Abstract
The main cofactors involved in the oxygen evolution activity of Photosystem II (PSII) are located in two proteins, D1 (PsbA) and D2 (PsbD). In Thermosynechococcus elongatus, a thermophilic cyanobacterium, the D1 protein is encoded by either the psbA1 or the psbA3 gene, the expression of which is dependent on environmental conditions. It has been shown that the energetic properties of the PsbA1-PSII and those of the PsbA3-PSII differ significantly (Sugiura, M., Kato, Y., Takahashi, R., Suzuki, H., Watanabe, T., Noguchi, T., Rappaport, F., and Boussac, A. (2010) Biochim. Biophys. Acta 1797, 1491–1499). In this work the structural stability of PSII upon a PsbA1/PsbA3 exchange was investigated. Two deletion mutants lacking another PSII subunit, PsbJ, were constructed in strains expressing either PsbA1 or PsbA3. The PsbJ subunit is a 4-kDa transmembrane polypeptide that is surrounded by D1 (i.e. PsbA1), PsbK, and cytochrome b559 (Cyt b559) in existing three-dimensional models. It is shown that the structural properties of the PsbA3/ΔPsbJ-PSII are not significantly affected. The polypeptide contents, the Cyt b559 properties, and the proportion of PSII dimer were similar to those found for PsbA3-PSII. In contrast, in PsbA1/ΔPsbJ-PSII the stability of the dimer is greatly diminished, the EPR properties of the Cyt b559 likely indicates a decrease in its redox potential, and many other PSII subunits are lacking. These results shows that the 21-amino acid substitutions between PsbA1 and PsbA3, which appear to be mainly conservative, must include side chains that are involved in a network of interactions between PsbA and the other PSII subunits.
Keywords: Electron Transfer, Photosynthesis, Protein Stability, Protein Structure, Site-directed Mutagenesis, Photosystem II, Thermosynechococcus elongatus, Water Oxidation
Introduction
Photosystem II (PSII)2 catalyzes the light-driven water oxidation and plastoquinone reduction in cyanobacteria, algae, and plants. In cyanobacteria, its minimum structural unit capable of oxygen evolution consists of a membranous complex with 20 protein subunits, 17 of which are membrane-spanning proteins and 3 of which are extrinsic proteins. In the recent refined three-dimensional x-ray structures from 3.5 to 2.9 Å resolution using PSII core complex isolated from the thermophilic cyanobacterium Thermosynechococcus elongatus (1–3) the PSII complex is organized in dimers. A PSII monomer involves 35 chlorophyll molecules, 2 pheophytin molecules, 2 hemes, 1 non-heme iron, 4 manganese ions, 1 calcium ion, 2 (+1) quinones, and at least 12 carotenoid molecules and 25 lipids. One or two chloride-binding sites were also localized (3–5). All cofactors in charge of the photosynthetic electron transport involving P680 chlorophylls (PD1 (P680 Chl on D1), PD2 (P680 Chl on D2), and ChlD1 and ChlD2 (monomeric Chls bound to D1 and D2, respectively)), PheoD1, and the plastoquinones QA and QB are bound to amino acid residues of the D1 and D2 subunits. The redox active Tyr residues, TyrZ and TyrD, correspond to amino acids of D1–161 and D2–160, respectively. The catalytic center responsible for water oxidation, a Mn4Ca cluster, interacts with amino acid residues from D1 and CP43.
PSII is often exposed to photo-oxidative stress. PSII has several protection systems against photo-oxidative damage, and these include activation of the xanthophyll cycle in higher plants (for review, see Refs. 6 and 7) and quenching of singlet oxygen by cofactors of PSII (for review, see Refs. 8–10). In addition to these protection systems, it is known that the turnover of the D1 protein is accelerated under the light stress conditions (for review, see Refs. 11–14). For the assembly and synthesis of the de novo D1 protein in PSII complex, the old D1 polypeptide is digested by specific proteases such as FtsH and Deg/HtrA (14), and then the newly synthetized D1 is assembled into the PSII complex (for review, see Refs. 15–18).
Under the conditions in which the D1 turnover is increased, both the rate of transcription and translation from the D1 gene, psbA, must be important for maintaining the PSII complex with a functional structure. For that, cyanobacterial species have multiple psbA variants, the promoter of each being “sensitive,” for example, to high light and UV light illumination (19–22) or low oxygen conditions (23, 24). For instance, the mesophilic cyanobacterium, Synechocystis PCC 6803, which is widely used in research on the structure-function relationships in PSII, has three psbA genes in its genome. Two of them (psbAII and psbAIII) produce an identical D1. Nevertheless, psbAII is expressed under the normal cultivation conditions, whereas psbAIII is expressed when the cells are exposed to high light or UV light (21). Although it has been thought that transcription of the remaining psbAI gene is silent, Sicora et al. (24) found recently that this gene is expressed under microaerobic conditions. In experiments on PSII mutants in Synechocystis PCC 6803, the gene on which the mutations are made is either psbAII or psbAIII (after the deletion of either psbAI and psbAIII or psbAI and psbAII, respectively).
The thermophilic cyanobacterium, T. elongatus, has also three different psbA in its genome (25). The amino acid sequences of these D1 variants are not identical, so that PSII complex composed of different D1 variants must have different conformations. Of the known genomes of photosynthetic organisms, T. elongatus seems to be the only case in which the multiple genes for D1 are all different. Kós et al. (26) have shown that psbA1 was constitutively expressed under normal conditions, whereas the transcription of the psbA3 gene was induced by high light or UV light. In T. elongatus, in contrast to Synechocystis PCC 6803, the processed PsbA3 (344 amino acid residues) differs by 21 residues from the processed PsbA1 (see supplemental Fig. S1). Although most of the amino acid differences between the PsbA1 and the PsbA3 seem to be small, such as Val to Ile and Leu to Val, other substitutions have been shown to modify the molecular structure and function (27). For instance, the substitution of Gln-130 in PsbA1 into Glu in PsbA3 creates a hydrogen bond between Glu-130 and the 131-keto C O group of PheoD1, e.g. Refs. 28 and 29). Hence, the redox potential of PheoD1/PheoD1− in PsbA3-PSII is more positive by 17 mV than in that of PsbA1-PSII (27, 30).
O group of PheoD1, e.g. Refs. 28 and 29). Hence, the redox potential of PheoD1/PheoD1− in PsbA3-PSII is more positive by 17 mV than in that of PsbA1-PSII (27, 30).
In addition to affecting the energetics of PSII, the PsbA1 to PsbA3 exchange is also expected to modify the interactions between the subunits that constitute the PSII complex. Among the changes already discussed (27, 31), two other substitutions have been proposed to be important. First, the exchange of Ser-270 in PsbA1 for Ala-270 in PsbA3 was suggested to influence the stabilization of the sulfoquinovosyl-diacylglycerol, which locates between QB and non-heme iron (3, 27, 31). Second, there is an essential substitution at position 310, which is Lys and Gln in PsbA1 and PsbA3, respectively. Indeed, in the three-dimensional x-ray structures of the PsbA1-PSII, the loop bearing this amino acid seems to interact with the base of an extrinsic protein, PsbV (Cyt c550), at a distance of 3–4 Å. In addition, the N-terminal region of PsbJ polypeptide also seems to be in contact with this domain. The PsbJ subunit is a 4-kDa transmembrane polypeptide that is surrounded by D1 (PsbA1), PsbK, and Cyt b559 in the three-dimensional structural models (1–3).
To our knowledge little is known about the influence of PsbA variants on the PSII assembly and stability. Given the role of stress conditions in the expression of PsbA variants, this seems worthy of investigation.
As stated before, the interaction among the internal loop of D1, the base of Cyt c550, and the N-terminal region of PsbJ is expected to be a key domain for maintaining the structure of the PSII complex. Because PsbJ is expected to interact differently with PsbA1 and PsbA3, we constructed a PsbJ deletion mutant in two different T. elongatus strains that express either psbA1 or psbA3, and the protein composition of the isolated PSII complexes was analyzed.
EXPERIMENTAL PROCEDURES
Construction of ΔPsbJ Strains
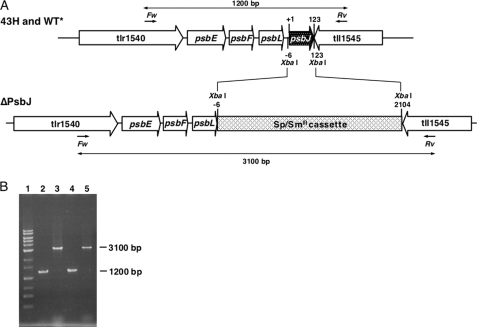
DNA fragments of ≈930 bp of the 5′-flanking region of psbJ (gene number tsr1544) were cloned from T. elongatus wild-type genomic DNA by PCR amplification and subcloned into a plasmid vector pBluescript II SK+ between KpnI and XbaI sites. Then, separately amplified ≈1010-bp DNA fragments of the 3′-flanking region of psbJ were ligated to the downstream part of the 5′-flanking region in the subcloned plasmid vector at XbaI and SacI. A spectinomycin/streptomycin resistance gene cassette (≈2100 bp) (32) was inserted between the 5′-flanking region and the 3′-flanking region of psbJ at XbaI (Fig. 1A). The constructed plasmid DNA was introduced into both the T. elongatus 43H, which has a His6 tag on the C terminus of CP43 (33), and the WT*, which was obtained by deleting psbA1 and psbA2 from the 43H strain (34) by electroporation (Bio-Rad gene pulser) as described in Ref. 33. The 43H/ΔPsbJ transformants were selected as single colonies on DTN agar plates containing 25 μg of spectinomycin ml−1, 10 μg of streptomycin ml−1, and 40 μg of kanamycin ml−1 as previously described in Refs. 33–37. The WT*/ΔPsbJ transformants were selected as single colonies on DTN agar plates containing 5 μg of chloramphenicol ml−1, 25 μg of spectinomycin ml−1, 10 μg of streptomycin ml−1, and 40 μg of kanamycin ml−1. Segregation of all genome copies was confirmed by the difference in length of DNA amplified by PCR using the forward primer (5′-GTCCAGCCAGAGGATTTGCTCCGGCATGGC-3′) that locates 1200 bp upstream from the initial codon of psbJ and the reverse primer (5′-CTGCAGCAACGCTACTTTTGGGGGTTACCC-3′) that locates 200 bp downstream of the termination codon of psbJ as shown in Figs. 1, A and B.
FIGURE 1.
A, shown is the map around the psbJ gene (gene number tsr1544) in 43H and WT* and deletion of psbJ gene from either the 43H or the WT* genome to produce 43H/ΔPsbJ or WT*/ΔPsbJ strains, respectively. A 129-bp fragment including the open reading frame of psbJ was replaced by a 2109-bp fragment containing spectinomycin/streptomycin-resistant cassette at XbaI sites. In both the WT* and the WT*/ΔPsbJ genomes, psbA1 and psbA2 have been deleted (34). In the 43H, the WT*, the 43H/ΔPsbJ, and the WT*/PsbJ, psbC has been extended with an additional DNA fragment encoding six consecutive His (33). Forward (Fw) and reverse (Rv) show positions of PCR primers to confirm the length of the psbJ and/or spectinomycin/streptomycin resistant cassette. B, shown is an agarose gel (1%) electrophoresis of amplified products by PCR using forward and reverse primers. Lanes 1, 1-kb ladder markers (Toyobo, Japan); lane 2, 43H strain; lane 3, 43H/ΔPsbJ strain; lane 4, WT* strain; lane 5, WT*/ΔPsbJ strain. In lane 1, the band labeled with an asterisk originates from contamination by allophycocyanin B. Such a contamination sometimes occurs in some samples but does not mask any protein from PSII.
Purification of PSII Core Complexes
The transformed cells were grown in 1-liter cultures of DTN in 3-liter Erlenmeyer flasks in a rotary shaker with a CO2-enriched atmosphere at 45 °C under continuous light (≈60 μmol of photons m−2 s−1). Thylakoids and PSII core complexes were prepared as described earlier in Refs. 33–38. The thylakoid membranes were solubilized by using 1% n-dodecyl-β-d-maltoside at a Chl concentration of 1 mg ml−1. The PSII core complexes bound to the Ni2+ resin were eluted with 200 mm imidazole. PSII was concentrated by using Amicon Ultra-15 concentrator devices (Millipore) with a 100-kDa cut-off. Routinely, the total amount of Chl before breaking the cells was ≈150 mg, and the yield after PSII purification in terms of Chl was ≈3–5%. PSII was stored in liquid nitrogen at a concentration of about 2 mg ml−1 in a medium containing 10% glycerol, 1 m betaine, 15 mm CaCl2, 15 mm MgCl2, and 40 mm MES (pH 6.5).
Oxygen Evolution Measurements
Oxygen evolution of isolated PSII complexes and of whole cells under continuous illumination was measured at 25 °C by polarography using a Clark-type oxygen electrode (Hansatech) with saturating white light at a Chl concentration of 5 μg of Chl ml−1 in 40 mm MES buffer (pH 6.5) containing 15 mm CaCl2, 15 mm MgCl2, 100 mm NaCl, and 1 m betaine in both isolated PSII complexes and whole cells. A total of 0.5 mm dichloro-p-benzoquinone (dissolved in dimethyl sulfoxide) was added as an electron acceptor. The PSII activity was measured immediately after the addition of dichloro-p-benzoquinone.
MALDI-TOF Mass Spectrometric Measurements
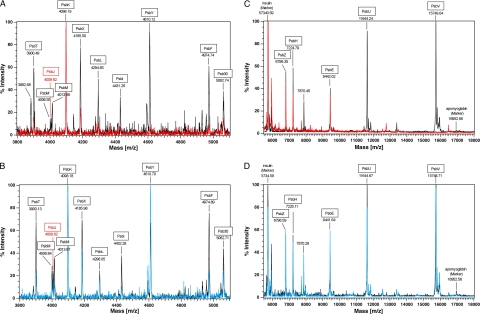
The isolated PSII complexes (30 or 150 μg of Chl ml−1 for linear mode or reflector mode, respectively) were mixed with the same volume of a saturated matrix (sinapic acid, Fluka) solution that consists of 60% acetonitrile and 0.1% trifluoroacetic acid. The mixed sample were loaded onto the stainless steel target plate and dried at ambient atmosphere on a clean-bench. MALDI-TOF mass analysis was performed using a Voyager-DE PRO MALDI-TOF mass spectrometer (Applied Biosystems). The instrument was operated in reflector mode at a 20-kV accelerating voltage and 100-ns ion extraction delay with the nitrogen laser working at 337 nm and 3 Hz. Two hundred laser flashes were accumulated per spectrum. The traces in Fig. 3 result from the average of 10–15 spectra. Operation and measurement conditions in linear mode were as described in Ref. 38. Internal calibration was performed on the samples premixed with adrenocorticotropic hormone fragment (from human, m/z of average = 2466.7200, m/z of resolved = 2465.1989, Sigma), insulin (from bovine, m/z of average = 5734.5900, m/z of resolved = 5730.6087, Sigma) and apomyoglobin (from bovine heart, m/z of average = 16952.5600, Sigma).
FIGURE 3.
MALDI-TOF mass spectra of isolated PSII complexes. Shown is the analysis of molecular range from 3800 to 5500 in reflector mode (A and B) and the range from 5,500 to 18,000 in linear mode (C and D). Numbers under the names of PSII polypeptides indicate m/z values. The values in A and B are monoisotopic molecular weight, and the values in C and D are average molecular weight. The peaks of m/z 5,734.59 and 16,952.56 in C and D correspond to bovine insulin (Sigma) and horse skeletal apomyoglobin (Sigma), respectively, as calibration markers. The spectra in A and B were also calibrated with adrenocorticotropic hormone fragment (m/z 2465.1989 in monoisotopic) and bovine insulin (m/z 5730.6087 in monoisotopic). In A and C: black, PsbA1-PSII; red, PsbA1/ΔPsbJ-PSII. In B and D: black, PsbA3-PSII; blue, PsbA3/ΔPsbJ-PSII.
Gel Permeation Chromatography
For size separation, the PSII complex was further treated with 1.5% n-dodecyl-β-d-maltoside at a Chl concentration of 1.5 mg ml−1 for 30 min at 4 °C in the dark. Then, the sample was loaded onto a Superdex-200 column (PC 3.2/30) chromatography (SMARTTM System, GE Healthcare) at a flow rate of 10 μl min−1 as described in Refs. 33 and 39.
SDS-Polyacrylamide Gel Electrophoresis
PSII complexes suspended in 40 mm MES/NaOH (pH 6.5), 10 mm NaCl, 10 mm CaCl2, 10 mm MgCl2, 0.03% n-dodecyl-β-d-maltoside were solubilized with 2% lithium lauryl sulfate and then analyzed by SDS-polyacrylamide gel electrophoresis with a 16–22% gradient gel containing 7.5 m urea as described in Ref. 40.
Blue Native Polyacrylamide Gel Electrophoresis
Thylakoids were solubilized with 1% n-dodecyl-β-d-maltoside at 1 mg of Chl ml−1 for 30 min at 4 °C. The non-solubilized fraction was first removed by precipitation by centrifugation at 23,000 × g for 40 min. Blue native polyacrylamide gel electrophoresis was performed according to Ref. 41.
EPR Spectroscopy
Cw-EPR spectra were recorded using a standard ER 4102 (Bruker) X-band resonator with a Bruker Elexsys 500 X-band spectrometer equipped with an Oxford Instruments cryostat (ESR 900). Thylakoids samples at ≈3–6 mg of Chl ml−1 were loaded in the dark into quartz EPR tubes and further dark-adapted for 1 h at room temperature. Then, the samples were frozen in the dark to 198 K, degassed at 198 K, and then transferred to 77 K in liquid N2. Illumination of the samples was done at 77 K in liquid nitrogen with a 1000-watt Tungsten lamp from which infrared light was filtered with water and infrared filters. Samples were illuminated for ∼2 min, long enough to induce the oxidation of Cyt b559 in a proportion of the centers but short enough to avoid the warming of the sample (even at 77 K) and, therefore, to prevent the relaxation of the non-relaxed into the relaxed state of the light-induced oxidized Cyt b559.
RESULTS
Construction of ΔPsbJ Mutants and Cell Growth
To compare the structural interactions in PsbA1-PSII and PsbA3-PSII between either PsbA1 or PsbA3 and other small subunits, PsbJ was deleted either from the T. elongatus 43H strain, which assembles PSII with PsbA1 (34), or from WT* strain, which assembles PSII with PsbA3. For the deletion of the psbJ gene from the T. elongatus 43H and WT* genomes, a 129-bp DNA fragment including the open reading frame of psbJ (tsr1544) was replaced by a spectinomycin/streptomycin-resistant cassette gene (2109 bp). Complete segregation of the psbJ deletion mutants (strains 43H/ΔPsbJ and WT*/ΔPsbJ) was confirmed by PCR amplification as shown in Fig. 1B. In 43H and WT*, a 1200-bp DNA fragment including the 123 bp of the psbJ open reading frame was amplified by forward and reverse primers (lanes 2 and 4). In contrast, a 3100-bp fragment including a spectinomycin/streptomycin-resistant cassette was amplified without the 1200-bp band in both 43H/ΔPsbJ and WT*/ΔPsbJ genomes (lanes 3 and 5).
The 43H/ΔPsbJ and the WT*/ΔPsbJ cells grew photosynthetically under the usual light conditions (60 μmol of photons m−2 s−1). In both the 43H and WT* cells, the deletion of PsbJ lengthened the lag phase. In the exponential phase, the doubling time of 43H/ΔPsbJ cells was 1.6 times longer than that of 43H cells (≈17 h for the doubling time of 43H cells). In contrast, the cell growth of WT*/ΔPsbJ was similar to that of WT* (≈17 h for the doubling time of WT*) (not shown, but see supplemental Fig. S2).
Water Oxidation Activity
It was reported that water oxidation activity of purified PSII was generally found to be higher (≈1.5–1.7 times) in PsbA3-PSII than in PsbA1-PSII (34) and that most of this increase likely originates from changes on the electron acceptor side (27). Table 1 shows that the oxygen evolution activity measured under saturating continuous light illumination in WT* cells was also slightly higher than in 43H cells. In both strains the deletion of PsbJ did not strongly affect the activity. Nevertheless, because the concentration of the cells was adjusted on a Chl concentration basis, the activity of whole cells is dependent on the PSI/PSII ratio, and this could vary significantly depending on the strain and the cultivation conditions. Therefore, the activity was also measured in purified PSII. The isolated PSII complex from 43H/ΔPsbJ (PsbA1/ΔPsbJ-PSII) showed only ≈30% that of the activity from 43H (PsbA1-PSII). However, the isolated PSII complex from WT*/ΔPsbJ (PsbA3/ΔPsbJ-PSII) exhibited an oxygen-evolving activity equal to 3500–4500 μmol of O2 (mg of Chl)−1 h−1, which was similar to that measured in WT* (PsbA3-PSII). These results indicate that the purified PsbA1-PSII complex is functionally unstable without PsbJ, whereas the purified PsbA3-PSII lacking PsbJ is fully stable.
TABLE 1.
Oxygen-evolving activities of cells and purified PSII complexes from 43H (PsbA1-PSII), 43H/ΔPsbJ (PsbA1/ΔPsbJ-PSII), WT* (PsbA3-PSII), and WT*/ΔPsbJ (PsbA3/ΔPsbJ-PSII)
| Strain | Cells | PSII complex |
|---|---|---|
| μmol of O2 (mg Chl)−1h−1 | μmol O2 (mg Chl)−1h−1 | |
| 43H | ∼250 (100%) | 2000–3000 (100%) |
| 43H/ΔPsbJ | ∼250 (100%) | 800–900 (30%) |
| WT* | ∼300 (100%) | 3500–4500 (100%) |
| WT*/ΔPsbJ | ∼330 (110%) | 3500–4500 (100%) |
Effects of PsbJ Deletion on Polypeptide Composition
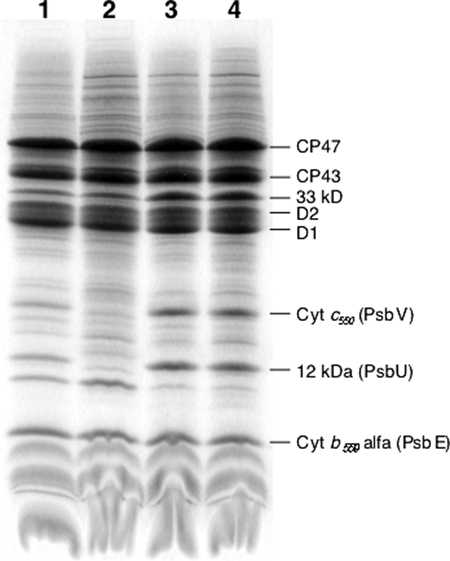
Fig. 2 shows the results of an SDS-polyacrylamide gel electrophoresis done on PsbA1-PSII (lane 1), PsbA1/ΔPsbJ-PSII (lane 2), PsbA3-PSII (lane 3), and PsbA3/ΔPsbJ-PSII (lane 4). In the molecular mass range higher than that of the α subunit of Cyt b559 (PsbE), most of the PSII subunits are detected in the four PSII complexes with the exception of Cyt c550 (PsbV) and 12-kDa extrinsic protein (PsbU), which are not detected in the PsbA1/ΔPsbJ-PSII (lane 2). Resolution of small polypeptides with a molecular mass lower than 10 kDa is difficult with SDS-polyacrylamide gels. Therefore, the polypeptides content of the purified PSII complexes was also analyzed by using MALDI-TOF mass spectroscopy. Fig. 3, A and B, show the spectra of PsbA1-PSII (black spectrum in Fig. 3A), PsbA1/ΔPsbJ-PSII (red spectrum in Fig. 3A), PsbA3-PSII (black spectrum in Fig. 3B), and PsbA3/ΔPsbJ-PSII (blue spectrum in Fig. 3B) in the m/z range from 3800 to 5500. In both the isolated PsbA1/ΔPsbJ-PSII and PsbA3/ΔPsbJ-PSII complexes, the PsbJ subunit band (m/z = 4009.62) was not detected as expected. In the PsbA1/ΔPsbJ-PSII, in addition to PsbJ, other polypeptides were lacking as PsbM (m/z = 4013.68), PsbI (m/z = 4431.26), and PsbY (m/z = 4610.12). Furthermore, the PsbT content (m/z = 3900.49), PsbX content (m/z = 4185.50), PsbL content (m/z = 4294.83), and PsbF content (β-subunit of Cyt b559 with a m/z = 4974.74) in the PsbA1/ΔPsbJ-PSII complex were all lower than in the other PSII samples if we use the amplitude of PsbK band (m/z = 4098.19) as the reference (note that in PsbA3/ΔPsbJ-PSII the PsbI content seemed lower when compared with the other subunits, which could suggest that PsbI does not interact strongly with the other subunits in PSII despite the PsbA form). In the higher molecular mass range from 5500 to 18000, PsbU and PsbV polypeptides were also lacking in purified PsbA1/ΔPsbJ PSII as shown in Fig. 3C. Like for SDS-polyacrylamide gel electrophoresis (Fig. 2, lane 2), PsbU and PsbV were not detected. In contrast, the PsbA3/ΔPsbJ-PSII maintained all the polypeptides except PsbJ, as shown in Fig. 3, B and D. Tables 2 and 3 are summary of the assignment of these polypeptides in PSII complexes from 43H (PsbA1-PSII), 43H/ΔPsbJ (PsbA1/ΔPsbJ-PSII), WT* (PsbA3-PSII), and WT*/ΔPsbJ (PsbA3/ΔPsbJ-PSII).
FIGURE 2.
Analysis by SDS-polyacrylamide gel electrophoresis of isolated PSII complexes. Lane 1, PsbA1-PSII; lane 2, PsbA1/ΔPsbJ-PSII; lane 3, PsbA3-PSII; lane 4, PsbA3/ΔPsbJ-PSII. The amount of PSII loaded was 10 μg of Chl for each lane.
TABLE 2.
Assignment, observed, and calculated molecular masses of small polypeptides of photosystem II complexes from MALDI-TOF MS of T. elongatus 43H (PsbA1-PSII) and 43H/ΔPsbJ (PsbA1/ΔPsbJ-PSII)
The data correspond to Fig. 3, A and C.
| Identification | MMOBSa | MMCALCb average | MMCALCc monoisotopic | ΔMMd | Number of amino acidse | Putative post-translational modifications | 43H | 43H/ΔPsbJ |
|---|---|---|---|---|---|---|---|---|
| Da | Da | Da | Da | |||||
| ? | 3881.68* | Yes | No | |||||
| PsbT | 3899.49* | 3874.67 | 3872.0968 | +27.39 | 32 (32) | Formylation | Yes | Yes |
| PsbM | 4005.55* | 3980.67 | 3978.1788 | +27.37 | 36 (36) | Formylation | Yes | No |
| 4012.68* | 3980.67 | 3978.1788 | +34.50 | 36 (36) | Acetylation? | Yes | No | |
| PsbJ | 4008.62* | 3973.70 | 3971.1085 | +37.51 | 40 (41) | Acetylation, −Met1 | Yes | No |
| PsbK | 4097.19* | 4099.88 | 4097.3157 | −0.12 | 37 (46) | −Met-1 ∼ Ala-9 | Yes | Yes |
| PsbX | 4184.50* | 4188.02 | 4185.4539 | −0.95 | 40 (50) | −Met-1 ∼ Leu-10 | Yes | Yes |
| PsbL | 4293.83* | 4297.02 | 4294.3220 | −0.49 | 37 (37) | Yes | Yes | |
| PsbI | 4430.26* | 4405.18 | 4402.4163 | +27.84 | 38 (38) | Formylation | Yes | No |
| PsbY | 4609.12* | 4584.61 | 4581.7224 | +27.40 | 41 (43) | Formylation, −Met-1, −Gly-2 | Yes | No |
| PsbF | 4973.74* | 4933.65 | 4930.5996 | +43.14 | 44 (45) | Acetylation, −Met-1 | Yes | No |
| Psb30 | 5061.74* | 5037.15 | 5033.7495 | +27.99 | 46 (46) | Formylation | Yes | Yes |
| PsbZ | 6795.35 | 6764.18 | 6759.8105 | +31.17 | 62 (62) | Formylation | Yes | Yes |
| PsbH | 7223.79 | 7222.49 | 7217.9362 | +1.3 | 65 (66) | −Met-1 | Yes | Yes |
| PsbE | 9441.02 | 9441.53 | 9435.8711 | −0.51 | 83 (84) | −Met-1 | Yes | Yes |
| PsbU | 11643.24 | 11644.81 | 11638.0103 | −1.57 | 104 (134) | −Met-1 ∼ Ala-30 | Yes | No |
| PsbV | 15748.04 | 15130.01 | 15120.8165 | +618.03 | 137 (66) | −Met-1, +haem | Yes | No |
a MMOBS = (peak m/z) − 1, assuming the protein molecules are singly charged. Asterisks correspond to monoisotopic molecular mass.
b Molecular mass calculated average molecular weight.
c Molecular mass calculated from monoisotopic weight.
d ΔMM = MMOBS − MMCALC.
e Number of amino acids is from mature protein, and the number in parentheses indicates the number of amino acids deduced from the open reading sequence.
TABLE 3.
Assignment, observed and calculated molecular masses of small polypeptides of Photosystem II complexes from MALDI-TOF MS of T. elongatus WT* (PbsA3-PSII) and WT*/ΔPsbJ (PsbA3/ΔPsbJ-PSII)
The data correspond to Fig. 3, B and D.
| Identification | MMOBSa | MMCALC averageb | MMCALC monoisotopicc | ΔMMd | Number of amino acidse | Putative post-translational modifications | WT* | WT*/ΔPsbJ |
|---|---|---|---|---|---|---|---|---|
| Da | Da | Da | Da | |||||
| PsbT | 3899.13* | 3874.67 | 3872.0968 | +27.03 | 32 (32) | Formylation | Yes | Yes |
| PsbM | 4005.84* | 3980.67 | 3978.1788 | +27.66 | 36 (36) | Formylation | Yes | Yes |
| 4012.67* | 3980.67 | 3978.1788 | +34.49 | 36 (36) | Acetylation? | Yes | Yes | |
| PsbJ | 4008.62* | 3973.70 | 3971.1085 | +37.51 | 40 (41) | Acetylation, −Met-1 | Yes | No |
| PsbK | 4097.18* | 4099.88 | 4097.3157 | −0.13 | 37 (46) | −Met-1 ∼ Ala-9 | Yes | Yes |
| PsbX | 4184.98* | 4188.02 | 4185.4539 | −0.47 | 40 (50) | −Met-1 ∼ Leu-10 | Yes | Yes |
| PsbL | 4295.05* | 4297.02 | 4294.3220 | +0.72 | 37 (37) | Yes | Yes | |
| PsbI | 4431.28* | 4405.18 | 4402.4163 | +28.86 | 38 (38) | Formylation | Yes | Yes |
| PsbY | 4609.70* | 4584.61 | 4581.7224 | +27.98 | 41 (43) | Formylation, −Met-1, −Gly-2 | Yes | Yes |
| PsbF | 4973.89* | 4933.65 | 4930.5996 | +43.29 | 44 (45) | Acetylation, −Met1 | Yes | Yes |
| Psb30 | 5061.71* | 5037.15 | 5033.7495 | +27.96 | 46 (46) | Formylation | Yes | Yes |
| PsbZ | 6795.59 | 6764.18 | 6759.8105 | +31.41 | 62 (62) | Formylation | Yes | Yes |
| PsbH | 7224.11 | 7222.49 | 7217.9362 | +1.95 | 65 (66) | −Met-1 | Yes | Yes |
| PsbE | 9440.64 | 9441.53 | 9435.8711 | −0.89 | 83 (84) | −Met-1 | Yes | Yes |
| PsbU | 11643.67 | 11644.81 | 11638.0103 | −1.14 | 104 (134) | −Met1 ∼ Ala30 | Yes | Yes |
| PsbV | 15747.71 | 15130.01 | 15120.8165 | +617.70 | 137 (66) | −Met-1, +heme | Yes | Yes |
a MMOBS = (peak m/z) − 1, assuming the protein molecules are singly charged. Asterisks correspond to monoisotopic molecular mass.
b Molecular mass calculated average molecular weight.
c Molecular mass calculated from monoisotopic weight.
d ΔMM = MMOBS − MMCALC.
e The number of amino acids is from mature protein and the number in parentheses indicates the number of amino acids deduced from the open reading sequence.
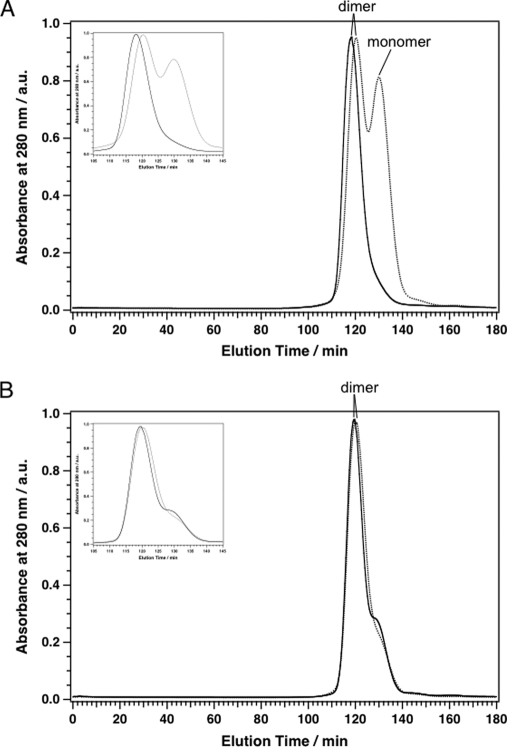
PSII Size in the ΔPsbJ Mutants
The MALDI-TOF mass spectra and the SDS-polyacrylamide gel electrophoresis showed that the isolated PsbA1/ΔPsbJ-PSII lost several polypeptide subunits in contrast to purified PsbA3/ΔPsbJ-PSII. To examine the size of the PSII complexes, these four PSII complexes were separated in terms of their size by gel permeation chromatography (Fig. 4). In the conditions in which the gel permeation was done, i.e. in the presence of 1.5% n-dodecyl-β-d-maltoside for resolubilization just before loading the samples onto the column, both the PsbA1-PSII and PsA3-PSII were essentially in the dimer form, and a very minor proportion of monomer was detected in both samples, although possibly slightly more in PsbA3-PSII. Whereas all the PsbA1-PSII was present as a dimer, about half of the PsbA1/ΔPsbJ-PSII was monomeric (Fig. 4A). The elution time, which depends on the molecular size, of the dimer form of PsbA1/ΔPsbJ-PSII appeared longer than that of the dimer form of PsbA1-PSII. This is in agreement with a lower molecular mass as expected from the MALDI-TOF and SDS-polyacrylamide gel electrophoresis data showing the lack of several subunits in PsbA1/ΔPsbJ-PSII. In contrast, almost all PsbA3/ΔPsbJ-PSII was found in a dimer form, as is the case for PsbA3-PSII. The elution time of PsbA3/ΔPsbJ-PSII was slightly longer than that of PsbA3-PSII, very likely as a consequence of the lack of PsbJ.
FIGURE 4.
Gel permeation elution patterns of PSII complexes with a column Superdex 200 (PC 3.2/30). A: solid line, PSII complex from 43H (PsbA1-PSII); dashed line, PSII complex from 43H/ΔPsbJ (PsbA1/ΔPsbJ-PSII). B: solid line, PSII complex from WT* (PsbA3-PSII); dashed line, PSII complex from WT*/ΔPsbJ (PsbA3/ΔPsbJ-PSII). Insets in A and B are the pattern from 105 to 145 min in the elution time. The intensities of absorbance at 280 nm were normalized with the intensities of the dimer form PSII. a.u., absorbance units.
To examine if the monomerization detected in the purified PsbA1/ΔPsbJ-PSII already occurred in the thylakoid membranes, the thylakoids were solubilized with 1% n-dodecyl-β-d-maltoside just after the breaking of the cells, and the solubilization mixture was analyzed by blue native polyacrylamide gel electrophoresis (Fig. 5). Surprisingly, almost all PsbA1/ΔPsbJ-PSII (lane 3) appeared in a dimer form with a molecular mass of 550 kDa, as was the case for PsbA1-PSII (lane2), PsbA3-PSII (lane 4) and PsbA3/ΔPsbJ-PSII (lane 5). A few diffuse bands from 400 to 380 kDa were present in PsbA1/ΔPsbJ-PSII, but this was also true for PsbA3-PSII and PsbA3/ΔPsbJ-PSII.
FIGURE 5.
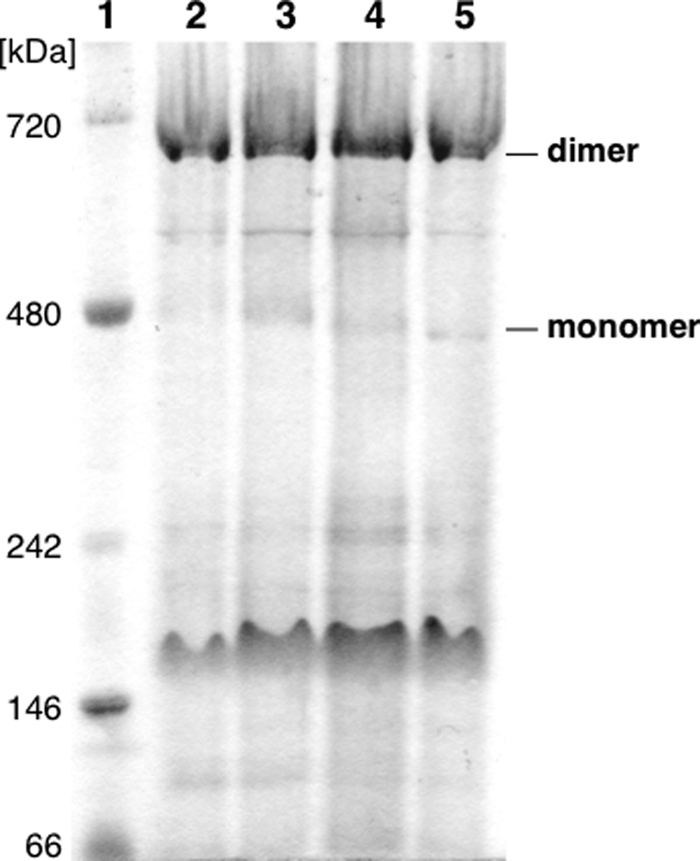
Analysis by blue native polyacrylamide gel electrophoresis of solubilized thylakoids. Lane 1, molecular size marker (Native Mark, Invitrogen); lane 2, PsbA1; lane 3, PsbA1/ΔPsbJ; lane 4, PsbA3; lane 5, PsbA3/ΔPsbJ. The amount of solubilized thylakoids loaded was 10 μg of Chl for each lane.
Cyt b559 in the Mutants
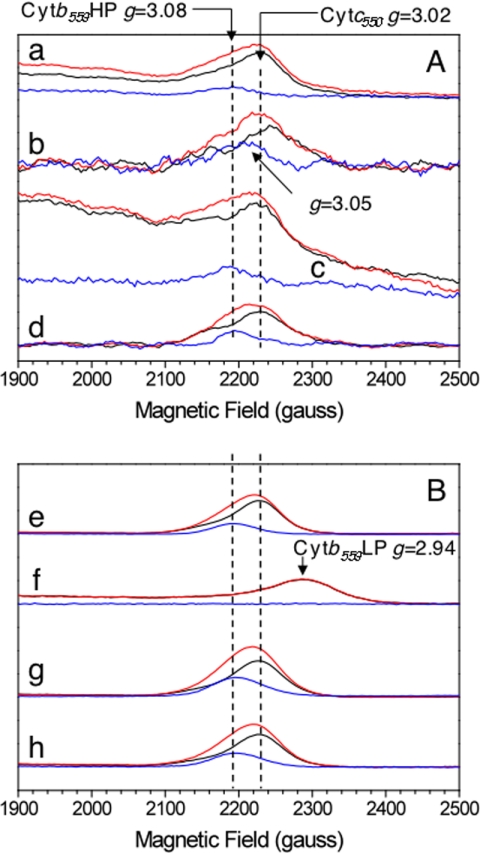
Cyt b559 is known to have at least two states that differ in terms of their redox potential. In intact PSII, the high potential (HP) form is predominant, whereas the low potential (LP) form is found when the PSII structure is affected, e.g. Refs. 42–45. Each of these two redox forms exhibits two different EPR signals corresponding to (i) a non-relaxed state (when the Cyt b559 is oxidized by illumination at temperatures ≤77 K) and (ii) to a relaxed state when the illuminated sample is warmed up to allow the sample to relax to a less constraint geometry. The nature of this structural change is not yet fully understood, but FTIR studies were consistent with at least a change in environment of one histidine ligand and a propionic group of the heme for Cyt b559 in the HP to LP conversion (43).
EPR spectroscopy can be used to monitor the HP and LP forms of Cyt b559, as each of the two oxidized forms exhibits a characteristic EPR signal, e.g. Refs. 42–44. In addition to Cyt b559, T. elongatus has a second cytochrome (Cyt c550) (1–3) which is oxidized at the ambient potential and, therefore, detected by EPR in the conditions used in the present study, e.g. Refs. 44, 46, and 47). It is shown in this work that the PsbA1/ΔPsbJ-PSII is less stable than PsbA3/ΔPsbJ-PSII. Therefore, the EPR measurements were done first in thylakoids to determine the consequences of the PsbJ deletion in the most intact material. In this material only the gz feature is easily detectable in thylakoids, but fortunately this resonance is sensitive to the state of Cyt b559. Subsequently, similar experiments were performed with the purified PSII.
Panels A and B in Fig. 6 show the gz EPR spectra recorded in thylakoids and PSII, respectively. Spectra were recorded in dark-adapted samples (black spectra) and after illumination at 77 K (red spectrum). The light-minus-dark spectra are shown in blue. In dark-adapted PsbA1-thylakoids (black spectrum in a), the signal originates mainly from Cyt c550, which is expected to be fully oxidized, and possibly from Cyt b559 in the centers in which it is oxidized. The signal increased upon the illumination at 77 K (red spectrum), a protocol known to result in the oxidation of Cyt b559 instead of the Mn4Ca cluster, which undergoes oxidation at higher temperatures. The light-minus-dark spectrum (blue spectrum) exhibits a gz value of 3.08 that is characteristic of the non-relaxed form of the HP form of Cyt b559 (42–43). Similar results were obtained in the PsbA3 thylakoids (spectra c). In PsbA3/ΔPsbJ-thylakoids (spectra d), the spectra were similar to those in PsbA3 thylakoids. This indicates that in this sample Cyt b559 was mainly in the reduced HP form before the 77 K illumination.
FIGURE 6.
EPR spectra in the gz spectral range of cytochromes in purified membrane fragments (thylakoids) (panel A) and in purified PSII (panel B). EPR spectra were recorded in PsbA1 (spectra a and e), in PsbA1/ΔPsbJ (spectra b and f), in PsbA3 (spectra c and g), and in PsbA3/ΔPsbJ (spectra d and h). Spectra were first recorded in dark-adapted sample (black spectra) and after illumination at 77 K (red spectra). Blue spectra are the light-minus-dark spectra. Other instrument settings: modulation amplitude, 25 gauss; microwave power, 5 milliwatts; microwave frequency, 9.4 GHz; modulation frequency, 100 kHz. Temperature, 15 K. Amplitude of the spectra was normalized to same reaction center concentration by using the TyrD• EPR signal as a probe. The apparent lower signal-to-noise ratio of spectra b in panel A is due to the use of a lower sample concentration.
In PsbA1/ΔPsbJ thylakoids (spectra b), the amplitude of the light-induced signal was comparable with that in the two other samples. This indicates that Cyt b559 was also mainly in a reduced form before the 77 K illumination. Nevertheless, the gz value of the light-induced signal (g = 3.05) was slightly lower than in the two other samples, and this indicates a small structural perturbation and may suggest that the redox potential of Cyt b559 is already shifted to a slightly lower value in PsbA1/ΔPsbJ-thylakoids when compared with PsbA3/ΔPsbJ thylakoids.
In purified PSII (panel B, Fig. 6), similar results were obtained in PsbA1-PSII (spectra e), PsbA3-PSII (spectra g), and PsbA3/ΔPsbJ-PSII (spectra h). The 77 K illumination induced the characteristic EPR spectrum of the non-relaxed HP form of Cyt b559. In contrast, in PsbA1/ΔPsbJ-PSII (spectra f) the spectrum recorded in the dark-adapted sample clearly shows that the signal from Cyt c550 was missing and that the gz signal from Cyt b559 recorded in the dark-adapted sample was indicative of a relaxed oxidized LP form. The 77 K illumination induced no additional signal, indicating that all the Cyt b559 was oxidized before the illumination.
DISCUSSION
The D1 (PsbA) protein provides binding sites for almost all of the cofactors involved in the photosynthetic electron transfer in PSII. The thermophilic cyanobacterium T. elongatus has three genes (psbA1, psbA2, and psbA3) encoding the D1 protein. Of the 344 amino acids that constitute D1, the processed PsbA1 differs from PsbA2 and PsbA3 by 37 and 21 amino acids, respectively (supplemental Fig. S3). Although many cyanobacteria are known to have multiple psbA genes, T. elongatus is unique among those with sequenced genomes in possessing structurally different PsbA for each of the gene copies. Other species with multiple copies of psbA have duplicates of the same gene. Under the cultivation conditions commonly used in laboratories, T. elongatus produces PsbA1-PSII due to the transcription and translation of the psbA1 gene, because transcription of psbA3 is induced and that of psbA1 is suppressed under high light conditions (26). The PsbA3-PSII complex purified from the T. elongatus mutant, in which both psbA1 and psbA2 have been deleted, exhibited an ≈1.5–1.7 times higher water oxidation activity than the isolated PsbA1-PSII complex (34). Recently, the comparison of energetic properties between PsbA1-PSII and PsbA3-PSII revealed that the redox potential of PheoD1 is increased by 17 mV from −522 mV in PsbA1-PSII to −505 mV in PsbA3-PSII in addition to modification of the redox potential of QA/QA− and QB/QB− in PsbA3-PSII (27). All these changes were discussed in the context of PsbA3 providing an advantage over PsbA1 for T. elongatus cells to survive under particular conditions.
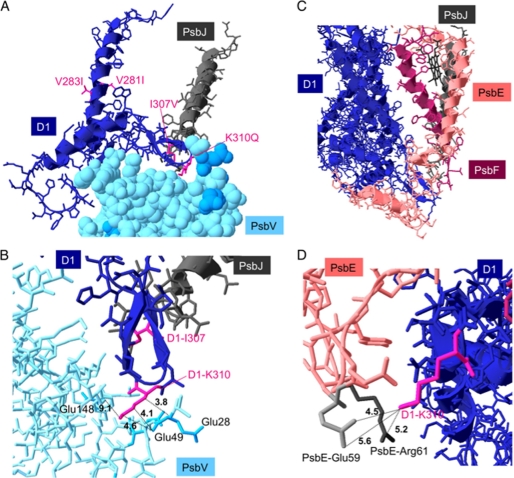
Although PsbJ is close to the periphery of PSII, its N-terminal region approaches a loop of the D1 protein on the lumenal side (Fig. 7) in the x-ray crystal structures of PsbA1-PSII (1–3). Some amino acid residues in this contact region between D1 and PsbJ differ in PsbA3 and PsbA1. To study the influence of these differences between PsbA1 and PsbA3, we analyzed some properties of PsbA1-PSII and PsbA3-PSII in which the PsbJ subunit was deleted. The present experimental data give information that can be added to a previous theoretical study (31).
FIGURE 7.
A and B, shown are structures around the soluble loop of PsbA1 (navy blue), PsbJ (gray), and PsbV (pale blue). C and D, shown are structures around PsbA1 (navy blue), PsbJ (gray), PsbE (α-subunit of Cyt b559) (pale pink), and PsbF (β-subunit of Cyt b559) (magenta). Amino acid residues colored with magenta (V281I, V283I, I307V, and K310Q) correspond to different residues between PsbA1 and PsbA3 (alphabets left and right of the number are residues of PsbA1 and PsbA3, respectively). Blue-colored residues in A and B are negatively charged amino acids in PsbV. In panel D, negatively charged Glu and positively charged Arg of PsbE are colored with pale gray and dark gray, respectively. The numbers on dotted lines indicate the distance (Å). The figures were drawn with Swiss Pdb Viewer with PDB 3BZ1 (PsbA1-PSII).
The oxygen-evolving activity in whole cells was slightly higher in WT* cells, which have only PsbA3 as D1 protein, than in 43H cells, which have PsbA1 as D1. Upon the deletion of PsbJ, the activity was hardly affected in both strains (Table 1). In contrast, the oxygen-evolving activity in purified PSII was strongly decreased upon the deletion of PsbJ in PsbA1-PSII, whereas the deletion of PsbJ in PsbA3-PSII has almost no effect. These results strongly suggest that the absence of PsbJ destabilizes PsbA1/ΔPsbJ-PSII, making it more susceptible to damage during the isolation procedure. In agreement with this, the analysis of the protein composition of the purified PsbA1/ΔPsbJ-PSII revealed that in addition to PsbJ, several other subunits, PsbV (Cyt c550), PsbU (extrinsic 12-kDa protein), PsbY, PsbI, and PsbM, were also lacking. Furthermore, analysis of gel permeation chromatography indicated that half of the PsbA1/ΔPsbJ-PSII was in a monomer form, although isolated PsbA3/ΔPsbJ-PSII was essentially in dimeric form as was the case for PsbA1-PSII and PsbA3-PSII (Fig. 4).
The results in Fig. 5 show that PSII monomers are virtually absent in thylakoids from 43H/ΔPsbJ cells. This indicates that the monomer form in PsbA1/ΔPsbJ-PSII is due to the addition of detergent to a weakened PSII complex. Because the growth rate of 43H/ΔPsbJ cells was half that of the 43H cells, WT* (see supplemental Fig. S2), and WT*/ΔPsbJ cells, it is tempting to propose that the same structural changes are responsible for monomer formation in PsbA1/ΔPsbJ-PSII and for the slower growth of 43H/ΔPsbJ cells. Because this destabilization is not observed in the other three samples studied here, this approach may help identify the molecular interactions between either PsbA1 or PsbA3 with the other subunits that are important for maintaining the dimeric form.
Bentley et al. (48) and Iwai et al. (49) reported that the lack of both PsbM and PsbT might cause a complete monomerization of PSII complex, although in Synechocystis PCC 6803, the absence of PsbT destabilized PSII to a greater extent than that of PsbM. In addition to PsbT, PsbM is also present in the hinge region of PSII dimer (1–3). It is shown here that the PsbA1/ΔPsbJ-PSII contained ≈25% PsbT (Fig. 3A) but completely lacked PsbM. Therefore, both PsbM and PsbT could be indeed involved in the stabilization of the dimer. Nevertheless, in the PsbA1/ΔPsbJ-PSII mutant, other subunits are also lacking, and this makes it difficult to draw a definitive conclusion on which subunit is essential for maintaining the dimer. The striking observation is that PsbJ is not required to maintain the dimer form, when PsbA3 is the D1 protein.
Why did the isolated PsbA1/ΔPsbJ-PSII lose several protein subunits in addition to PsbJ despite missing the PsbJ and lower content of the PsbI in PsbA3/ΔPsbJ-PSII? As shown in Fig. 7A, the N-terminal region of PsbJ approaches the soluble loop of D1 (PsbA1), which is in contact with the pocket of the extrinsic protein, PsbV (Cyt c550). The tip of the PsbA1 loop includes the positively charged amino acid residue, Lys-310. In the pocket of PsbV, the Glu residues (Glu-28 and Glu-49), which are likely negatively charged, are located at 3–5 Å from PsbA1-Lys-310 (Fig. 7B). Therefore, PsbA1 might be structurally stabilized by an electrostatic interaction(s) with PsbV. As the electrostatic interaction is relatively weak, elimination of PsbJ subunit located 3–5 Å from the D1 loop might destabilize the electrostatic interaction, and consequently, the binding of PsbV to PsbA1/ΔPsbJ-PSII could be weakened.
When we look at the interaction between PsbA1 and PsbE in the structure, the tip of the internal loop of D1, PsbA1-Lys-310, also seems to make an electrostatic interaction with Glu-59 of PsbE (Fig. 7, C and D). However, a positively charged amino acid residue, PsbE-Arg-61, is also located close to the D1-Lys-310 (within 5.2 Å). Surprisingly, the replacement of Lys-310 in PsbA1, which is assumed to interact electrostatically with the pocket of PsbV and the lumenal region of PsbE with an uncharged Gln in PsbA3 increased the stability of the PsbA3/ΔPsbJ-PSII complex. Furthermore, PsbA1-Ile-307, which is also located in the internal loop of D1, is substituted by Val in PsbA3 (Fig. 7A). In addition to these substitutions, PsbA1-Val-281 and PsbA1-Val-283, which are located in helix V, close to the internal the loop of D1, are also substituted by Ile residues in PsbA3 (Fig. 7A). In the HIV protease, for instance, the Val to Ile substitution has been shown to modify the van der Waals interaction between the protein and inhibitors (50, 51). Therefore, we cannot rule out a role of the Ile to Val substitution in the hydrogen-bond network in PSII complex. To clarify the importance of the possible electrostatic interaction between PsbA1-Lys-310 and its neighbors in PsbV and PsbE, a site-directed mutant PsbA3-Lys-310 is required in addition to the PsbJ deletion.
Isolated PsbA1/ΔPsbJ-PSII lost not only PsbJ but also several polypeptides including PsbF, whereas isolated PsbA3/ΔPsbJ-PSII lost only PsbJ (Figs. 2 and 3). Recently, it was shown that the removal of PsbK, which is close to PsbJ, in PsbA1-PSII resulted in the complete loss of Psb30 and partial loss of PsbZ (49). In the present work it is shown that the removal of PsbJ has no effect on the PsbK content, and maybe as a consequence, it has no effect on the content of Psb30 and PsbZ.
In Synechocystis PCC 6803, the removal of PsbJ has been shown to disrupt the electron transfer on the acceptor side of PSII, e.g. Ref. 52. This is in agreement with PsbJ as an element making the Qc channel as recently identified (3). It was also recently shown that there was probably a relationship between the redox potential of Cyt b559 and the filling of the Qc site (53). The lack of PsbJ is, therefore, expected to modify the properties of Cyt b559. In the EPR spectra, the gz value of Cyt b559 was found here to be similar in PsbA1 thylakoids, PsbA3 thylakoids, and PsbA3/ΔPsbJ thylakoids but lower in PsbA1/ΔPsbJ thylakoids (Fig. 6). These results suggest that the redox potential of Cyt b559 is lower in PsbA1/ΔPsbJ-PSII than in the three other samples. Preliminary data also shows that the rate of QA− oxidation after several flashes is not modified in PsbA3/ΔPsbJ-PSII when compared with PsbA3-PSII.3
PsbF, one of the two subunits of Cyt b559, was also lost during the isolation of PsbA1/ΔPsbJ-PSII. In the native membrane even though the PSII still maintains the PsbF subunit, the conformation of Cyt b559 appears be modified. This situation is different from that in which Cyt b559 was altered before PSII assembly, for example by mutation (54). In this case, PSII is fully inactive. In our work, the consequences of the PsbJ deletion occurs mainly in the purified PSII rather than in the living cell. Data in Fig. 6 (spectra f) show that in purified PsbA1/ΔPsbJ-PSII, the Cyt b559 exhibits an EPR spectrum that is characteristic of a low spin state. This shows that the sixth axial ligand of the heme, the His24 of PsbF, was substituted for another ligand, likely a water molecule. Nevertheless, for the moment we cannot rule out the ligation by another amino acid residue.
Of the three extrinsic proteins, PsbU, PsbV, and PsbO, only the latter was maintained in purified PsbA1/ΔPsbJ-PSII. This is expected to significantly change the structural interactions between PsbO and PSII. Therefore, it is not surprising that because the N-terminal region of PsbI is in contact with PsbO, this subunit was released from the PSII complex.
The following hypothesis can be proposed to explain the cascade of events in PsbA1/ΔPsbJ during the PSII purification procedure. First, the removal of PsbJ modifies the structure of the internal loop and helix V of PsbA1. Second, this modification triggers the release of the extrinsic PsbV and then PsbU proteins. As a consequence, PsbO is expected to be less tightly bound to the intrinsic proteins. Finally, because the binding site of PsbO to the intrinsic proteins approaches the N-terminal region of PsbM, the PsbA1/ΔPsbJ-PSII could release PsbM subunit (see supplemental Fig. S4). These modifications are expected to favor the monomer formation.
All the points mentioned above suggest that the observed decrease in the stability of PsbA1/ΔPsbJ-PSII, when compared with PsbA3/ΔPsbJ-PSII, is a result of a weaker stability of PsbA1-PSII relatively to PsbA3-PSII. Although many amino acid substitutions could be at the origin of this instability, it is likely that a change in the general hydrogen bond network could be also involved. The x-ray crystal structure analysis of PsbA3-PSII would also be an important step in the understanding of the relationship between the molecular structure, the stability, and the energetics.
Supplementary Material
Acknowledgments
We thank Jian-Ren Shen for kindly analyzing the PSII complexes with a gel-permeation chromatography. We are grateful to Jim Barber, James Murray, Yuki Kato, Hiroshi Ishikita, and Bill Rutherford for helpful discussions and Takashi Manabe for technical assistance with MALDI-TOF MS analyses. Bill Rutherford is acknowledged for careful reading of the manuscript.
This study was supported by Grant-in-aid for scientific research 21612007 from the Ministry of Education, Science, Sports, Culture, and Technology (to M. S.) and the Japan Society for the Promotion of Science and CNRS under the Japan-France Research Cooperative Program. This work was also supported in part by the European Union/Energy Network project SOLAR-H2 (FP7 contract 212508).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
F. Rappaport, A. Boussac, and M. Sugiura, unpublished data.
- PSII
- photosystem II
- Chl
- chlorophyll
- Cyt
- cytochrome
- Pheo
- pheophytin
- P680
- primary electron donor
- QA
- primary quinone acceptor
- QB
- secondary quinone acceptor
- 43H
- T. elongatus strain with a His6 tag on the C terminus of CP43
- WT*
- His6 tag T. elongatus strain in which the psbA1 and psbA2 genes were deleted
- WT*/ΔPsbJ
- His6-tag T. elongatus strain in which the psbA1, psbA2, and psbJ genes were deleted (producing PsbA3/ΔPsbJ-PSII)
- 43H/ΔPsbJ
- His6 tag T. elongatus strain in which the psbJ gene was deleted (producing PsbA1/ΔPsbJ-PSII)
- HP
- high potential
- LP
- low potential.
REFERENCES
- 1.Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 2.Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 3.A. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 4.Murray J. W., Maghlaoui K., Kargul J., Ishida N., Lai T.-L., Rutherford A. W., Sugiura M., Boussac A., Barber J. (2008) Energy and Environmental Science 1, 161–166 [Google Scholar]
- 5.Kawakami K., Umena Y., Kamiya N., Shen J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8567–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton P., Ruban A. (2005) J. Exp. Bot. 56, 365–373 [DOI] [PubMed] [Google Scholar]
- 7.Horton P., Johnson M. P., Perez-Bueno M. L., Kiss A. Z., Ruban A. V. (2008) FEBS J. 275, 1069–1079 [DOI] [PubMed] [Google Scholar]
- 8.Krieger-Liszskay A. (2004) J. Exp. Bot. 56, 337–346 [DOI] [PubMed] [Google Scholar]
- 9.Vass I., Cser K., Cheregi O. (2007) Ann. N.Y. Acad. Sci. 1113, 114–122 [DOI] [PubMed] [Google Scholar]
- 10.Krieger-Liszkay A., Fufezan C., Trebst A. (2008) Photosynth. Res. 98, 551–564 [DOI] [PubMed] [Google Scholar]
- 11.Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S. I. (2007) Biochim. Biophys. Acta 1767, 414–421 [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S., Murata N. (2008) Trends Plant Sci. 13, 178–182 [DOI] [PubMed] [Google Scholar]
- 13.Edelman M., Mattoo A. K. (2008) Photosynth. Res. 98, 609–620 [DOI] [PubMed] [Google Scholar]
- 14.Huesgen P. F., Schuhmann H., Adamska I. (2009) Res. Microbiol. 160, 726–732 [DOI] [PubMed] [Google Scholar]
- 15.Baena-González E., Aro E. M. (2002) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohanty P., Allakhverdiev S. I., Murata N. (2007) Photosynth. Res. 94, 217–224 [DOI] [PubMed] [Google Scholar]
- 17.Mulo P., Sirpiö S., Suorsa M., Aro E. M. (2008) Photosynth. Res. 98, 489–501 [DOI] [PubMed] [Google Scholar]
- 18.Kato Y., Sakamoto W. (2009) J. Biochem. 146, 463–469 [DOI] [PubMed] [Google Scholar]
- 19.Golden S. S. (1995) J. Bacteriol. 177, 1651–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tichý M., Lupínková L., Sicora C., Vass I., Kuviková S., Prásil O., Komenda J. (2003) Biochim. Biophys. Acta 1605, 55–66 [DOI] [PubMed] [Google Scholar]
- 21.Sicora C. I., Appleton S. E., Brown C. M., Chung J., Chandler J., Cockshutt A. M., Vass I., Campbell D. A. (2006) Biochim. Biophys. Acta 1757, 47–56 [DOI] [PubMed] [Google Scholar]
- 22.Sicora C. I., Brown C. M., Cheregi O., Vass I., Campbell D. A. (2008) Biochim. Biophys. Acta 1777, 130–139 [DOI] [PubMed] [Google Scholar]
- 23.Summerfield T. C., Toepel J., Sherman L. A. (2008) Biochemistry 47, 12939–12941 [DOI] [PubMed] [Google Scholar]
- 24.Sicora C. I., Ho F. M., Salminen T., Styring S., Aro E. M. (2009) Biochim. Biophys. Acta 1787, 105–112 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura Y., Kaneko T., Sato S., Ikeuchi M., Katoh H., Sasamoto S., Watanabe A., Iriguchi M., Kawashima K., Kimura T., Kishida Y., Kiyokawa C., Kohara M., Matsumoto M., Matsuno A., Nakazaki N., Shimpo S., Sugimoto M., Takeuchi C., Yamada M., Tabata S. (2002) DNA Res. 9, 123–130 [DOI] [PubMed] [Google Scholar]
- 26.Kós P. B., Deák Z., Cheregi O., Vass I. (2008) Biochim. Biophys. Acta 1777, 74–83 [DOI] [PubMed] [Google Scholar]
- 27.Sugiura M., Kato Y., Takahashi R., Suzuki H., Watanabe T., Noguchi T., Rappaport F., Boussac A. (2010) Biochim. Biophys. Acta 1797, 1491–1499 [DOI] [PubMed] [Google Scholar]
- 28.Shibuya Y., Takahashi R., Okubo T., Suzuki H., Sugiura M., Noguchi T. (2010) Biochemistry 49, 493–501 [DOI] [PubMed] [Google Scholar]
- 29.Hughes J. L., Cox N., Rutherford A. W., Krausz E., Lai T. L., Boussac A., Sugiura M. (2010) Biochim. Biophys. Acta 1797, 11–19 [DOI] [PubMed] [Google Scholar]
- 30.Kato Y., Sugiura M., Oda A., Watanabe T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17365–17370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loll B., Broser M., Kós P. B., Kern J., Biesiadka J., Vass I., Saenger W., Zouni A. (2008) Biol. Chem. 389, 609–617 [DOI] [PubMed] [Google Scholar]
- 32.Golden J. W., Wiest D. R. (1988) Science 242, 1421–1423 [DOI] [PubMed] [Google Scholar]
- 33.Sugiura M., Inoue Y. (1999) Plant Cell. Physiol. 40, 1219–1231 [DOI] [PubMed] [Google Scholar]
- 34.Sugiura M., Boussac A., Noguchi T., Rappaport F. (2008) Biochim. Biophys. Acta 1777, 331–342 [DOI] [PubMed] [Google Scholar]
- 35.Sugiura M., Rappaport F., Brettel K., Noguchi T., Rutherford A. W., Boussac A. (2004) Biochemistry 43, 13549–13563 [DOI] [PubMed] [Google Scholar]
- 36.Un S., Boussac A., Sugiura M. (2007) Biochemistry 46, 3138–3150 [DOI] [PubMed] [Google Scholar]
- 37.Sugiura M., Rappaport F., Hillier W., Dorlet P., Ohno Y., Hayashi H., Boussac A. (2009) Biochemistry 48, 7856–7866 [DOI] [PubMed] [Google Scholar]
- 38.Sugiura M., Harada S., Manabe T., Hayashi H., Kashino Y., Boussac A. (2010) Biochim. Biophys. Acta 1797, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 39.Shen J. R., Kamiya N. (2000) Biochemistry 39, 14739–14744 [DOI] [PubMed] [Google Scholar]
- 40.Ikeuchi M., Inoue Y. (1988) Plant Cell Physiol. 29, 1233–1239 [Google Scholar]
- 41.Takahashi T., Inoue-Kashino N., Ozawa S., Takahashi Y., Kashino Y., Satoh K. (2009) J. Biol. Chem. 284, 15598–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart D. H., Brudvig G. W. (1998) Biochim. Biophys. Acta 1367, 63–87 [DOI] [PubMed] [Google Scholar]
- 43.Berthomieu C., Boussac A., Mäntele W., Breton J., Nabedryk E. (1992) Biochemistry 31, 11460–11471 [DOI] [PubMed] [Google Scholar]
- 44.Roncel M., Boussac A., Zurita J. L., Bottin H., Sugiura M., Kirilovsky D., Ortega J. M. (2003) J. Biol. Inorg. Chem. 8, 206–216 [DOI] [PubMed] [Google Scholar]
- 45.Kaminskaya O., Kurreck J., Irrgang K. D., Renger G., Shuvalov V. A. (1999) Biochemistry 38, 16223–16235 [DOI] [PubMed] [Google Scholar]
- 46.Shen J. R., Inoue Y. (1993) J. Biol. Chem. 268, 20408–20413 [PubMed] [Google Scholar]
- 47.Kerfeld C. A., Krogmann D. W. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 397–425 [DOI] [PubMed] [Google Scholar]
- 48.Bentley F. K., Luo H., Dilbeck P., Burnap R. L., Eaton-Rye J. J. (2008) Biochemistry 47, 11637–11646 [DOI] [PubMed] [Google Scholar]
- 49.Iwai M., Suzuki T., Kamiyama A., Sakurai I., Dohmae N., Inoue Y., Ikeuchi M. (2010) Plant Cell. Physiol. 51, 554–560 [DOI] [PubMed] [Google Scholar]
- 50.Johnson V. A., Brun-Vezinet F., Clotet B., Gunthard H. F., Kuritzkes D. R., Pillay D., Schapiro J. M., Richman D. D. (2009) Top. HIV Med. 17, 138–145 [PubMed] [Google Scholar]
- 51.Ala P. J., Huston E. E., Klabe R. M., McCabe D. D., Duke J. L., Rizzo C. J., Korant B. D., DeLoskey R. J., Lam P. Y., Hodge C. N., Chang C. H. (1997) Biochemistry 36, 1573–1580 [DOI] [PubMed] [Google Scholar]
- 52.Regel R. E., Ivleva N. B., Zer H., Meurer J., Shestakov S. V., Herrmann R. G., Pakrasi H. B., Ohad I. (2001) J. Biol. Chem. 276, 41473–41478 [DOI] [PubMed] [Google Scholar]
- 53.Kaminskaya O., Shuvalov V. A., Renger G. (2007) Biochemistry 46, 1091–1105 [DOI] [PubMed] [Google Scholar]
- 54.Hung C. H., Huang J. Y., Chiu Y. F., Chu H. A. (2007) Biochim. Biophys. Acta 1767, 686–693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.