Abstract
Bacterial periplasmic binding proteins (PBPs) and eukaryotic PBP-like domains (also called as Venus flytrap modules) of G-protein-coupled receptors are involved in extracellular GABA perception. We investigated the structural and functional basis of ligand specificity of the PBP Atu2422, which is implicated in virulence and transport of GABA in the plant pathogen Agrobacterium tumefaciens. Five high-resolution x-ray structures of Atu2422 liganded to GABA, Pro, Ala, and Val and of point mutant Atu2422-F77A liganded to Leu were determined. Structural analysis of the ligand-binding site revealed two essential residues, Phe77 and Tyr275, the implication of which in GABA signaling and virulence was confirmed using A. tumefaciens cells expressing corresponding Atu2422 mutants. Phe77 restricts ligand specificity to α-amino acids with a short lateral chain, which act as antagonists of GABA signaling in A. tumefaciens. Tyr275 specifically interacts with the GABA γ-amino group. Conservation of these two key residues in proteins phylogenetically related to Atu2422 brought to light a subfamily of PBPs in which all members could bind GABA and short α-amino acids. This work led to the identification of a fingerprint sequence and structural features for defining PBPs that bind GABA and its competitors and revealed their occurrence among host-interacting proteobacteria.
Keywords: Amino Acid Transport, Crystal Structure, GABA Receptors, Plant, Transport Amino Acids, Host-Pathogen Relationship, Periplasmic Binding Protein, Quorum Sensing, Solute-binding Protein, Virulence
Introduction
The non-protein amino acid GABA acts as an intercellular signal during development and differentiation of eukaryotes (including animals, plants, and amebas) as well as an interspecies signal for the establishment of host-microbe interactions (1–3). GABA is proposed to be an ancient messenger in cellular and organismic communication (3, 4). In the central nervous system of mammals, GABA is the main inhibitory neurotransmitter that is sensed by ionotropic GABAA and metabotropic GABAB proteins (5). In the ameba Dictyostelium, GABA binds to the protein GrlE, which controls terminal steps of sporulation (4). In plants, GABA accumulates in response to abiotic and biotic stresses (6), including wind, injury, and infection by fungi (7) and symbiotic and pathogenic bacteria, such as Rhizobium leguminosarum (8) and Agrobacterium tumefaciens (9). In A. tumefaciens, GABA acts as a signal that triggers gene expression (10, 11). GABA-regulated functions contribute to moderate development of plant tumors and encompass the lactonase AttM, which inactivates the bacterial signal 3-oxo-octanoylhomoserine lactone (OC8HSL)3 (12, 13). However, free l-Pro, which also accumulates in infected plant tissues, blocks importation of GABA in the bacterial cells and hence blocks the GABA-induced degradation of OC8HSL and enhances the emergence of plant tumors (14, 15). GABA and Pro import from the plant host into the A. tumefaciens cytoplasm requires Atu2422, which is coupled to the ATP-binding cassette transporter Bra.
Atu2422 belongs to the large family of periplasmic binding proteins (PBPs), also named solute-binding proteins. In eukaryotes, proteins sensing GABA, such as GABAB and GrlE, are class C G-protein-coupled proteins (GPCRs). They possess a PBP-like domain (also termed “Venus flytrap module”) that exhibits a sequence identity of ∼20% to actual PBPs (4, 5, 16–18). The PBP fold consists of two flexible lobes with an α/β-fold separated by a cleft in which the ligand binds. The ligand-free PBPs adopt an open conformation. In bacteria, their closed liganded conformation is stable and can transfer the cognate ligand to an ATP-binding cassette transporter. So far, there is no available structure of a PBP or a eukaryotic GPCR in complex with GABA.
Here, we report five structures at high resolution (from 1.35 to 1.8 Å) of the mature PBP Atu2422 in the presence of four ligands, GABA, Pro, l-Ala, and l-Val. Structural analysis of the ligand-binding site of Atu2422 and that of the point mutant Atu2422-F77A in combination with in vivo mutational and functional studies demonstrated that the two residues Phe77 and Tyr275 contribute to ligand specificity and GABA binding and hence to the modulation of virulence of A. tumefaciens. Phylogenetic analysis supports Atu2422 as a structural basis for a PBP subfamily, whose members, which occur in several host-interacting bacteria, can bind GABA and protein amino acids with short lateral chains only.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
The A. tumefaciens strains used in this work were derivatives of A. tumefaciens C58. They were cultivated at 30 °C in LB medium and Agrobacterium broth (AB) medium (19) in the presence of mannitol (2 g/liter) and ammonium chloride (1 g/liter). Escherichia coli strains were cultivated in LB medium at 37 °C. Antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; gentamycin, 25 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 10 μg/ml.
Site-directed Mutagenesis of atu2422
The pGEM-T Easy vector (Promega) harboring the atu2422 gene was used as a template to generate directed mutations with the QuikChange® II XL site-directed mutagenesis kit (Stratagene). The synthetic forward primer 5′-TTCGTTGTCGGTCACGCCAACTCGGGTGTTTCC (with the altered nucleotides underlined) and the reverse complement primer were designed for mutation of the codon Phe to Ala at position 77. For mutation of the codon Tyr275 to Lys, Ala, and Phe, primers 5′-TTCAACCGGGAAGCCAAGACGCTCTACTCCTAT, 5′-TTCAACCGGGAAGCCGCCACGCTCTACTCCTAT, and 5′-TTCAACCGGGAAGCCTTCACGCTCTACTCCTAT and their appropriate reverse primers were used. The atu2422 gene and mutated alleles atu2422-F77A, atu2422-Y275K, atu2422-Y275A, and atu2422-Y275F were subcloned into the host wide-range plasmid pME6000 (20). The resulting plasmids were introduced into the A. tumefaciens defective mutant atu2422 (14) by electroporation as described previously (10). All constructed alleles and strains were verified by PCR and DNA sequencing.
Purification of Mature Atu2422
Purification under classical conditions of mature (without peptide signal) Atu2422 binding Ala as a fortuitous ligand has been described previously (21). To replace Ala with amino acids of interest (Pro, Val, and GABA), Atu2422 was purified under denaturing/renaturing conditions. After cell lysis in denaturing buffer containing 25 mm Tris-HCl (pH 7.5), 10 mm imidazole, and 7 m urea, the supernatant was loaded on nickel-nitrilotriacetic acid resin equilibrated in the same buffer. Low-affinity binding contaminants were washed from the column with 25 mm Tris-HCl (pH 7.5), 20 mm imidazole, and 7 m urea. A buffer of 25 mm Tris-HCl (pH 7.5), 20 mm imidazole, and 300 mm NaCl was passed through the column to return the protein to renaturing conditions prior to its elution with 25 mm Tris-HCl (pH 7.5), 300 mm imidazole, and 300 mm NaCl. The eluted sample was further purified by size-exclusion chromatography using a Superdex 200 HiLoad column (Amersham Biosciences) equilibrated in 25 mm Tris-HCl (pH 7.5) and 150 mm NaCl. Atu2422-His was concentrated to 11 mg/ml using Vivaspin 10 centrifugal concentrators (Vivascience) and stored at −80 °C. The Atu2422-F77A mutant was purified as the wild-type protein (21) and concentrated to 15 mg/ml.
Crystallization and Data Collection
Crystallization of Atu2422 under PEG conditions was as described (21). Ammonium sulfate (AS) conditions were screened using the AS kit from Qiagen. One condition manually optimized with homemade solutions in hanging drops composed of a 1:1 volume ratio of crystallization solution (0.1 m Tris-HCl (pH 8.5), 2.5 m ammonium sulfate, and 6% glycerol) and protein solution with or without the addition of amino acids led to crystals. After purification either in the presence of Ala or under denaturing conditions, protein was incubated with different ligands (such as GABA, proline, and valine) for 30 min at concentrations ranging from 40 to 100 mm to promote exchange of amino acids. Atu2422-F77A crystallized under the same AS conditions as the wild-type protein. Crystals were transferred to a cryoprotectant solution (mother liquor supplemented with 20% glycerol for PEG conditions or oil for AS conditions) and flash-frozen in liquid nitrogen. Diffraction data at high resolution were collected at 100 K on the PROXIMA 1 beamline (SOLEIL Synchrotron, Saint-Aubin, France). Diffraction intensities were integrated with the program XDS (22). Data collection and processing statistics are given in Table 1.
TABLE 1.
Crystallographic data and refinement parameters
Values for the highest resolution shell are in parentheses. PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
| WT-Ala | WT-Ala | WT-Pro | WT-Val | WT-GABA | F77A-Leu | |
|---|---|---|---|---|---|---|
| PDB code | 3IP5 | 3IPA | 3IP6 | 3IP7 | 3IP9 | 3IPC |
| Precipitant | PEG | AS | AS | PEG | AS | AS |
| Space group C2 cell parameters | a = 116.2, b = 38.9, c = 71.6 Å; β = 95.7° | a = 119, b = 43, c = 66.2 Å; β = 109.6° | a = 119.2, b = 43, c = 66.3 Å; β = 109.6° | a = 116.6, b = 39.1, c = 70.7 Å; β = 94.4° | a = 119.7, b = 43.1, c = 66.5 Å; β = 109.8° | a = 119.8, b = 43, c = 66.5 Å; β = 109.8° |
| Resolution (Å) | 30-1.35 (1.42-1.35) | 30-1.55 (1.64-1.55) | 30-1.4 (1.48-1.4) | 30-1.7 (1.8-1.7) | 30-1.8 (1.91-1.8) | 30-1.3 (1.39-1.3) |
| No. of observed reflections | 283,328 (38,572) | 154,581 (24,403) | 203,587 (31,467) | 133,087 (19,630) | 108,719 (17,021) | 244,845 (11,695) |
| No. of unique reflections | 69,760 (9671) | 45,887 (7312) | 61,219 (9581) | 34,935 (5500) | 29,021 (4526) | 68,599 (6206) |
| Rsym (%)a | 10.4 (50.9) | 10.9 (66) | 8.4 (50.4) | 6.5 (51.6) | 9.7 (51) | 4.5 (33.7) |
| Completeness (%) | 99 (98.2) | 99.5 (99) | 97.6 (95.1) | 98.6 (97.2) | 97 (94.9) | 89 (50) |
| I/σ | 13 (2.1) | 9.4 (2.1) | 9.1 (2.5) | 12.5 (2.3) | 10.9 (3.1) | 17.3 (2.6) |
| No. of reflections for refinement | 69,735 | 45,849 | 61,197 | 34,905 | 29,018 | 68,579 |
| Rcryst (%)b | 16.2 | 17 | 19.8 | 23 | 16.9 | 15.8 |
| Rfree (%)c | 18.6 | 20.2 | 23 | 27.1 | 21.5 | 18.6 |
| Bond r.m.s.d. (Å) | 0.005 | 0.004 | 0.007 | 0.008 | 0.005 | 0.006 |
| angle r.m.s.d. | 1.0° | 0.9° | 1.0° | 1.1° | 0.9° | 1.0° |
| Average B (Å2) | ||||||
| Protein | 12.1 | 11.5 | 11 | 26.4 | 16.4 | 12.6 |
| Ligand | 6.2 | 7.8 | 6 | 14.2 | 17.6 | 9.5 |
| Solvent | 23.8 | 22.8 | 24.4 | 32.4 | 26.4 | 24.7 |
a Rsym = ΣhklΣi|Ii(hkl) − 〈Ihkl〉|/ΣhklΣIi(hkl), where Ii(hkl) is the ith observed amplitude of reflection hkl, and 〈Ihkl〉 is the mean amplitude for all observations i of reflection hkl.
b Rcryst = Σ‖Fo| − |Fc‖/Σ|Fo|.
c 5% of the data were set aside for free R factor calculation.
Molecular Replacement and Refinement
Refinement details of the six structures are shown in Table 1. Molecular graphics images were generated using PyMOL. Structure determination of mature Atu2422 from PEG crystals using molecular replacement was as described (21). Visual investigation of the electron density maps using COOT (23) revealed that Atu2422 had a bound ligand, subsequently identified as an alanine residue, which fits unambiguously into the electron density maps. The structure of Atu2422 crystals from AS conditions was solved by molecular replacement with PHASER (24) using the coordinates of Atu2422-Ala from PEG conditions. Inspection of the resulting model and density maps using COOT confirmed that an endogenous Ala was bound to Atu2422.
As the co-crystals of Atu2422 (purified under classical conditions) with 50 mm valine are isomorphous to those of Atu2422-Ala from PEG conditions (21), preliminary phases were calculated using the coordinates of Atu2422-Ala (PEG conditions). The resulting electron density maps showed the presence of a valine in the ligand-binding site. As the co-crystals of Atu2422 (purified under classical conditions) with 50 mm proline and those of Atu2422 (purified under denaturing/renaturing conditions) with 100 mm GABA as well as those of Atu2422-F77A are isomorphous to those of Atu2422-Ala from AS conditions, preliminary phases were calculated using the coordinates of Atu2422-Ala (AS conditions). The resulting electron density maps revealed a proline and GABA bound to Atu2422 and a leucine bound to the F77A mutant. Refinement of each structure was performed with REFMAC5 (25) and Phenix (26). In all structures, the last two residues (positions 349 and 350) and the C-terminal His6 tag are disordered.
Isothermal Titration Microcalorimetry Measurements
Isothermal titration calorimetry experiments were performed with an iTC200 isothermal titration calorimeter (MicroCal LLC, Northampton, MA). The experiments were carried out at 20 °C. The unfolded/refolded Atu2422 concentration in the microcalorimeter cell (0.2 ml) varied from 10 to 50 μm. 19 injections of 2 μl of amino acid (Ala, GABA, Pro, or Val) solution at concentrations of 120–300 μm were performed at intervals of 240 s with stirring at 1000 rpm. The experimental data were fitted to theoretical titration curves with software supplied by MicroCal (Origin®). This software uses the relationship between the heat generated by each injection and ΔH (enthalpy change in kcal/mol), Ka (the association binding constant in m−1), n (the number of binding sites), total protein concentration, and free and total ligand concentrations (27). The values of associated stoichiometry to calculated Kd showed a protein/ligand ratio of 1:0.4, revealing that only 40% of Atu2422 was ligand-free during isothermal titration calorimetry experiments. Although different procedures of denaturation/renaturation were attempted, the purification of fully ligand-free Atu2422 was unsuccessful.
Measurement of Free Amino Acids in E. coli BL21 Cell Extracts
The abundance of individual amino acids (including GABA) in cell-free extracts of E. coli BL21 grown in 2×Tryptone yeast medium was determined by ion-exchange chromatography using an AminoTac JLC-500V amino acid analyzer (28).
Uptake of 3H-Labeled Amino Acids
Cultures of A. tumefaciens grown in AB minimal medium were washed with fresh AB medium and adjusted to A600 = 0.7. Transport assays were initiated by mixing cell suspension (100 μl) with 3H-labeled amino acids (GABA or Pro) at a final concentration of 1 μm. Just before starting assays, the competing unlabeled amino acids were added at a final concentration of 10 μm. After an incubation of 15 min, the extracellular 3H-labeled amino acids were eliminated by rapid filtration of cell suspensions (50 μl) on ultrafiltration membrane (0.22-μm Millipore HA) and three washings of filters with 3 ml of sterile water. Filter-bound radioactivity was quantified in a Packard TriCarb 2200 CA liquid scintillation counter. The 3H-labeled derivatives of GABA ([2,3-3H]GABA at 76 Ci/mmol) and Pro (l-[2,3-3H]Pro at 49 Ci/mmol) were provided by PerkinElmer Life Sciences.
Quantification of OC8HSL
Quantification of OC8HSL was performed on TLC silica plates (C18 reverse phase; Whatman) in the presence of OC8HSL bioindicator strain A. tumefaciens NT1 (pZLR4) as described previously (29). Tested samples were compared with a calibration curve of pure OC8HSL that was a generous gift from Prof. P. Williams (University of Nottingham, Nottingham, United Kingdom). In OC8HSL degradation assay, A. tumefaciens strains were grown in 5 ml of AB minimal medium supplemented with pure OC8HSL at 400 nm and containing GABA (0.1 mm) and/or other amino acids (1 mm). At the end of exponential growth, residual levels of OC8HSL in culture supernatants were measured in at least four replicates.
Plant Assays
In virulence assays, 10-week-old tobacco plants (Nicotiana tabacum L. cv. Samson) and 6-week-old tomato plants (Solanum lycopersicum L. cv. Dona) were grown under greenhouse conditions on compost and were automatically supplied with water three times/day (14). Each stem was incised (4 cm) with a scalpel and infected with ∼106 colony-forming units of A. tumefaciens strains that were picked up from 48-h cultures on LB plates. In each incision, the number of A. tumefaciens-induced tumors was counted 3 and 5 weeks post-inoculation. Virulence assay was performed on 10 plants per bacterial genotype. The number of tumors per incision was compared using Student's t test (α = 0.05).
Bioinformatics Analysis
Sequences were analyzed by BLAST (www.ncbi.nlm.nih.gov/BLAST/) and BioEdit Sequence Alignment Editor. Alignments of Atu2422 and related sequences were conducted using Clustal (30). The signal peptide of all PBPs was removed for phylogenetic analysis. Relationship tree building was conducted using MEGA Version 4 (31). The phylogeny was inferred using the neighbor-joining method (32). The bootstrap consensus tree inferred from 2000 replicates is taken to represent the evolutionary history of the taxa analyzed (33). The evolutionary distances were computed using the Poisson correction method (34) and are in units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set.
RESULTS
Structures of Atu2422 in Complex with Ala, Pro, Val, and GABA
We determined five x-ray structures of liganded Atu2422 at high resolution (Table 1). Two of them were obtained from crystals grown either in PEG (21) or AS with Atu2422 copurified with Ala. Ala is one of the most abundant free amino acids in E. coli BL21 used for Atu2422 expression (supplemental Table S1). The binding of endogenous amino acids to PBPs during the production/purification procedure is a common phenomenon (35, 36). Three other structures were obtained by replacing Ala with Pro, Val, and GABA prior to crystallization as described under “Experimental Procedures.”
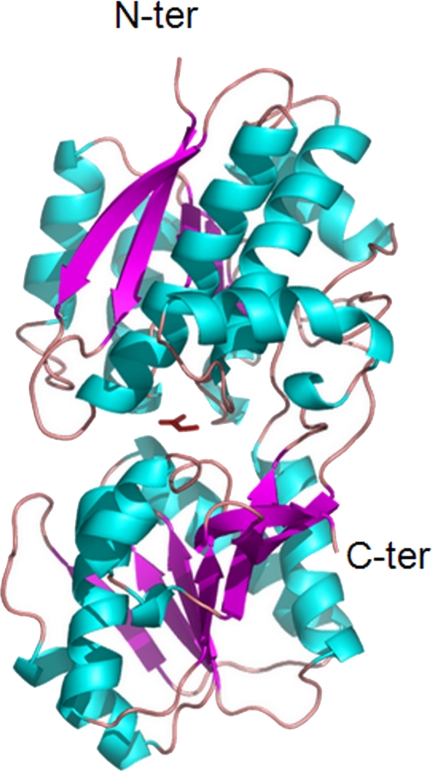
The mature Atu2422 structure is a monomer of 350 residues composed of two domains, residues 1–120 and 251–329 for domain 1 and residues 121–250 and 330–350 for domain 2, both consisting of a central β-sheet flanked by α-helices (Fig. 1). The five liganded Atu2422 structures adopt a closed conformation due to the presence of a single molecule of Ala, GABA, Pro, or Val bound to the central cleft between the two lobes (Fig. 1). The overall root mean square deviations of Cα atoms between all liganded structures are from 0.12 to 0.44 Å, indicating that they are all very similar.
FIGURE 1.
Ribbon representation of Atu2422 liganded with alanine. The Ala ligand located in the cleft between the two lobes is shown as a red stick.
A structural comparison of Atu2422 with all Protein Data Bank entries using SSM-EBI (www.ebi.ac.uk/mrd-srv/ssm) (37) indicated that the most similar overall structures are the liganded structures of E. coli Leu/Ile/Val-binding protein (LIVBP; codes 1Z16, 1Z17, and 1Z18) (35, 38) and E. coli Leu-binding protein (LBP; codes 1USI and 1USK) (39, 40) with root mean square deviations ranging from 1.1 to 1.2 Å over 339 and 443 Cα atoms, respectively. Sequence identity between Atu2422 and LBP/LIVBP is 40%. All other Protein Data Bank entries, including that of the Venus flytrap module of the GPCR metabotropic glutamate receptor (41), led to a root mean square deviation of >2 Å for approximately half of the backbone matching, as expected from their sequence identity, which is below 20%.
Ligand-binding Site of Atu2422 and Comparison with LBP/LIVBP
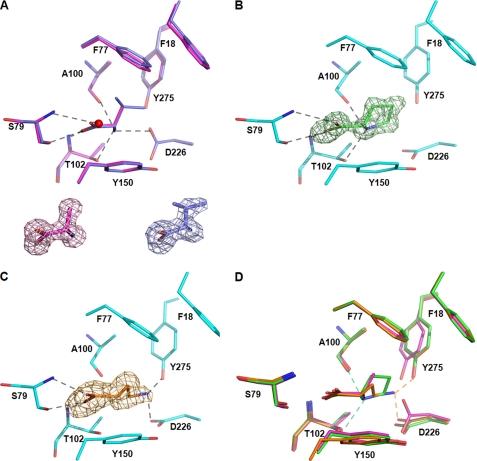
Each ligand, Ala, GABA, Pro, and Val, was well defined in the electron density (Fig. 2) and bound between the two closed lobes. The ligand-binding cleft is defined by the side chains or main chains of Phe18, Phe77, Asn78, Ser79, Ala100, Ala101, Thr102, Tyr150, Leu202, Asp226, Gly227, and Tyr275.
FIGURE 2.
Atu2422-ligand interactions and structural comparison of the ligand-binding site. A, superposition of Ala (magenta) and Val (purple) bound to the ligand-binding site. A conserved water molecule involved in ligand binding is shown as a red sphere. Ala and Val are shown in their Fo − Fc omit map contoured at 4σ. B, Pro (green) bound to the ligand-binding site in an Fo − Fc omit map contoured at 4σ. C, GABA (orange) bound to the ligand-binding site in an Fo − Fc omit map contoured at 4σ. D, Superposition of Ala (magenta), Pro (green), and GABA (orange) ligands. Hydrogen bonds are shown as black dashed lines in A–C and in color-coded ligand between the ligand amino group and Atu2422 in D (distances are <3.0 Å). All ligands are shown in stick representation.
The α-amino acids Ala, Val, and Pro bind in a very similar manner with their side chains located in the same protein pocket and with both hydrogen-bonding and non-polar contributions coming mainly from the N-terminal domain (Fig. 2, A and B). Their carboxylate group makes the same protein interactions: one oxygen atom forms hydrogen bonds with the main chain NH of Thr102 and the Oγ of Ser79, whereas the other oxygen atom interacts with the main chain NH of Ser79 and a conserved structural water molecule (Fig. 2A). The amino group of Ala and Val ligands make the same contacts with the main chain carbonyl of Ala100, the Oγ of Thr102, and the Oδ1 of Asp226 (Fig. 2A). In contrast, the amino group of the Pro ligand loses the interaction with the side chain of Asp226, which has moved ∼3 Å, providing room for the pyrrolidine ring (Fig. 2, B and D).
The carboxylate group of GABA makes the same interactions as that of α-amino acids. However, its amino group is placed 2 Å away from that of α-amino acid ligands and retains only the interaction with the Oδ1 of Asp226, the side chain having the same conformation as observed for proline binding (Fig. 2, C and D). The interactions with Thr102 and Ala100 are lost. Instead, the γ-amino group contacts the OH of Tyr275, which, in comparison with its position upon Ala and Val binding, has moved to accommodate the GABA (Fig. 2D). Interestingly, a similar displacement of Tyr275 was observed to accommodate the bulkier Pro ligand.
There are several van der Waals contacts between ligands and Atu2422 (58, 64, 69, and 53 for Ala, Pro, Val, and GABA, respectively). Asn78 and Ala101 make main chain contacts with all ligands, whereas Leu202 and Gly227 make contacts with Val only. Phe77 and Tyr150 provide half of the van der Waals interactions. All ligands are wedged between the keto ring of Tyr150, which stacks on the planar ligand main chain group, and the phenyl ring of Phe77. Phe77 makes strong non-polar contacts with the hydrophobic side chains of the ligands, notably with Val. In contrast, the Phe77 side chain has no van der Waals contacts with GABA. The Phe77 side chain seems to prevent the binding of a ligand with a larger side chain than a valine. Indeed, modeling different large amino acids, such as Leu or Glu, showed steric clashes between both side chains.
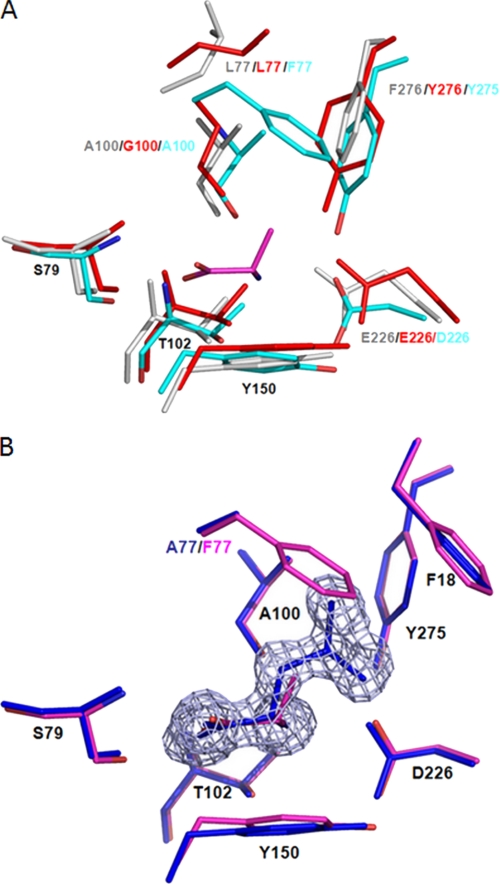
The ligand-binding sites of Atu2422, LBP, and LIVBP are similar (Fig. 3A). Among the 12 residues forming the ligand-binding cleft, Ser79, Ala101, Thr102, Tyr150, and Gly227 are strictly conserved. Moreover, Phe18 and Tyr275 of Atu2422 are conserved aromatic residues, being equivalent to Trp/Tyr and Tyr/Phe in LBP/LIVBP, respectively. Ala100 and Asp226 are homologous residues corresponding to Gly/Ala and Glu/Glu in LBP/LIVBP. In contrast, the last three residues, including Phe77 and Leu202, which make side chain contacts with the side chain of the ligand, are not conserved. They correspond to Leu77 and Tyr202 in LBP and LIVBP. Interestingly, a conserved water molecule close to Leu202 in Atu2422 structures mimics the OH of Tyr202 in LBP and LIVBP and thus restores the hydrogen bond with the carboxylate group of a ligand. Therefore, all polar and non-polar interactions that position the main chain atoms of the protein amino acid ligand are conserved between Atu2422 and LBP/LIVBP except those of residue 77. This strongly suggests that Phe77 has an important role in ligand selectivity of Atu2422.
FIGURE 3.
A, comparison of the ligand-binding site between Atu2422, LBP, and LIVBP. Atu2422 is shown in cyan, and bound Ala in magenta. LBP and LIVBP in their closed form are in red and gray, respectively. Conserved residues are labeled in black. B, comparison of the ligand-binding site between Atu2422 and Atu2422-F77A. Shown is a superposition of Ala bound to the ligand-binding site of Atu2422 (magenta) and Leu bound to Atu2422-F77A (blue) in an Fo − Fc omit map contoured at 4σ.
Affinity and α-Amino Acid Transport
The dissociation constant (Kd) between Atu2422 and Ala, GABA, Pro, and Leu was measured using isothermal titration calorimetry. As expected from modeling of Leu in the Atu2422-binding site, no interactions were found between Leu and Atu2422. In contrast, the mean Kd values were 0.25, 0.6, and 2.4 μm for Pro, Ala, and GABA respectively. Thus, they were in the low micromolar range commonly observed for PBP ligands (35, 40). Interestingly, Atu2422 had a markedly lower affinity for GABA and showed a preference for Pro, in line with their contacts with Atu2422 (58, 64, and 69 contacts of <4 Å for GABA, Ala, and Pro respectively).
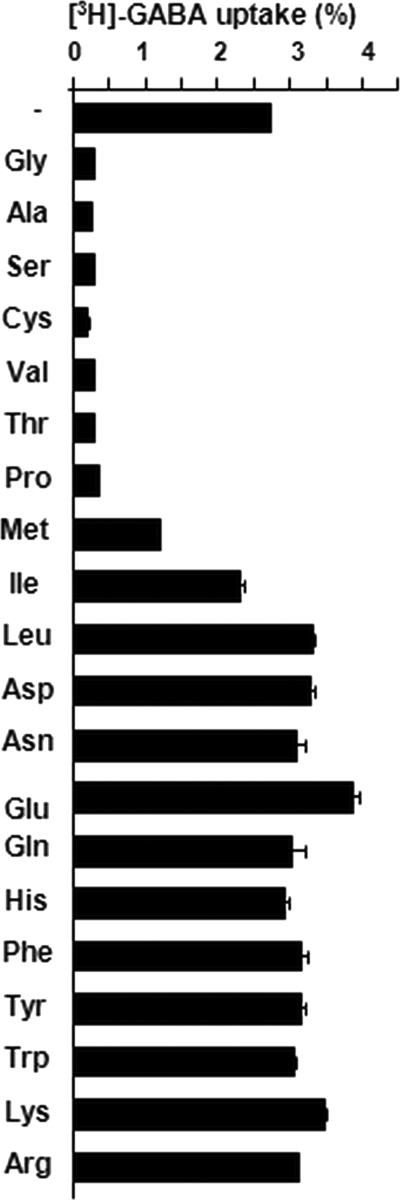
Pro and Val have previously been identified as antagonists of the transport of GABA (14). In this work, the structure of Ala-liganded Atu2422 and modeling of the interactions between Atu2422 and several amino acids suggested that other small α-amino acids could bind Atu2422 and therefore act as GABA antagonists. To validate this proposition in vivo, a set of amino acids were tested for their capacity to block the transport of GABA (Fig. 4). In addition to Pro and Val, five amino acids with a small side chain, namely Gly, Ala, Ser, Cys, and Thr, dramatically affected the transport of exogenous GABA in A. tumefaciens cells. This proved that it is the size of the amino acids in contrast to other characteristics, such as polarity or hydrophobicity, that determines if they can be accommodated in the ligand-binding site of Atu2422.
FIGURE 4.
Spectrum of amino acid uptake by wild-type Atu2422. Uptake of 1 μm [3H]GABA was assayed by the rapid filtration method in cultures of A. tumefaciens C58 cells grown in AB medium. Competing amino acids were added to a final concentration of 10 μm. Amino acids are classified by the lateral chain size from smaller to larger. The reference for the percentage (100%) corresponds to the total radioactivity of the added [3H]GABA.
Structural and Functional Studies of the Atu2422-F77A Mutant
We determined the x-ray structure of the Atu2422-F77A point mutant (Table 1). In contrast to Atu2422, which copurified with a fortuitous Ala, Atu2422-F77A copurified with an endogenous ligand identified as Leu according to the electron density maps (Fig. 3B). Leu is the most abundant free amino acid in E. coli BL21 (supplemental Table S1). The structure of Atu2422-F77A is identical to the liganded wild-type structures as the root mean square deviation between 0.12 and 0.4 Å for all Cα atoms indicates. The carboxyl and amino groups of Leu and Ala are identically positioned and thus make the same seven hydrogen bonds. Leu also makes a similar number of van der Waals contacts (60) despite its larger side chain. This is due to the F77A mutation, which causes the loss of several side chain van der Waals contacts. Phe18 brings an additional van der Waals contact compared with Atu2422-ligand complexes.
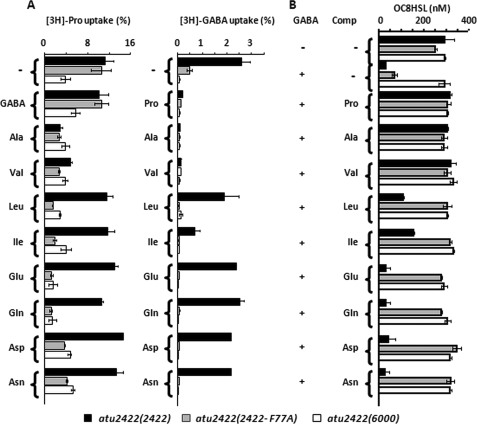
In parallel with the structural study, we evaluated the role of Phe77 in vivo. Two plasmids expressing Atu2422 or Atu2422-F77A, as well as the empty vector pME6000, were introduced into the A. tumefaciens defective mutant atu2422, which does not produce wild-type Atu2422. The constructed strains atu2422(atu2422), atu2422(atu2422-F77A), and atu2422(6000) were compared for uptake of radiolabeled GABA and Pro in the presence of different amino acids, as well as for OC8HSL levels (Fig. 5). In the absence of competitors, transport of Pro was similar in the atu2422 mutants expressing Atu2422 and Atu2422-F77A. However, Pro uptake in both strains was not affected by the addition of a 10-fold excess of GABA, but it decreased in the presence of small hydrophobic amino acids, such as Ala and Val. These data are in agreement with the lower Kd value of GABA compared with those of Pro and Val. Notably, in the presence of larger amino acids, Leu, Ile, Glu, Gln, Asn, and Asp (Fig. 5A), the transport of Pro decreased only in mutant atu2422 expressing Atu2422-F77A compared with that expressing Atu2422. The spectrum of Pro competitors was therefore enlarged by mutating Phe77 into an alanine.
FIGURE 5.
Effect of amino acid competitors on Pro and GABA uptake and OC8HSL degradation. A, uptake of 1 μm l-[3H]proline (left) and [3H]GABA (right) was assayed by the rapid filtration method in cultures of A. tumefaciens mutants atu2422(atu2422-F77A), atu2422(atu2422), and atu2422(6000). Competing solutes were added to a final concentration of 10 μm. B, the OC8HSL concentration (added at 400 nm) was measured in cell cultures grown in AB medium supplemented with GABA (0.1 mm) in the presence of competing solutes added at final concentration of 1 mm. Experiments were done in duplicate.
The key role of Phe77 was confirmed in GABA uptake and OC8HSL experiments (Fig. 5B). Although a reduced uptake of GABA was observed in the atu2422 mutant expressing Atu2422-F77A compared with that expressing Atu2422, it was sufficient to induce a significant degradation of OC8HSL. These data suggested that, in the absence of competitors, mutation of Phe77 showed a negligible effect on GABA signaling. In the presence of a large spectrum of competitors, including Leu and Glu, the uptake of GABA and the biodegradation of OC8HSL were blocked in strain atu2422(atu2422-F77A). In contrast, only competitors such as Pro, Val, and Ala affected uptake of GABA and OC8HSL degradation in strain atu2422 expressing Atu2422. Phe77 of Atu2422 contributes to reducing the number of α-amino acids that are able to block GABA uptake and therefore GABA-induced degradation of the OC8HSL signal in A. tumefaciens.
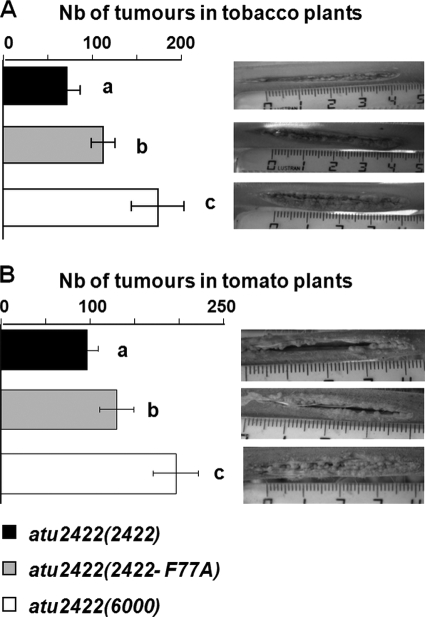
The role of Phe77 in the virulence of A. tumefaciens was also evaluated. We previously demonstrated that strain atu2422(6000) induces more symptoms than a complemented strain with Atu2422, revealing that GABA signaling down-regulates the aggressiveness of A. tumefaciens (14). We hypothesized that, in atu2422(atu2422-F77A), an increase of the number of potential GABA competitors would decrease the efficiency of GABA signaling and would therefore enhance the emergence of plant tumors. Notably, Asn, Asp, Gln, and Glu together represent up to 70% of the total free amino acids in plant tumors (14). To test this hypothesis, virulence assays were performed on two plant hosts, tomato and tobacco plants (Fig. 6). We observed that strain atu2422(atu2422-F77A) was more virulent than strain atu2422(atu2422), but it remained less virulent than a mutant defective for Atu2422.
FIGURE 6.
Effect of mutant atu2422(atu2422-F77A) on virulence. Virulence symptoms (number (Nb) of emerging tumors) induced by mutants atu2422(atu2422-F77A), atu2422(atu2422), and atu2422(6000) were measured in tobacco plants (A) and tomato plants (B). Bacterial strains showed statistical differences (Student's test; p < 0.01) in their aggressiveness.
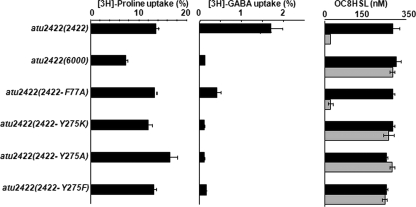
Effect of Tyr275 Mutations on GABA Transport, OC8HSL Degradation, and Virulence
Because the Tyr275 side chain in Atu2422 makes a short hydrogen bond with the amino group of GABA, this residue should play a key role in GABA binding. To test this hypothesis in vivo, the A. tumefaciens defective mutant atu2422 was complemented by a plasmid expressing each modified protein, Atu2422-Y275A, Atu2422-Y275K, or Atu2422-Y275F. The constructed strains, atu2422(atu2422-Y275A), atu2422(atu2422-Y275K), and atu2422(atu2422-Y275F), were compared with strain atu2422(atu2422) for the uptake of radiolabeled GABA and Pro, as well as for the ability to degrade OC8HSL in the presence of GABA (Fig. 7). Whereas uptake of Pro was not affected in any of the complemented strains, uptake of GABA decreased dramatically in the Tyr275 mutants compared with that in strain atu2422(atu2422). As expected, the Tyr275 mutants impaired in GABA transport were not able to degrade the OC8HSL signal. Virulence assays were also performed on two plant hosts, tomato and tobacco plants (supplemental Fig. S1). As expected, strains atu2422(atu2422-Y275A), atu2422(atu2422-Y275K), and atu2422(atu2422-Y275F) were as virulent as the mutant defective for Atu2422.
FIGURE 7.
Effect of Tyr275 mutations on GABA uptake and OC8HSL degradation. Left and middle, uptake of 1 μm l-[3H]proline and [3H]GABA (as described in the legend to Fig. 5) was assayed in cultures of mutants atu2422(atu2422-F77A), atu2422(atu2422-Y275K), atu2422(atu2422-Y275A), atu2422(atu2422-Y275F), atu2422(atu2422), and atu2422(6000). Right, the OC8HSL concentration (added at 400 nm) was measured in cultures of these same strains grown in AB medium supplemented with (0.1 mm; gray bars) and without (black bars) GABA in the presence of competing solutes added at a final concentration of 1 mm. Experiments were done in duplicate.
Atu2422 as an Identifier of a Subfamily of PBPs
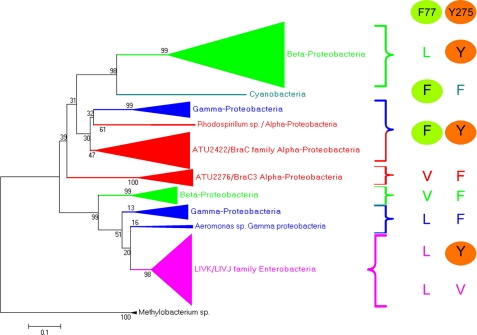
A search of the A. tumefaciens genome for Atu2422 homologs yielded seven predicted proteins (Atu2276, Atu1838, Atu5531, Atu1413, Atu4519, Atu2414, and Atu4123) with sequence identities of between 20 and 47%. Only two of them, Atu2422 and its closest homolog, Atu2276, possess the α-amino acid-binding signature ([S]-x(22)-[TS]-x(13,14)-[R]-x(4)-[D]-x(2)-[Q]-x(24,25)-[Y]-[GA]-x(74,84)-[ED]) identified (16) in LBP/LIVBP and in the Venus flytrap module of some GPCRs. However, the two key residues Phe77 and Tyr275 of Atu2422 are not conserved in Atu2276, corresponding to Val and Phe, respectively. Notably, we have previously demonstrated in vivo that Atu2276 is not involved in GABA transport and degradation of the OC8HSL signal (14). Consequently, Atu2276 should have a distinct physiological role in A. tumefaciens C58.
A search in eukaryotic, archaeal, and bacterial genomes for Atu2422 homologs using blastp and tblastn led to relatives only in Bacteria and Archaea. The closest archaeal predicted PBP homolog showed 29% identity to Atu2422, whereas 500 bacterial relatives with a threshold of 35% identity were retrieved from the NCBI Database. We then analyzed these 500 homologs using a bacterial Venus flytrap-specific signature modified from Ref. (16). This consensus signature ([S]- x(21,22)-[T]-x(13,14)-[R]-x(4)-[D]-x(2)-[Q]-x(24,25)-[Y]-[G]-x(74)-[ED]) was found in 404 proteins, whereas altered signatures were identified in 33 others (supplemental Table S2). These 437 proteins correspond to 386 unique proteins, which are encoded by 155 different bacterial species.
The relation tree of the 386 unique proteins revealed several subgroups of PBPs. In each subgroup except the LBP/LIVBP cluster and Rhodospirillum centenum strain SW, residues corresponding to positions 77 and 275 in Atu2422 are strictly conserved (see Fig. 8 and supplemental Fig. S2 for details). In the particular case of R. centenum SW, two highly close proteins (57% identity) show a conserved Tyr at position 275 but Leu or Phe at position 77 (supplemental Fig. S2). This suggests a recent duplication/specialization of these two Atu2422 orthologs. The phylogenetic analysis also revealed that Atu2422 belongs to a large subfamily of PBPs exhibiting the essential Phe77 and Tyr275 residues involved in the binding of short α-amino acids and GABA, respectively. The members of this subfamily belong to different genera of α- and γ-proteobacteria, including the animal pathogens Brucella and Pseudomonas; the plant symbionts Rhizobium, Sinorhizobium, and Bradyrhizobium; and the photosynthetic bacteria Rhodospirillum.
FIGURE 8.
Evolutionary relationship of Atu2422 relatives. Left, relation tree of 386 Atu2422-related proteins collected from 155 bacterial species. The branches are collapsed when a clade is composed of proteins from the same taxonomic group: α-proteobacteria in red, β-proteobacteria in green, γ-proteobacteria in blue, and enterobacteria in pink. An extended version of this tree is presented in supplemental Fig. S2. Right, residues at equivalent positions of Phe77 and Tyr275 in Atu2422 are shown for each clade.
DISCUSSION
The five high-resolution x-ray structures of A. tumefaciens liganded Atu2422 show explicit protein-ligand interactions that discriminate the binding mode of GABA versus that of α-amino acids. As expected, the carboxylate groups of GABA and α-amino acids make the same interactions in the ligand-binding site, and this binding mode is conserved in all amino acid-binding PBPs. In contrast, comparison of the amino group bound to Atu2422 between GABA and protein amino acids revealed that Tyr275 makes a short polar interaction with the γ-amino group of GABA only. We further proved by site-directed mutagenesis and functional studies that Tyr275 is specific for GABA binding. Indeed, our work demonstrated that Tyr275 is essential for GABA transport and expression of GABA-regulated functions, such as degradation of the OC8HSL signal in A. tumefaciens. We believe that all PBPs related to Atu2422 that possess this tyrosine residue, such as E. coli LBP and R. leguminosarum Rl3745, would similarly be able to bind GABA. This was confirmed with Rl3745, which is involved in GABA transport (42). Hence, our work, which combined structural and in vivo mutational-functional results, offers an opportunity for identifying bacterial GABA-binding PBPs in databases. In contrast, because of a very low level of sequence identity between bacterial PBPs and the Venus flytrap module of class C GPCRs, as well as the dimeric structure of GPCR (43), modeling the GABA-binding site of GABAB from Atu2422 would be speculative (44). Remarkably, the observed GABA extended conformation in Atu2422 is so far unique, as it does not resemble any GABA conformations available in the Protein Data Bank (codes 2OKJ, 2OKK, and 6JDW), suggesting that the ligand-binding site of PBPs exerts a specific constraint on the highly flexible molecule (45).
Atu2422 displays higher affinity (up to 10-fold) for protein amino acids, such as Pro and Ala, than for GABA as shown by isothermal titration calorimetry and inferred by structural analysis in terms of number of protein-ligand contacts. Among a set of α-amino acids exhibiting a small lateral chain, such as Ala, Val, and Pro, we correlated capacity of Atu2422 binding with the antagonism of GABA transport. Notably, because all GABA competitors but Pro remain at a low level in plant tumors, Pro was proposed as the most probable GABA antagonist (14). Phe77 plays a crucial role in restricting the spectrum of the Atu2422-binding α-amino acids and hence the number of antagonists of GABA transport and signaling. The liganded Atu2422-F77A structure revealed a higher affinity for Leu than for Ala in contrast to the wild-type protein, which does not bind Leu. Moreover, in A. tumefaciens cells expressing Atu2422-F77A instead of the wild-type protein, the range of GABA antagonists was expanded to Asn, Asp, Glu, Gln, and Leu and correlated with a decrease in the GABA-regulated degradation of the quorum-sensing signal OC8HSL. Our work led to the designation of this residue, which contributes to the restriction of the number of GABA antagonists, as a novel key structural characteristic of PBPs that bind amino acids with a limited lateral chain.
Atu2422-related PBPs, which exhibit the two key residues Tyr275 and Phe77 and should accommodate GABA and a few α-amino acids, are clustered in a well defined phylogenetic group of proteins that are present in α- and γ-proteobacteria. Most of these bacteria, including Brucella, Pseudomonas, Bradyrhizobium, Methylobacterium, Rhizobium, and Sinorhizobium, interact with animal and plant hosts. The role of GABA in animal-microbe interactions remains to be explored. In R. leguminosarum, the Atu2422 ortholog Rl3745 is involved in the transport of GABA, which accumulates in the nitrogen-fixing nodules induced on the host plant (42), but the role of GABA in the symbiosis is still unclear (8). In A. tumefaciens, the ratio between exogenous GABA and GABA antagonists controls GABA signaling and modulation of the emergence of plant tumors (10, 14). By expressing the mutant protein Atu2422-F77A in A. tumefaciens, we correlated an increase in the number of potential GABA competitors with a variation of the virulence symptoms in two plant hosts. As a conclusion, the combination of structural and functional analyses designates Atu2422 as a pivotal sensor of the relative level of GABA and its competitors in A. tumefaciens and, probably, in several other host-interacting bacteria.
Supplementary Material
Acknowledgments
We are grateful to B. Guimaraes, P. Legrand, and A. Thompson for help in data collection on PROXIMA 1 at the SOLEIL Synchrotron; Sylvie Citerne and Grégory Mouille for quantification of free amino acids at the Green Chemistry Platform (Institut National de la Recherche Agronomique, Versailles, France); and F. Acher for encouraging discussions at the beginning of the project.
This work was supported by the French Research Minister (Ecole Doctorale Sciences du Végétal ED145, to S. P.) and by CNRS (to A. V., S. Mondy, M. N., D. F., and S. Moréra).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- OC8HSL
- 3-oxo-octanoylhomoserine lactone
- PBP
- periplasmic binding protein
- GPCR
- G-protein-coupled protein
- AB
- Agrobacterium broth
- AS
- ammonium sulfate
- LIVBP
- Leu/Ile/Val-binding protein
- LBP
- Leu-binding protein.
REFERENCES
- 1.Bouché N., Lacombe B., Fromm H. (2003) Trends Cell Biol. 13, 607–610 [DOI] [PubMed] [Google Scholar]
- 2.Shelp B. J., Bown A. W., Faure D. (2006) Plant Physiol. 142, 1350–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Ari Y., Gaiarsa J. L., Tyzio R., Khazipov R. (2007) Physiol. Rev. 87, 1215–1284 [DOI] [PubMed] [Google Scholar]
- 4.Anjard C., Loomis W. F. (2006) Development 133, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 5.Galvez T., Parmentier M. L., Joly C., Malitschek B., Kaupmann K., Kuhn R., Bittiger H., Froestl W., Bettler B., Pin J. P. (1999) J. Biol. Chem. 274, 13362–13369 [DOI] [PubMed] [Google Scholar]
- 6.Shelp B. J., Bown A. W., McLean M. D. (1999) Trends Plant Sci. 4, 446–452 [DOI] [PubMed] [Google Scholar]
- 7.Solomon P. S., Oliver R. P. (2002) Planta 214, 414–420 [DOI] [PubMed] [Google Scholar]
- 8.Prell J., Bourdès A., Karunakaran R., Lopez-Gomez M., Poole P. (2009) J. Bacteriol. 191, 2177–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeken R., Engelmann J. C., Efetova M., Czirjak T., Müller T., Kaiser W. M., Tietz O., Krischke M., Mueller M. J., Palme K., Dandekar T., Hedrich R. (2006) Plant Cell 18, 3617–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevrot R., Rosen R., Haudecoeur E., Cirou A., Shelp B. J., Ron E., Faure D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Z. C., Haudecoeur E., Faure D., Kerr K. F., Nester E. W. (2008) Cell. Microbiol. 10, 2339–2354 [DOI] [PubMed] [Google Scholar]
- 12.Haudecoeur E., Tannières M., Cirou A., Raffoux A., Dessaux Y., Faure D. (2009) Mol. Plant-Microbe Interact. 22, 529–537 [DOI] [PubMed] [Google Scholar]
- 13.Carlier A., Chevrot R., Dessaux Y., Faure D. (2004) Mol. Plan- Microbe Interact. 17, 951–957 [DOI] [PubMed] [Google Scholar]
- 14.Haudecoeur E., Planamente S., Cirou A., Tannières M., Shelp B. J., Moréra S., Faure D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haudecoeur E., Faure D. (2010) Commun. Integr. Biol. 3, 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acher F. C., Bertrand H. O. (2005) Biopolymers 80, 357–366 [DOI] [PubMed] [Google Scholar]
- 17.Kuang D., Yao Y., Maclean D., Wang M., Hampson D. R., Chang B. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J., Huang S., Qian J., Huang J., Jin L., Su Z., Yang J., Liu J. (2009) BMC Evol. Biol. 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilton M. D., Currier T. C., Farrand S. K., Bendich A. J., Gordon M. P., Nester E. W. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 3672–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurhofer M., Reimmann C., Schmidli-Sacherer P., Heeb S., Haas D., Défago G. (1998) Phytopathology 88, 678–684 [DOI] [PubMed] [Google Scholar]
- 21.Moréra S., Gueguen-Chaignon V., Raffoux A., Faure D. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 1153–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 26.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27.Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 28.Diaz C., Purdy S., Christ A., Morot-Gaudry J. F., Wingler A., Masclaux-Daubresse C. (2005) Plant Physiol. 138, 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cha C., Gao P., Chen Y. C., Shaw P. D., Farrand S. K. (1998) Mol. Plant-Microbe Interact. 11, 1119–1129 [DOI] [PubMed] [Google Scholar]
- 30.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 32.Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. (1985) Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 34.Zuckerkandl E., Pauling L. (1965) Genes Proteins, 97–166 [Google Scholar]
- 35.Trakhanov S., Vyas N. K., Luecke H., Kristensen D. M., Ma J., Quiocho F. A. (2005) Biochemistry 44, 6597–6608 [DOI] [PubMed] [Google Scholar]
- 36.Huvent I., Belrhali H., Antoine R., Bompard C., Locht C., Jacob-Dubuisson F., Villeret V. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 38.Sack J. S., Saper M. A., Quiocho F. A. (1989) J. Mol. Biol. 206, 171–191 [DOI] [PubMed] [Google Scholar]
- 39.Sack J. S., Trakhanov S. D., Tsigannik I. H., Quiocho F. A. (1989) J. Mol. Biol. 206, 193–207 [DOI] [PubMed] [Google Scholar]
- 40.Magnusson U., Salopek-Sondi B., Luck L. A., Mowbray S. L. (2004) J. Biol. Chem. 279, 8747–8752 [DOI] [PubMed] [Google Scholar]
- 41.Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., Morikawa K. (2000) Nature 407, 971–977 [DOI] [PubMed] [Google Scholar]
- 42.Hosie A. H., Allaway D., Galloway C. S., Dunsby H. A., Poole P. S. (2002) J. Bacteriol. 184, 4071–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumpost J., Syrova Z., Kulihova L., Frankova D., Bologna J. C., Hlavackova V., Prezeau L., Kralikova M., Hruskova B., Pin J. P., Blahos J. (2008) Neuropharmacology 55, 409–418 [DOI] [PubMed] [Google Scholar]
- 44.Schreier B., Stumpp C., Wiesner S., Höcker B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18491–18496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crittenden D. L., Chebib M., Jordan M. J. (2005) J. Phys. Chem. A 109, 4195–4201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.