sucrose/phosphate/glutamic acid buffer (SPG). Mouse polyclonal sera to whole EBs were used as positive control of neutralization, whereas SPG buffer alone was used as negative control of neutralization (control of infection). Purified infectious EBs from the serotype-D CT strain were diluted in SPG buffer to contain 3 × 105 IFU/μl, and 10 μl of EBs suspension were added to each serum dilution in a final volume of 100 μl. Antibody-EB interaction was allowed to proceed for 30 min at 37 °C on a slowly rocking platform. The 100 μl of reaction mixture from each sample was used to inoculate PBS-washed LLC-MK2 confluent monolayers (in triplicate for each serum dilution), in a 96-well tissue culture plate, and centrifuged at 805 × g for 1 h at 37 °C. After centrifugation Eagle's minimal essential medium containing Earle's salts, 20% fetal bovine serum, and 1 μg/ml of cycloheximide was added. Infected cultures were incubated at 37 °C in 5% CO2 for 72 h. The monolayers were fixed with methanol and the chlamydial inclusions were detected by staining with a mouse anti-Chlamydia fluorescein-conjugated monoclonal antibody (Merifluor Chlamydia, Meridian Diagnostics, Inc.) and quantified by counting 5 fields per well at a magnification of ×40. The inhibition of infectivity due to EB interaction with the immune sera was calculated as percentage reduction in mean IFU number as compared with the SPG (buffer only)/EBs control. In this calculation the IFU counts obtained with immune sera were corrected for background inhibition of infection due to the corresponding preimmune mouse serum. Experimental variability was evaluated by calculating the S.E. of measurement from three titration experiments for each recombinant antigen.
Flow Cytometry
Gradient purified serotype D EBs were rendered not infectious by either heat or UV treatments and flow cytometry analysis was carried out on EB samples inactivated in the two ways. Inactivated EBs (2 × 105 cells) from CT resuspended in PBS, 0.1% bovine serum albumin (BSA), were incubated for 30 min at 4 °C with the specific mouse antisera (standard dilution 1:400). After centrifugation and washing with 200 μl of PBS, 0.1% BSA, the samples were incubated for 30 min at 4 °C with goat anti-mouse IgG, F(ab′)2-specific conjugated with R-phycoerythrin (Jackson ImmunoResearch Laboratories Inc.). The samples were washed with PBS, 0.1% BSA, resuspended in 150 μl of PBS, 0.1% BSA, and analyzed by Flow Cytometry using a FACSCalibur apparatus (BD Biosciences). Control samples were similarly prepared. Positive control antibodies were: (i) a commercial anti-CT specific monoclonal antibody (Argene Biosoft, Varilhes, France) and (ii) a mouse polyclonal serum prepared by immunizing mice with gradient purified EBs. Sera raised against E. coli contaminant proteins were used as background control sera. FACS data were analyzed using the Cell Quest Software (BD Biosciences). The significance of the FACS assay data has been elaborated by calculating the Kolmogorov-Smirnov statistic (K-S score) (22).
Peptide Spot Synthesis
Spot synthesis of 15-mer peptides, overlapping by 10, was performed on amino-polyethylene glycol-cellulose membranes by an automated spot synthesizer (MultiSynTech, Bochum, Germany) using 9-fluorenylmethoxycarbonyl chemistry and O-(benzotriazol-1-yl)-N,N,N,N-tetramethyluronium hexafluorophosphate-1,3-diisopropylethylamine activation. After the final synthesis cycle, the side chain protective groups were removed using a mixture of trifluoroacetic acid/triisobutylsilane/water/dichloromethane.
Peptide Binding Assay
Cellulose-bound peptides were soaked in ethanol to prevent hydrophobic interaction between the peptides. Nonspecific binding was blocked by incubating cellulose membranes overnight at 4 °C with 10 ml of 5% milk in PBS containing 0.05% Tween 20. The membranes were incubated for 2 h at 37 °C with mouse anti-CT ARTJ and CPn ArtJ polyclonal antibodies at a 1/1000 dilution, followed by 1/1000 dilution of HRP-conjugated goat anti-mouse immunoglobulin G (IgG). The membranes were developed with Opti 4CN-substrate kit (Bio-Rad). The analysis was repeated twice with identical results. The response of alkaline phosphatase-conjugated goat anti-mouse IgG on the same panel of peptides was negligible. Membranes incubated with an unrelated antiserum were used as negative controls.
Epitope Mapping with Monoclonal Antibodies
Immunocapturing of Peptides from Antigen Partial Digestion
Peptide mixtures were obtained by digestion with trypsin and Lys-C (independently) in 50 mm ammonium bicarbonate buffer in a 10/1 ratio, at 37 °C for 3 h. Each peptide mixture was analyzed separately. To capture the epitope-containing peptide, a 25-μl suspension of Dynabeads Pan Mouse IgG (uniform, superparamagnetic polystyrene beads of 4.5 μm diameter coated with monoclonal human anti-mouse IgG antibodies) was used. The beads were washed twice with PBS using a magnet and resuspended in the initial volume. 1 μg of the probe (murine) mAb was added and incubated for 30 min at room temperature, after which the beads were washed twice with PBS to remove excess mAb. 0.5 μl of Protease Inhibitor Mixture (GE Healthcare) was added before the peptide mixture to avoid potential degradation of the antibodies. On addition of the peptide mixture, the sample was incubated for 30 min at room temperature with gentle tilting and rotation. After incubation, the beads were washed three times with 1 ml of PBS, and the bound peptide was then eluted with 50 μl of 0.2% TFA. The elute fraction was concentrated and washed with C18 ZipTips (Millipore) and eluted in 3 μl of 50% acetonitrile and 0.1% TFA. For MALDI-MS analysis, 1 μl of sample was mixed with the same volume of a solution of α-cyano-4-hydroxytranscinnamic acid matrix (0.3 mg/ml in H2O/CAN/TFA at 6/3/1), spotted onto the MALDI target plate (Bruker), and allowed to air dry at room temperature. MALDI-mass spectra were recorded in the positive ion mode on an Ultraflex TOF instrument. Ion acceleration was set to 25 kV. All mass spectra were externally calibrated using a standard peptide mixture. For MS/MS analysis, the MASCOT search engine (Matrix Science, London, UK) was used with the following parameters: one missed cleavage permission, 50 ppm measurement for MS, and 0.1 Da for MS/MS tolerance. Positive identifications were accepted with p values lower than 0.05. In the searches, methionine residues modified to methionine sulfoxide were allowed.
Partial Digestion of Immunocaptured Antigens
To allow the capture of conformational epitopes, the order of the steps in the previous protocol was inverted. The intact protein (20 μg) was added to the beads, allowing binding to the immobilized mAbs. The protease was then added to the sample in a 50/1 ratio, with incubation at 37 °C for 3 h. After proteolysis, the beads were washed 10 times with 1 ml of PBS, and the bound peptide was then eluted as previously described. To discard the presence of proteolyzed fragments of the antibody in the elute fraction, negative controls were carried out where PBS was used instead of protein samples.
Dot Blot of Reverse Phase HPLC-separated Peptides from Antigen Partial Digestion
100 μg of CPn ArtJ and 100 μg of CT ArtJ were digested with trypsin and Lys-C, respectively. The peptides from each digestion were separated by reverse phase HPLC in a Waters Alliance apparatus: the peptide mixture was loaded onto a 5-μm, 4.6-mm inner diameter × 250-mm C18 column (Vydac Grace) with a linear gradient from 10 to 80% acetonitrile with 0.1% TFA at a flow rate of 0.5 ml/min for 70 min. Each collected fraction was vacuum-dried and re-dissolved in 10 μl of H2O. Nitrocellulose membranes were pre-treated with 1% glutaraldehyde during 10 min. 1 μl of each fraction was spotted onto the membrane. Nonspecific binding was blocked by incubating nitrocellulose membranes overnight at 4 °C with 10 ml of 1% Blocking reagent (Roche) in PBS containing 0.05% Tween 20. The membranes were incubated for 1 h at room temperature with anti-CT ArtJ or anti-CPn ArtJ mAbs at a 1/100 dilution, followed by a 1/3000 dilution of AP-conjugated rabbit anti-mouse IgG. The membranes were developed with nitro blue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate. Positive fractions were analyzed with an Ultraflex MALDI-TOF (Bruker Daltonics).
Fluorescence-activated Cell Sorter Analysis of the Binding of Recombinant CT ArtJ and CPn ArtJ to Epithelial Cells
LLC-MK2 epithelial cells were mixed with 1 and 50 μg/ml of recombinant CT ArtJ and CPn ArtJ and incubated for 1 h at 4 °C. Cells were subsequently incubated for 45 min at 4 °C with the specific mouse antisera in 5% FCS and binding revealed by R-phycoerytrin-conjugated secondary antibodies (Jackson ImmunoResearch Inc.) at 4 °C. Cell-bound fluorescence was analyzed by a FACSscan flow cytometer (BD Biosciences). Cells incubated with or without proteins were compared for Δ mean of fluorescence intensity values.
RESULTS
Recombinant CT ArtJ Does Not Induce Neutralizing Antibodies
Finco and co-workers (6) have recently reported that antibodies against CPn ArtJ, an EB surface-exposed protein, neutralize CPn infectivity in vitro (2). Prompted by this observation and the fact that CT also expresses ArtJ and the protein shares 61% identity with its CPn ortholog, we asked whether antibodies against recombinant CT ArtJ could similarly interfere with CT EB infectivity.
Before addressing this matter, we carried out two important preliminary experiments. First, we verified whether CT ArtJ is expressed on the surface of EBs as is the case for its CPn counterpart. Absence of surface expression would in fact make anti-ArtJ antibody neutralization unlikely. As shown in Fig. 1A, flow cytometry analysis of heat-inactivated CT EBs revealed that anti-ArtJ antibodies were able to bind purified EBs, as judged by the significant fluorescence shift with respect to the negative control serum (Ks 59.1). Protein accessibility to the surface was not determined by heat inactivation because a similar fluorescent intensity shift was observed when the infectivity of EBs was inactivated by UV irradiation (data not shown). These data were also confirmed by immunoblotting analysis on EB extracts (Fig. 1A). Second, we assessed whether ArtJ is involved in the interaction of Chlamydia with host cells. The recombinant forms of both CT and CPn ArtJ were incubated with the LLC-MK2 monkey kidney cell line and binding to the epithelial cells was followed by flow cytometry using anti-ArtJ antibodies. Both proteins bound the cell surface in a dose-dependent fashion, with binding reaching a plateau at a concentration of 50 μg/ml (approximately 1 μm) (Fig. 1C). Negative control cells were incubated with the respective antiserum in the absence of recombinant proteins, followed by fluorescent-conjugated secondary antibodies. BibA, a known group B streptococcus adhesion protein, was used as positive control.
FIGURE 1.
Recombinant ArtJ binds to human epithelial cells. A and B, CT and CPn ArtJ are exposed on chlamydial EBs. Flow cytometry analysis of gradient purified heat-inactivated EBs (2 × 105 cells) from CT incubated with polyclonal mouse anti-CT and CPn ArtJ antibodies and stained with R-phycoerytrin (PE)-conjugated anti-mouse IgG antibody (black histogram). The white histogram indicates bacteria treated with a preimmune control serum. Left panels to histograms represent immunoblotting analysis on EB extracts for each of the investigated antigens. C, binding of recombinant CT and CPn ArtJ to epithelial cells. LLC-MK2 epithelial cells were incubated for 1 h at 4 °C with increasing concentrations of recombinant ArtJ (red histograms, 1 μg/ml; blue histograms, 10 μg/ml; and green histograms, 50 μg/ml). Cells were incubated with mouse anti-ArtJ antibodies followed by R-phycoerytrin-conjugated secondary antibodies and analyzed by flow cytometry. Orange histograms represent binding to control cells incubated with the anti-ArtJ antiserum followed by fluorescent-conjugated secondary antibodies. Binding of a recombinant streptococcal cell-adhesion protein (BibA) was used as positive control.
Having demonstrated that CT ArtJ is exposed on the surface of EBs and that the protein specifically interacts with epithelial cells, we next assessed the effect of CT ArtJ antisera on the capability of CT EBs to infect in vitro cultures of LLC-MK2 cells. Infectious EBs were preincubated with either anti-ArtJ mouse polyclonal sera or preimmune sera and then used to infect LLC-MK2 cell monolayers. The number of inclusions was finally counted under the microscope for both EB preparations. The data indicate that anti-ArtJ antibodies do not affect inclusion formation in LLC-MK2 cells. Similar results were obtained when different Art-J antisera preparations were used (data not shown). These results suggest that, although very similar at the amino acid sequence level, CT and CPn ArtJ have different antigenic properties that ultimately result in the elicitation of antibody repertoires recognizing different regions (epitopes) of ArtJ.
C. trachomatis and C. pneumoniae ArtJ Share a Conserved Fold Consisting of Two Domains with an Arginine-binding Site at Their Interface
To investigate the presence of structure-dependent factors that could partly account for the different antigenic properties of the two orthologs, we determined the crystallographic structure of CT and CPn ArtJ by x-ray crystallography at 1.9 and 2.1 Å, respectively (Fig. 2, A and B, and Table 1). As expected, the structures of these two proteins are very similar to that of Geobacillus stearothermophilus ArtJ, recently solved in its arginine-bound state (PDB code 2Q2A)(23) (Fig. 2C), despite a sequence identity of only 21% between G. stearothermophilus and C. pneumoniae artJ. Like other periplasmic-binding proteins, CT and CPn ArtJ adopt a type II periplasmic-binding protein-fold (24) featuring two α-β domains with the arginine-binding site positioned at their interface. The N-C-terminal domain (D1) comprises the N terminus to residue ∼120 and from residue ∼215 to the C terminus (residue numbering corresponding to CT ArtJ). This domain shares the same structural features in CT and CPn ArtJ. The central domain (D2) encompasses residues ∼121 to ∼214 and displays only minor structural differences between the two proteins, mainly the presence of two additional β strands along residues 121–124 and 141–144 in CPn ArtJ (residue numbering corresponding to CT ArtJ) (Fig. 2B, S8 and S9). This high structural similarity between CPn and CT ArtJ is confirmed by α-carbon root mean square differences of 0.91 (D1) and 0.76 Å (D2) (Fig. 2D). In CPn ArtJ, an extra density was observed in the binding region. We attributed this density to a bound arginine molecule for the following reasons: (i) this density appeared despite the absence of ligand in the model used in refinement; (ii) there is an excellent fit of an Arg molecule in the experimental density that would not fit Lys or His; (iii) its position is identical to the one occupied by Arg in 2Q2A (23) (Fig. 2C). This amino acid was captured by the protein during expression and retained during purification despite extensive washes in nickel affinity and gel filtration steps. The Arg-binding site from CPn ArtJ is similar to that described for 2QA2 and is also made of two polar regions flanking a hydrophobic region (see Fig. 2E)(23). The arginine is held by hydrogen bonds with residues Asn-38, Thr-40, Ala-96, Gly-97, Ser-99, Arg-104, Phe-149, and Glu-186 and further enclosed in the binding site by the aromatic side chains of Tyr-41, Phe-79, Phe-149, and Met-98, Thr-148. There is no arginine density in CT ArtJ but, in this case, the C-terminal His tag folded back into the ligand-binding region and a density was observed for all His residues.
FIGURE 2.
Structures of CT ArtJ (A), CPn ArtJ (B), and G. stearothermophilus ArtJ (PDB code 2Q2A (23)) (C). Schematic representation of structures with α-helices colored red, β strands colored blue, and unstructured regions shown as gray tubes. Secondary structure elements (according to DSSP (32)) are labeled H1 to H8 for helices and S1 to S12 for β strands, following the polypeptide chain from the N to C terminus. The His tag occupying the binding site in the CT ArtJ structure is labeled and colored orange. The arginine ligand co-crystallized with CPn and G. stearothermophilus ArtJ is labeled and displayed as van der Waals spheres (C, orange; N, blue; O, red). Domains D1 and D2 are identified and separated by a dashed line. D, independent superposition of domains D1 and D2 from CT (red) and CPn (blue) ArtJ. For D1, the C-α atoms of residues Glu-34 to Glu-120 and Val-215 to Asn-256 (CT ArtJ residue numbering) were used for superposition and calculation of the reported C-α root mean square deviation. For D2, residues Ile-121 to Lys-129 and Pro-135 to Trp-214 were used. E, arginine binding site in the crystallographic structure of CPn ArtJ. The arginine ligand and the binding site residues interacting with it are represented with sticks (C, orange for ligand, gray for binding-site residues; N, blue; O, red).
TABLE 1.
X-ray crystallography data
MD Simulation Indicates That CT and CPn ArtJ Isolated Domains Are Stable
To guide the design of constructs for the experimental analysis of domain antigenicity, a MD simulation study of CT ArtJ and its isolated domains was performed. Several linkers were modeled and evaluated for the connection of the two polypeptide chains forming the terminal domain (D1). A single glycine residue was found to be sufficient for an energetically favorable linker solution. A total of five MD simulations of 50 ns were performed for each system in an aqueous environment. The isolated domains were found to be stable with dynamic properties nearly undistinguishable from the full-length protein.
Computational and Experimental Epitope Mapping of CT and CPn ArtJ Shows Differential Epitopes
Computational Analysis
To include information on ArtJ dynamics in the computational prediction of epitope regions, the dynamics of the apo and Arg-bound forms of CT and CPn ArtJ were first studied by MD simulation, based on the crystallographic structures described in this paper. For each system, five individual simulations of 200 ns were performed in explicit water. The apo forms, as expected for ligand-free periplasmic-binding proteins, exhibited large scale (semi-rigid body) inter-domain movements, whereas the Arg-bound forms were relatively rigid. The structures sampled in these simulations were then clustered by conformation and used for energy-based and electrostatic desolvation-based predictions of epitope regions.
Energy Decomposition-based Predictions
Epitope predictions were performed by means of a new computational method based on the integration of the topological information available from the atomic-contact matrix with the energetic information attainable through the energy decomposition approach (17, 25), allowing the mapping of the principal energy couplings in a protein undergoing dynamics. This approach provides information on both the dynamic and energetic properties of antigens, focusing on the identification of non-optimized, low-intensity interaction energy networks in the protein structure isolated in solution (18). Antibody recognition sites may in fact correspond to localized regions with low intensity energetic couplings with the rest of the protein. This would allow them to undergo conformational changes, to be recognized by a binding partner, and to tolerate mutations with a minimal energetic expense. The analysis was applied to representatives of the main clusters from each simulation of the apo- and Arg-bound CT and CPn ArtJ. The results of the predictions are reported in supplemental Table S1 and Fig. S3A. The predictions overlap with experimentally determined epitope sequences (supplemental Table S2 and see experimental mapping below). Thus, for CT ArtJ, the five predicted sequences overlap with experimentally determined ones, three in D1 and two in D2, whereas for the CPn ArtJ, overlap with the experiment was observed for five of the eight predicted sequences, three in D1, one in D2, and one split over the two domains. The most relevant aspect of these results, besides the overlap with experimental data, is that they suggest qualitative differences between the two domains (supplemental Table S2) and predict distinct epitope maps for the two proteins (supplemental Fig. S3A), supporting a physicochemical basis for their different antigenic properties. In particular, the D1 domain of CPn ArtJ shows a higher population of possible antigenic regions than the corresponding domain in CT ArtJ (the predicted epitopes in CT ArtJ D1 are also present in CPn ArtJ D1, the latter having additional ones), whereas the distribution in the respective D2 domains is more similar. Globally, CPn ArtJ appears to have a higher immunogenic potential, with more predicted epitopes and a larger number of epitope residues (supplemental Table S2).
Electrostatic Desolvation-based Predictions
Similar to other protein-protein interactions, the formation of antibody-antigen complexes requires the removal of water molecules from the binding interface region (20). An approach to calculate the electrostatic energy change upon displacing water from the protein surface was used to systematically investigate the surface desolvation properties of CT and CPn ArtJ. The results of the epitope predictions on ArtJ are summarized in supplemental Table S1 and Fig. S3B and, as with the previous method, correlate well with the experimental analysis. For CT ArtJ, six of the seven predicted sequences overlap with experimentally determined ones, four in D1 and two in D2, whereas for CPn ArtJ, overlap with the experiment is observed for five of the nine predicted sequences, four in D1 and one in D2. As with the previous method, the predictions suggest qualitative differences between the two domains (supplemental Table S2) and predict distinct epitope maps for the two proteins (supplemental Fig. S3A). As a matter of fact, the results are remarkably consistent with those of the energy decomposition-based prediction, despite the different physical properties behind the two methods.
The prediction methods can also detect potential differences between the full molecule and the isolated domains, as indicated in supplemental Table S1, where some of the epitopes are predicted in the isolated domains and not in the complete x-ray structures. It should be also noted that the prediction of epitopes that have not been detected in the experimental mappings (three with the energy-based method and five with the desolvation-based method) may have a number of causes. Thus, the epitope might still exist but not be detected under the given experimental conditions or the binding-prone region identified as an epitope might instead participate in other types of interactions, for example, with membrane components.
Consensus Model from Computational Predictions
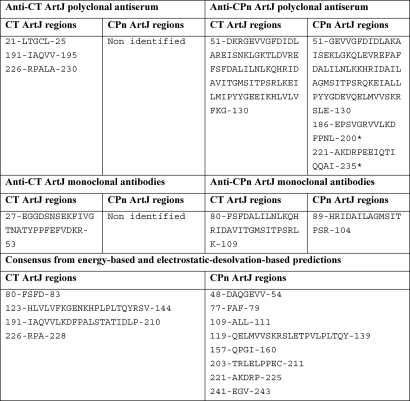
The results of the two computational methods were used to generate a consensus model for each protein, consisting in the strict intersection of the sets of epitopes predicted by the two approaches. The results are summarized in Table 2 and illustrated in Figs. 3 and 4. The consensus prediction shows the same basic features discussed after the independent methods, namely, that despite high sequence and structural similarity the two protein surfaces display local differences in their physicochemical properties that could potentially have an effect on the relative immunogenic properties of the two molecules. Again, CPn ArtJ appears to be more globally immunogenic (eight epitopes for four in CT ArtJ), with its D1 domain featuring epitopes that overlap with those in CT ArtJ D1 plus an additional three epitopes (Table 3). Predicted epitopes in D2 tend to be less numerous but longer in sequence and larger in surface, arguably indicating participation in less specific interactions (i.e. not with antibodies).
TABLE 2.
Epitope mapping by polyclonal antisera, monoclonal antibodies and computational predictions
Asterisk indicates a weak signal.
FIGURE 3.
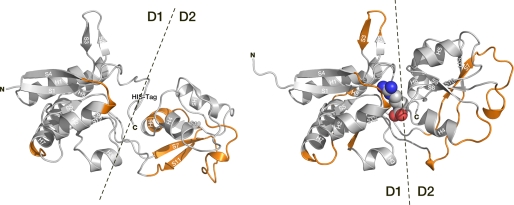
Mapping of the predicted epitopes on the x-ray structures of CT (left) and CPn (right) ArtJ. Orange segments correspond to the consensus between the energy-based and electrostatic desolvation-based epitope predictions. Secondary structural elements (according to DSSP (32)) are labeled as described in the legend to Fig. 2. Domains D1 and D2 are identified and separated by a dashed line.
FIGURE 4.
Graphical representation of the experimental and computational epitope mappings of CT and CPn ArtJ. Sequence alignment of the two proteins, with conserved residues in middle row. The beginning and end of D2 are marked with vertical lines. Experimentally determined epitopes are identified by the yellow background. Very low reactivity regions of CPn ArtJ are colored light blue. Epitope regions predicted by the two computational methods (consensus predicted epitopes) are shown in red, and highlighted in bold when they overlap with experimentally determined epitope regions.
TABLE 3.
Characterization of the epitopes predicted by the consensus sequences from the energy-based and electrostatic desolvation-based predictions
| CT ArtJ | CPn ArtJ | |
|---|---|---|
| Epitopes in D1 | 2 | 5 |
| Epitopes in D2 | 2 | 3 |
| Epitopes in D1 overlapping experimental ones | 2 | 4 |
| Epitopes in D2 overlapping experimental ones | 2 | 1 |
| Residues in D1 epitopes | 7 | 21 |
| Residues in D2 epitopes | 42 | 34 |
| Residues in D1 epitopes overlapping experimental ones | 7 | 15 |
| Residues in D2 epitopes overlapping experimental ones | 13 | 12 |
| Epitopes in D1 not overlapping with those in the ortholog protein | 0 | 3 |
| Epitopes in D2 not overlapping with those in the ortholog protein | 0 | 1 |
| Epitope residues in D1 not overlapping with those in the ortholog protein | 2 | 16 |
| Epitope residues in D2 not overlapping with those in the ortholog protein | 17 | 8 |
Experimental Analysis
Experimental mapping of epitopes was performed using both polyclonal sera and monoclonal antibodies. In either case, cross-recognition of the protein from one species by the serum or monoclonal antibodies generated against the other was also tested.
Polyclonal Mapping
To map epitopes recognized by sera from mice immunized with CT and CPn ArtJ, overlapping sequences of 15-residue peptides corresponding to both proteins were synthesized on a cellulose membrane and tested for binding of polyclonal antibodies by immunoblotting. As shown in Table 2 and Fig. 5A, anti-CPn ArtJ polyclonal antiserum recognized a number of different peptides within the N-terminal region of CPn ArtJ, spanning residues 51 to 130 and corresponding to peptides 11 to 26. No linear peptides were recognized from residues 131 to 259 (corresponding to peptides 27 to 52). A similar pattern was detected when anti-CPn ArtJ serum was incubated with CT ArtJ spotted membranes (Fig. 5B). As shown in Fig. 5C, when incubation was performed with anti-CT ArtJ polyclonal antiserum, the positive CT ArtJ peptides were found only within three restricted regions, with one located at the N terminus and corresponding to residues 16 to 30 (peptide 5) and the remaining two located at the C terminus (peptides 39 and 46). Interestingly, the anti-CT ArtJ serum did not strongly recognize any linear peptides of CPn ArtJ (Fig. 5D). In line with the predictions, these results show that the linear epitopes are partly common to both variants of ArtJ, but that the immunogenicity of the two antigens is different.
FIGURE 5.
Peptide recognition by Chlamydia ArtJ antisera. Two peptide arrays (8 peptides per row) were designed by synthesizing overlapping sequences of 15-residue peptides corresponding to CPn (A and D) and CT (B and C) ArtJ on a cellulose membrane and tested for binding by immunoblotting with mouse anti-CPn (A and B) and anti-CT (C and D) ArtJ polyclonal antibodies. Membranes incubated with an unrelated antiserum were used as negative controls. Numbers on the right of each panel mark the first peptide of each row. Red box in panel A indicates the CPn ArtJ region containing highly immunogenic peptides.
Monoclonal Mapping
mAbs against CT and CPn ArtJ were obtained and used to identify epitope regions by three mapping protocols based on limited antigen proteolysis. mAbs were selected according to their capability to recognize the antigen both by immunoblotting experiments (dot blot and Western blotting on recombinant ArtJ) and FACS analysis on chlamydial EBs. The analysis was performed both on the supernatant of hybridomas derived from the somatic fusion (first screening) and on the derived monoclones (second screening). Epitope mapping was performed for each mAb-antigen pair by three different protocols (see “Experimental Procedures” for details): 1) immunocapturing of peptides from antigen partial digestion, 2) partial digestion of immunocaptured antigens, and 3) dot blot of reverse phase HPLC-separated peptides from partial digestion of the antigen. In all three cases identification of the mAb-binding peptide was performed by mass spectrometry. Note that the second protocol should be able to identify structural epitopes, because antigen-mAb binding precedes digestion. Cross-recognition, between anti-CT ArtJ mAbs and CPn ArtJ as well as between anti-CT ArtJ mAbs and CPn ArtJ was also probed by each of the three methods. The monoclonal antibodies tested recognized the same peptides by any of the three protocols (see Table 2). The identified peptides belong in all cases to D1 and show overlap with regions identified by polyclonal antisera.
Both ArtJ D1 and D2 Domains Are Exposed on the Bacterial Surface and Bind to Epithelial Cells
After the finding that ArtJ is exposed on the bacterial surface and it is likely to be involved in the interaction of Chlamydia with host cells, we investigated whether the D1 and D2 domains were differently exposed on chlamydiae and maintained the capacity to bind to epithelial cells. Flow cytometry analysis of CT and CPn EBs showed that both D1 and D2 ArtJ antisera were able to recognize these domains on the surface of purified EBs from both chlamydiae species (Fig. 6A). The fact that both ArtJ domains may play a functional role in Chlamydia infectivity/adaptation to the host was further supported by evidence that both domains were able to bind to LLC-MK2 cells in a dose-dependent fashion (Fig. 6B).
FIGURE 6.
Individual ArtJ domains bind to human cells. A, D1 and D2 ArtJ domains are expressed on chlamydial EBs. Flow cytometry analysis of purified heat inactivated EBs (2 × 105 cells) incubated with polyclonal mouse anti-ArtJ domains antibodies. Experimental protocol was as described in the legend to Fig. 1A. The blue line histogram indicates bacteria treated with a preimmune serum. B, binding of recombinant D1 and D2 CT and CPn ArtJ domains to epithelial cells. LLC-MK2 epithelial cells were incubated for 1 h at 4 °C with 1 μg/ml (red histograms) or 10 μg/ml (blue histograms) of recombinant D1 and D2 domains. Staining of protein-bound cells was as described in the legend to Fig. 1B. Orange line histograms represent binding to control cells incubated with anti-ArtJ antiserum followed by fluorescent-conjugated secondary antibodies.
Sera Raised against CPn ArtJ D1 Partially Neutralize C. trachomatis Infectivity in Vitro
Our epitope mapping analysis showed a higher reactivity of the ArtJ D1 domain, compared with the D2 domain, when incubated with CPn ArtJ antiserum. Because no neutralizing activity on CT infectivity was observed using the low immunogenic CT ArtJ antiserum, we investigated the capacity of CPn ArtJ antisera (including both the sera against the whole protein and against the recombinant D1 and D2 domains) to neutralize CT EB infectivity in vitro. Antisera generated against both CT and CPn ArtJ recombinant proteins and the respective recombinant D1 and D2 domains were diluted 1/200 and incubated with CT EBs (all tested antisera showed comparable IgG titers, as estimated by ELISA on recombinant CT381). Opsonized EBs were then used to infect LLC-MK2 cell monolayers, using EB treated with preimmune sera as negative controls, at the same dilution. As shown in Fig. 7, antisera against the CPn ArtJ whole protein and CPn ArtJ D1 domain showed the highest activity, reducing EB infectivity by ∼30 and 45% (p value < 0.05), respectively, whereas the CPn D2 antiserum showed a negligible effect. The capacity of the CT ArtJ antisera to diminish CT EB infectivity was substantially lower, with values around 20%, in line with the immunogenicity data (Fig. 7).
FIGURE 7.
Antisera against CPn ArtJ neutralize C. trachomatis infectivity for LLC-MK2 cells. Infected CT EBs were opsonized with polyclonal mouse antisera raised against whole CT and CPn recombinant ArtJ proteins and their respective recombinant D1 and D2 domains (dilution 1/200) and used to infect LLC-MK2 cell monolayers, as compared with EBs treated with a corresponding dilution of preimmune sera. The antiserum neutralization activity was determined by measuring the reduction of the number of inclusions generated by antiserum-treated EBs after correction for background inhibition observed with preimmune sera. The histogram presents inclusion-forming unit values averaged from triplicate experiments, with standard deviations.
DISCUSSION
The advent of genomics and post-genomics has represented a turning point for antigen discovery. Starting from the knowledge of whole genome sequences, a brute force approach is now conceivable to which virtually all proteins of any given pathogen can be systematically tested for their capacity of eliciting protective immunity. “Reverse vaccinology” is a typical example of this, validated in different human pathogens (26, 27). The fact that the majority of antigens eliciting antibody-mediated protective immunity are surface-exposed proteins and secreted toxins have also promoted a number of proteomic strategies that allow the precise characterization, both in qualitative and quantitative terms, of membrane-associated and secreted proteins (28, 29). However, it remains intellectually challenging to understand which properties make these few proteins protective. To tackle this difficult question, we have recently proposed a novel approach based on a systematic and quantitative analysis of a number of molecular properties (including structural and dynamics analysis) within a set of protective and non-protective protein antigens from pathogenic bacteria (30). In this context, we found it interesting to elucidate the immunological properties of ArtJ, a surface-exposed chlamydial antigen annotated as a arginine-binding protein, which although well conserved among chlamydial species showed species-specific immunogenicity (2). Although Montigiani and co-workers (6), during their systematic analysis of the CPn genome, have identified in CPn ArtJ one of the EB surface-exposed antigens eliciting antibody-mediated neutralizing activity in vitro, we did not observe the induction of the same phenotype by using antisera against recombinant CT ArtJ homologues. Experimental mapping of antigenic regions in both CPn and CT ArtJ supported by structure-based computational analysis were crucial in unraveling the antigenic properties of Chlamydial ArtJ.
Indeed, analysis of the three-dimensional structural organization of the predicted epitope sequences suggests differences between the two proteins at the level of epitope presentation. In particular, both energy decomposition and electrostatic desolvation analyses show that putative interaction surfaces in CPn ArtJ are more extensive in CT ArtJ, suggesting a higher immunogenic character of the former. This is especially so in relation to the D1 domain, which features a larger number of putative epitopes and concentrates a majority of the differences between the epitope sets in the two ortholog proteins, with the CT ArtJ D1 epitopes overlapping with a subset of those predicted for CPn ArtJ. The D2 domain is characterized by a smaller number of predicted epitopes occupying a larger surface, which, combined with the weak immunogenic properties of D2 observed experimentally, could be indicative of a different, less-specific, type of interaction interface.
These results suggest that structural/dynamical differences, determined by relatively small differences in the primary and tertiary arrangements, may define the surface properties underlying differential epitope presentation and recognition by antibodies of the two antigens. It is important to note that the use of high resolution structures, proper characterization of the dynamics of the protein, and high resolution computational analyses can illuminate those small differences and give access into the structure-dynamic function relationships at the basis of antibody recognition.
In this context, it is worth underlining that both methods predict conformational epitopes, whereas experimental epitope mapping focuses on linear sequence stretches. This apparent contrast can be solved by noting that the majority of predicted epitopes are made up of amino acid stretches that are separated along the primary sequence. The latter can thus be conveniently expressed in terms of peptides as in epitope mapping experiments.
Another important result of the work presented here is that, contrary to what has been reported in the literature, according to which ArtJ-like transporters are restricted to the periplasmic space (31), both chlamydiae ArtJ appear to express domains exposed on the bacterial surface, as judged by the ability of anti-ArtJ polyclonal antibodies to bind EB. This fact was further corroborated by the capacity of recombinant ArtJ to bind to epithelial cells, suggesting a role for such a transporter in getting in contact with putative arginine-containing substrates expressed on the surface of host cells. In addition, preliminary results from our group7 showed that anti-CPn ArtJ antibodies, but not anti-CT ArtJ antibodies, are capable of interfering with the ability of recombinant ArtJ to bind to host cells, postulating a direct link between the adhesive properties of ArtJ and chlamydial infectivity.
An interesting aspect of this study is that it provides an additional piece of evidence on how adaptation can play a key role in survival and successful host colonization by microorganisms. One way that bacteria and viruses counteract host immune responses is by undergoing subtle changes of their exposed components to an extent that allows escape from the recognition of the immune system while preserving functional properties. Our data indicate that ArtJ, a protein well exposed on the surface of the EBs of both species and most likely required for EB internalization, slightly remodels its surface structure to diversify the immunodominant epitope profiles. This would confirm that epitope selection is a crucial step in bacterial adaptation to the host and that it is finely tuned to allow bacteria to successfully compete for the occupation of specific niches.
In conclusion, the combined analysis of structure, physicochemical determinants of antibody recognition, and epitope mapping allowed us to successfully identify and characterize immunogenic regions in ArtJ. A key aspect of this study was the analysis of two closely related antigens. Indeed, by this comparative approach we were able to provide a rational for the experimental differences in antibody recognition on the basis of the three-dimensional structure and surface properties derived from it. We therefore suggest that vaccine design may greatly benefit from this integrated approach.
Supplementary Material
Acknowledgments
We thank Giuliano Galli and Mauro Agnusdei for technical assistance and Oretta Finco for useful discussion.
This work was supported by Sixth Research Framework Programme of the European Union Ref. LSHB-CT-2006-037325, BacAbs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 3N26 and 3DEL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
R. Grifantini and G. Grandi, manuscript in preparation.
G. Gancitano, M. Soriani, R. Grifantini, and G. Grandi, unpublished results.
- CT
- Chlamydiae trachomatis
- CPn
- Chlamydiae pneumoniae
- EB
- elementary body
- MD
- molecular dynamics.
REFERENCES
- 1.Zügel U., Kaufmann S. H. (1999) Clin. Microbiol Rev. 12, 19–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finco O., Bonci A., Agnusdei M., Scarselli M., Petracca R., Norais N., Ferrari G., Garaguso I., Donati M., Sambri V., Cevenini R., Ratti G., Grandi G. (2005) Vaccine 23, 1178–1188 [DOI] [PubMed] [Google Scholar]
- 3.Makarova K. S., Mironov A. A., Gelfand M. S. (2001) Genome Biol. 2, RESEARCH0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klock H. E., Koesema E. J., Knuth M. W., Lesley S. A. (2008) Proteins 71, 982–994 [DOI] [PubMed] [Google Scholar]
- 5.Klock H. E., White A., Koesema E., Lesley S. A. (2005) J. Struct. Funct. Genomics 6, 89–94 [DOI] [PubMed] [Google Scholar]
- 6.Montigiani S., Falugi F., Scarselli M., Finco O., Petracca R., Galli G., Mariani M., Manetti R., Agnusdei M., Cevenini R., Donati M., Nogarotto R., Norais N., Garaguso I., Nuti S., Saletti G., Rosa D., Ratti G., Grandi G. (2002) Infect. Immun. 70, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 8.Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 9.Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Affentranger R., Tavernelli I., Di Iorio E. E. (2006) J. Chem. Theory Comput. 2, 217–228 [DOI] [PubMed] [Google Scholar]
- 11.Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2000) in Current Protocols in Bioinformatics, Suppl. 15, pp. 15.16.11–15.16.30, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiser A., Do R. K., Sali A. (2000) Protein Sci. 9, 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 14.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 15.Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W. F., Mark A. E. (1999) Angew. Chem. Int. Ed. 38, 236–240 [Google Scholar]
- 16.Daura X., van Gunsteren W. F., Mark A. E. (1999) Proteins 34, 269–280 [DOI] [PubMed] [Google Scholar]
- 17.Morra G., Colombo G. (2008) Proteins 72, 660–672 [DOI] [PubMed] [Google Scholar]
- 18.Scarabelli G., Morra G., Colombo G. (2010) Biophys J. 98, 1966–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorucci S., Zacharias M. (2010) Biophys. J. 98, 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meoni E., Faenzi E., Frigimelica E., Zedda L., Skibinski D., Giovinazzi S., Bonci A., Petracca R., Bartolini E., Galli G., Agnusdei M., Nardelli F., Buricchi F., Norais N., Ferlenghi I., Donati M., Cevenini R., Finco O., Grandi G., Grifantini R. (2009) Infect. Immun. 77, 4168–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young I. T. (1977) J. Histochem. Cytochem. 25, 935–941 [DOI] [PubMed] [Google Scholar]
- 23.Vahedi-Faridi A., Eckey V., Scheffel F., Alings C., Landmesser H., Schneider E., Saenger W. (2008) J. Mol. Biol. 375, 448–459 [DOI] [PubMed] [Google Scholar]
- 24.Fukami-Kobayashi K., Tateno Y., Nishikawa K. (1999) J. Mol. Biol. 286, 279–290 [DOI] [PubMed] [Google Scholar]
- 25.Tiana G., Simona F., De Mori G. M., Broglia R. A., Colombo G. (2004) Protein Sci. 13, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizza M., Scarlato V., Masignani V., Giuliani M. M., Aricò B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., Galeotti C. L., Luzzi E., Manetti R., Marchetti E., Mora M., Nuti S., Ratti G., Santini L., Savino S., Scarselli M., Storni E., Zuo P., Broeker M., Hundt E., Knapp B., Blair E., Mason T., Tettelin H., Hood D. W., Jeffries A. C., Saunders N. J., Granoff D. M., Venter J. C., Moxon E. R., Grandi G., Rappuoli R. (2000) Science 287, 1816–1820 [DOI] [PubMed] [Google Scholar]
- 27.Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., D'Agostino N., Miorin L., Buccato S., Mariani M., Galli G., Nogarotto R., Nardi Dei V., Vegni F., Fraser C., Mancuso G., Teti G., Madoff L. C., Paoletti L. C., Rappuoli R., Kasper D. L., Telford J. L., Grandi G. (2005) Science 309, 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Ortega M. J., Norais N., Bensi G., Liberatori S., Capo S., Mora M., Scarselli M., Doro F., Ferrari G., Garaguso I., Maggi T., Neumann A., Covre A., Telford J. L., Grandi G. (2006) Nat. Biotechnol. 24, 191–197 [DOI] [PubMed] [Google Scholar]
- 29.Berlanda Scorza F., Doro F., Rodríguez-Ortega M. J., Stella M., Liberatori S., Taddei A. R., Serino L., Gomes Moriel D., Nesta B., Fontana M. R., Spagnuolo A., Pizza M., Norais N., Grandi G. (2008) Mol. Cell. Proteomics 7, 473–485 [DOI] [PubMed] [Google Scholar]
- 30.Gourlay L. J., Colombo G., Soriani M., Grandi G., Daura X., Bolognesi M. (2009) Hum. Vaccin. 5, 1–4 [DOI] [PubMed] [Google Scholar]
- 31.Locher K. P. (2004) Curr. Opin. Struct. Biol. 14, 426–431 [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.