Abstract
Glycolipid glycosyltransferases (GGT) are transported from the endoplasmic reticulum (ER) to the Golgi, their site of residence, via COPII vesicles. An interaction of a (R/K)X(R/K) motif at their cytoplasmic tail (CT) with Sar1 is critical for the selective concentration in the transport vesicles. In this work using computational docking, we identify three putative binding pockets in Sar1 (sites A, B, and C) involved in the interaction with the (R/K)X(R/K) motif. Sar1 mutants with alanine replacement of amino acids in site A were tested in vitro and in cells. In vitro, mutant versions showed a reduced ability to bind immobilized peptides with the CT sequence of GalT2. In cells, Sar1 mutants (Sar1D198A) specifically affect the exiting of GGT from the ER, resulting in an ER/Golgi concentration ratio favoring the ER. Neither the typical Golgi localization of GM130 nor the exiting and transport of the G protein of the vesicular stomatitis virus were affected. The protein kinase inhibitor H89 produced accumulation of Sec23, Sar1, and GalT2 at the ER exit sites; Sar1D189A also accumulated at these sites, but in this case GalT2 remained disperse along ER membranes. The results indicate that amino acids in site A of Sar1 are involved in the interaction with the CT of GGT for concentration at ER exiting sites.
Keywords: Cell Compartmentation, Computer Modeling, Endoplasmic Reticulum (ER), Glycolipids, Golgi, Membrane Biogenesis, Membrane Trafficking, COPII Vesicles, Glycosyltransferase, Sar1
Introduction
Protein export from the endoplasmic reticulum (ER)5 is a selective process initiated by recruiting the small GTPase Sar1 to the ER membrane (2). Cytosolic Sar1 GDP is converted to membrane-bound Sar1 GTP by the GEF Sec12p, an ER membrane protein (2). In its GTP form, Sar1 exposes a hydrophobic domain at the N terminus that favors its membrane association (3, 4). Membrane-associated Sar1 GTP initiates further recruitment of the heterodimeric complex Sec23p-Sec24p forming the prebudding complex, with which the CT of integral membrane cargo proteins interact (5–8). Vesicle completion occurs with participation of the tetramer Sec13p-Sec31p (9–11) and of other cytosolic proteins constituent of the COPII complex (12, 13). This process induces the formation of ER exit sites from where prebudding COPII transport vesicles accommodate cargo molecules (cell surface proteins, secretory products, extracellular matrix components, etc.) for transport along the secretory pathway (14).
Cargo selection occurs by different mechanisms, with most transmembrane proteins binding directly to specific COPII subunits by interactions of particular amino acid motifs present within their CT. On the other hand, soluble cargos are concentrated in COPII vesicles by indirect association with transmembrane export receptors containing specific ER export signals (reviewed in Ref. 15). Golgi glycosyltransferases concentrate selectively into COPII vesicles by a direct interaction of their CTs with Sar1 through a (R/K)X(R/K) motif proximal to the transmembrane region (16–19). Both Sar1-GDP and Sar1-GTP interact with the CT sequence peptides, but only Sar1-GTP bound to peptides is able to bind Sec23 from a rat liver cytosol (16). In the present work we have carried out an in silico screening of regions in Sar1 responsible for the interaction with CTs bearing the (R/K)X(R/K) motif. Alanine replacements of amino acid residues in the best candidate region of interaction were generated, and the in vitro and in vivo consequences of the mutations were evaluated. The results revealed that certain amino acids in the region of Sar1 facing the ER membrane surface are critical for an efficient ER export and proper Golgi localization of glycolipid glycosyltransferases.
EXPERIMENTAL PROCEDURES
In Silico Assays
The LSLFRR peptide with sequence corresponding to the GalT2 CT and its noninteracting analog LSLFAA (16) were independently docked on the structure of mouse Sar1 (Protein Data Bank code 1F6B) (4). Missing loops between residues 49–54 and 79–82 were constructed with the program LOOPY (29), and the docking was performed with Autodock 3.0 (20). The search first spanned the whole protein surface and then concentrated in the binding zones detected in the global search. All of the docked peptide-protein complexes were reranked using the program STC (21). The complexes that ranked best (i.e. within 3 kcal from the minimum) with both algorithms plus the best of each method individually were subjected to cluster analysis. For the selection of the amino acid(s) to mutate to alanine in Sar1, we took into account the average interaction energy, as defined by STC, between the residues in the protein and the dibasic motif of the peptide LSLFRR. We also verified whether alanine substitution destabilized the protein structure using the ANOLEA server (22).
DNA Constructs
Expression vectors containing cDNA coding for the N-terminal domain of galactosyltransferase (GalT2), N-acetylgalactosaminyltransferase (GalNacT), and sialyltransferases (SialT2) fused to the N terminus of the enhanced cyan fluorescent protein have been previously described (30). Briefly, they are pEYFP-N1 (Clontech)-based vectors containing the N-terminal domains of the transferases (residues 1–52 for GalT2, residues 1–27 for GalNacT, and residues 1–57 for SialT2) fused to YFP. The chimeric construct containing the thermosensitive G protein of vesicular stomatitis virus (ts045) fused to YFP was kindly provided by P. Keller (Max-Planck Institute, Dresden, Germany) (31). Inserts were checked by sequencing both strands twice using flanking primers. Sar1-CFP was generated by fusion of Sar1 to the N terminus of CFP. The DNA fragment encoding Sar1 was generated by PCR with an EcoRI site and a consensus Kozak sequence at the 5′ end and a BamHI site at the 3′ end. The primers GCCGGAATTCCGCCACCATGTCCTTCCATATTTGACTGGATTTAC (forward) and CCGCGGATCCCGATCGATGTACTGTGCCATCCAGC (reverse) were used to amplify a full-length Sar1. The PCR fragments encoding full-length Sar1 were digested with EcoRI and BamHI and ligated into EcoRI-BamHI-digested pECFP-N1 from Clontech Laboratories (Palo Alto, CA) to generate Sar1-CFP. Sar1-CFP mutant constructs T39N, N94A, N126A, and D198A were generated using the QuikChange site-directed mutagenesis kit (Qiagen) and appropriate primer combinations.
Cell Culture and Transfection
CHO-K1 cells were grown in DMEM supplemented with 10% fetal calf serum, 100 mg/ml of penicillin, and 100 mg/ml of streptomycin. At ∼70% confluence, the cells were transfected with Lipofectamine (Invitrogen) and analyzed 18 h after transfection.
Fluorescence Microscopy
Cells grown on coverslips were fixed for 7 min in methanol at −20 °C, incubated with the specific antibody and fluorescent secondary antibodies. The coverslips were mounted with FluorSave (Calbiochem, EMD Biosciences, Inc., La Jolla, CA) and observed in an Olympus FV 1000 confocal microscope with a 100× planapochromat oil immersion objective and appropriate filters for CFP, YFP, rhodamine, and FITC. When indicated, 50 μm of the isoquinolinesulfonamide H89 was added 120 min before fixation. The quantification of GalT2 and GM130 present in ER upon co-transfection with Sar1 or Sar1 mutants was made using Metamorph 4.5 imaging system (Universal Imaging Corporation, West Chester, PA) software.
In Vitro Binding Assay
For Sar1-Sepharose bead preparation, recombinant His6-Sar1 (wt or D198A) in binding buffer (500 mm NaCl and 20 mm Tris/HCl, pH 7.9) was incubated with 60 μl of 50% Ni2+-charged Sepharose at 4 °C for 1 h and then washed three times with binding buffer to remove the unbound material. CHO-K1 cells harvested in lysis buffer (50 mm Tris/HCl, pH 7.5, 1% (v/v) Triton X-100, 150 mm NaCl, and 1 mm PMSF) were left to stand for 30 min at 4 °C and then passed 20 times through a 25-gauge needle. The lysates were cleared by centrifugation at 13,000 × g for 10 min, and the supernatant was used for immunoprecipitation and binding assays. Lysates of CHO-K1 cells expressing GalT2-HA-YFP, GalT2RR-AA-HA-YFP, or VSV-G-YFP were incubated with Sar1-Sepharose beads at 25 °C for 1.5 h, washed three times with binding buffer containing 1% (v/v) Triton X-100, and washed three times with washing buffer (500 mm NaCl, 20 mm Tris/HCl, pH 7.9, and 1% (v/v) Triton X-100). The bound proteins were eluted with 25 μl of Laemmli sample buffer. The eluted samples were subjected to SDS/PAGE and Western blot analysis using rabbit anti-GFP antibody.
RESULTS
Computational Docking Identified Potential Sites of Interaction between Sar1 and CTs Bearing the (R/K)X(R/K) Motif
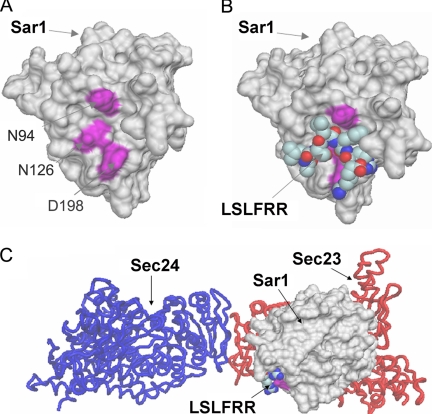
Crystallographic analysis of the yeast Sec23-Sec24-Sar1 prebudding ternary complex indicates a “bowtie-shaped” structure, with a concave membrane-proximal surface conforming an extensive interaction area with the ER membrane (3) (Fig. 1). A computational docking was performed with the peptide LSLFRR as ligand and the crystal structure of Sar1 as receptor. This peptide, which is the CT of GalT2, was shown to interact with Sar1 (16) and serves as a model peptide with the general basic cluster (R/K)X(R/K). The docking performed on the whole protein surface revealed the presence of three putative binding pockets for the peptide (sites A, B, and C). Sites B and C were not further analyzed because they were located in regions of Sar1, which are not likely to be in the reach of the dibasic motif, which in turn is presumed to be located near the membrane surface at which Sar1 binds (Fig. 1C). The remaining pocket (site A) was located in the C terminus of Sar1 and comprises residues Pro91, Asn94, Asn114, Thr123, Pro124, Asn126, Leu165, Pro172, Tyr196, and Asp198 (Fig. 1A). Site A is exposed to the ER membrane face when Sar1 is forming the ternary complex Sec23-Sec24-Sar1 (Fig. 1C). This site was further explored, and we found docked positions with free energy of binding between −8 and −10 kcal/mol using Autodock (20) and between −12 and −14 using STC (21). These binding energies suggest a high affinity constant, in the nanomolar order. The best position for the interaction Sar1-GalT2CT is shown in Fig. 1B. For control of the in silico approach, the peptide LSLFAA, which does not bind Sar1 (16), was used. In line with the experimental results, LSLFAA was found to have binding affinity constants 2–3 orders of magnitude lower than LSLFRR peptide for the candidate pockets. To test the predictions of the docking experiments, Sar1 mutants with replacements of Asn94, Asn126, and Asp198 by alanines were constructed. It is important to note that these changes do not destabilize the structure of Sar1 as predicted with ANOLEA (22).
FIGURE 1.
Docking simulation identified potential sites of interaction between Sar1 and the peptide LSLFRR corresponding to the CT of GalT2. A, the best interacting residues found for site A, namely Asn94, Asn126, and Asp198, are indicated in purple. B, the best docked pose of the peptide LSLFRR in site A is shown in van der Waal's representation. C, the ternary complex Sec23-Sec24-Sar1 (3) is shown indicating the position of site A with respect to the ER membrane.
Sar1D198A Shows Reduced Binding to GalT2CT
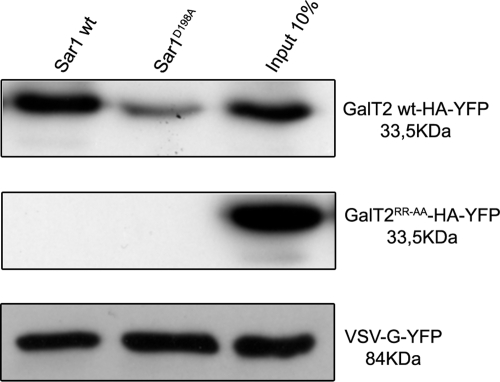
It is known that Sar1 immobilized on Sepharose beads binds to CT sequences of glycosyltransferases containing the (R/K)X(R/K) motif (16, 17). Amino acid substitutions in site A of Sar1, predicted by the computational docking as relevant in the interaction with the CT of GalT2, should affect the ability of GalT2 to bind the mutant Sar1 in vitro. To test this prediction, recombinant Sar1 or Sar1D198A was immobilized in Ni2+-charged Sepharose and incubated with lysates of cells expressing GalT2-HA-YFP or GalT2RR-AA-HA-YFP or VSV-G-YFP. After washing, bound proteins were analyzed by Western blot with anti-GFP antibody. It is clear from the experiment that the binding of GalT2 to Sar1D198A was reduced in comparison with the binding to Sar1 (Fig. 2) and that these bindings depended on the presence of the (R/K)X(R/K) motif in their CTs, because it was abrogated by RR-AA substitutions. To ascertain whether the effect of site A mutations in Sar1 was specific for the binding of the CT of glycosyltransferases, determination of the binding of the VSV-G protein to Sar1 (14) was also included in the experiment of Fig. 2. Importantly, the effect of the mutation in Sar1 was specific for GalT2 because the capacity of Sar1D198A to bind VSV-G was essentially unaltered with respect to Sar1. These results are in agreement with the prediction that site A in Sar1 is specifically involved in the interaction with the CT of GalT2.
FIGURE 2.
Sar1D198A shows reduced binding of GalT2 but not of VSV-G. Lysates of cells expressing GalT2-HA-YFP, GalT2RR-AA-HA-YFP, or VSV-G-YFP were incubated with His6-Sar1 (WT or D198A) bound to Ni2+-charged Sepharose beads. Bound proteins were analyzed by SDS/PAGE and Western blotted with an anti-GFP antibody.
Mutant Versions of Sar1 Affect ER-Golgi Distribution of GalT2 but Not of GM130
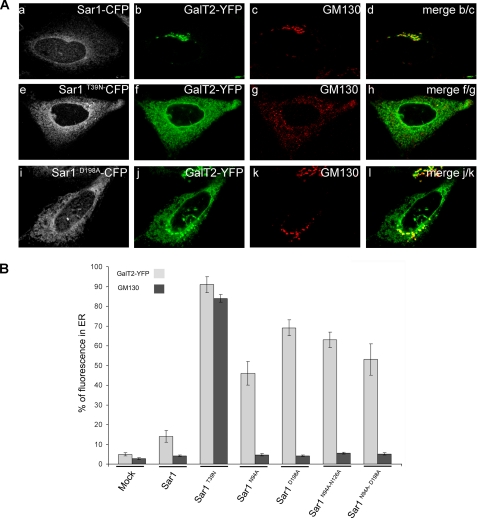
The substitution of two arginines by two alanines in the CT of GalT2 affect its binding to Sar1 and leads to changes in the subcellular localization of GalT2, from its typical concentration in the Golgi complex to a pattern of distribution along ER membranes (16). Alanine replacements of amino acids in the Sar1 region predicted to be involved in the Sar1-GalT2CT interaction (site A) should affect the subcellular localization of GalT2 in a similar manner. Sar1 and each of its mutants fused to CFP were co-expressed in CHO-K1 cells with the N-terminal domain of GalT2 fused to YFP. Cells expressing GalT2 and Sar1 showed Sar1 distributed all along the perinuclear contour and in ER membranes (Fig. 3A, panel a). GalT2, on the other hand, concentrates in a juxtanuclear structure defined as the Golgi complex by its abundant co-localization with the immunostained golgin GM130 (Fig. 3A, panels b–d). The dominant negative (GDP restricted) Sar1T39N displays a pattern of distribution similar to Sar1 but produced noticeable changes in the distribution of both GalT2 and GM130 (Fig. 3A, panels e–h). GalT2 redistribution followed an ER-like pattern, whereas GM130 acquired a punctuated pattern along the cytoplasm. The effect of Sar1T39N was expected, because it was reported that microinjection of Sar1T39N led to accumulation in the ER of Golgi resident proteins that retrotranslocate to the ER but cannot exit during the ER-Golgi cycling process (16, 23–25). The Sar1D198A mutant promoted a selective redistribution of a fraction of GalT2 to the cytoplasm (Fig. 3A, panels i–l) that co-localizes with the ER marker BiP (see supplemental Fig. S1). In comparison with Sar1T39N, Sar1D198A was less effective in relocating GalT2, because a fraction remained in the Golgi co-localizing with GM130. Importantly, Sar1D198A affects the subcellular distribution of GalT2 with high specificity and did not cause major changes in the subcellular localization of GM130. The localization of other proteins that, similarly to GM130, are not expected to interact with site A in Sar1 for exiting the Golgi, like Bet1, a SNARE that is included in COPII vesicles via interaction with Sec24, is not affected by Sar1D198A (see supplemental Fig. S2). The quantification of the effects described in Fig. 3 (panels a–l) and also of those of other Sar1 mutants examined in the same conditions is shown in Fig. 3B. Less than 10% of GalT2 and GM130 reside out of the Golgi in mock transfected cells, and this value was only slightly increased to ∼15% by co-transfection with Sar1. In contrast, co-transfection with different Sar1 mutants raised GalT2 values to 45–70% without changes in the GM130 values. Sar1D198A and Sar1N94A/N126A seem to be more effective than Sar1N94A and Sar1N94/D198A, but the differences were not statistically significant. Only Sar1T39N affects the ER/Golgi ratio of GM130. These results suggest that site A in Sar1, as identified by our docking analysis, is relevant for selectively recognizing the CT of GalT2 in its ER to Golgi transport process.
FIGURE 3.
Sar1D198A expression specifically redistributes GalT2 to the ER. A, top row, cells co-expressing Sar1-CFP (panel a, white) and GalT2-YFP (panel b, green) were immunostained for the Golgi marker GM130 (panel c, red); panel d is the merge of panels b and c. Middle row, cells co-transfected with Sar1T39N-CFP (panel e, white) and GalT2-YFP (panel f, green) were immunostained for the Golgi marker GM130 (panel g, red); panel h is the merge of panels f and g. Bottom row, cells co-expressing Sar1D198A-CFP (panel i, white) and GalT2-YFP (panel j, green) were immunostained for the Golgi marker GM130 (panel k, red); panel l is the merge of panels j and k. B, quantification of GalT2 in the ER. The fluorescence intensity of GalT2 (light gray) and of GM130 (black) in cells expressing GalT2 alone (mock) or co-expressing GalT2 with either Sar1 alone, Sar1T39N, or Sar1 with different alanine substitutions in site A as indicated on the abscissa was quantified with Metamorph™. For details see “Experimental Procedures.” The values express the percentages ± S.E. of the total fluorescence of GalT2 (n = 30) and of immunolabeled GM130 (n = 20) in ER membranes.
Mutant Versions of Sar1 Also Affect the Localization of GalNAcT and SialT2
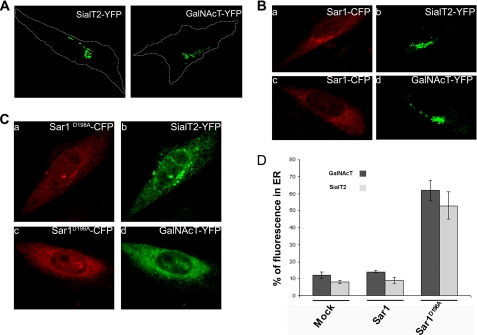
The basic motif (R/K)X(R/K) has been identified in the CT of many Golgi resident glycosyltransferases, including GalNAcT and SialT2 (16). This motif has also been shown to bind Sar1 and to have an essential role in ER-Golgi transport of proteins that present it on their CT (16–18). We investigated whether the expression of Sar1D198A also affects the subcellular localization of GalNAcT-YFP and SialT2-YFP as observed with GalT2. Both glycosyltransferases localize at the Golgi complex when they are expressed alone in CHO-K1 cells (Fig. 4A), and this localization does not change when they are co-expressed with Sar1 (Fig. 4B, panels a–d). However, when co-expressed with Sar1D198A, both SialT2 (Fig. 4C, panels a and b) and GalNAcT (Fig. 4C, panels c and d) present a broad redistribution to ER-like structures. Quantification of the fluorescence of SialT2 and GalNAcT in the ER of cells expressing Sar1D198A (Fig. 4D) gave redistribution values of ∼50%, slightly lower than those promoted by Sar1D198A on GalT2 (Fig. 3B). Similar results were obtained with Sar1N94A, Sar1N94A/N126A, and Sar1N94A/D198A (not shown). These results add evidence to the participation of site A of Sar1 in the ER-Golgi trafficking of glycolipid glycosyltransferases.
FIGURE 4.
Sar1D198A-CFP relocalizes SialT2 and GalNAcT to the ER. A, cells transfected with SialT2-YFP or GalNAcT-YFP show both constructs in a typical Golgi localization; the dashed line marks the cell boundaries. B, co-transfection with Sar1-CFP (panels a and c, red) does not affect either SialT2-YFP (panel b, green) or GalNAcT-YFP (panel d, green) localization. C, co-transfection with Sar1D198A-CFP (panels a and c, red) causes SialT2-YFP (panel b, green) and GalNAcT-YFP (panel d, green) redistribution to ER-like structures. D, quantification of SialT2 and GalNAcT fluorescence in the ER when expressed alone or when co-expressed with Sar1 or with Sar1D198A, as indicated. The percentage of total cell fluorescence in the ER was quantified with Metamorph™ as described under “Experimental Procedures.” The values are the means ± S.E. for n = 20.
GalT2 Fails to Concentrate in ER Exit Sites (ERES) When Co-expressed with Sar1D198A
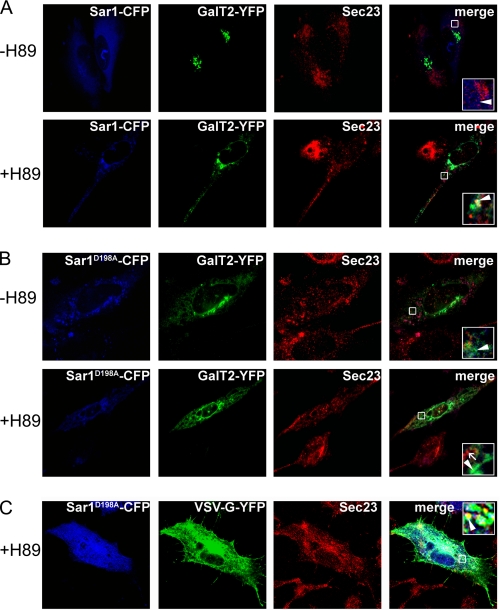
H89 is a serine/threonine kinase inhibitor that abolishes ER to Golgi transport, leading to accumulation of cargo in ER membranes (1, 26). Because Sar1D198A fails to bind the CT of GalT2, in cells expressing Sar1D198A in the presence of H89, the inclusion of GalT2 into the accumulated proteins at presumably ERES, as well as its co-localization with other components of the COPII ternary complex, should be less marked than in cells expressing Sar1. As shown in Fig. 5A, in the presence of Sar1 and the absence of H89 (−H89), cells show GalT2 with the typical localization in the Golgi complex and the endogenous Sec23 in a punctate pattern of cytoplasmic structures, characteristic of ERES; no GalT2 was observed at these sites (Fig. 5A, arrowhead in inset). In the presence of H89 (+H89) Sec23 acquires a more intense punctate pattern, and a fraction of GalT2 appeared in the cytoplasm that partially co-localized with Sec23. These ERES-like structures also include Sar1, showed by the co-localization of the three proteins observed at higher magnification (Fig. 5A, arrowhead in inset). In cells co-expressing Sar1D198A, partial co-localization between Sec23 and Sar1D198A was still observed, both in the absence (−H89) and in the presence (+H89) of H89 (Fig. 5B, arrowhead in inset). However, in no case did GalT2 (arrow in inset) co-localize with Sec23; rather it appeared spread along ER-like membranous structures without any obvious concentration in ERES. In summary, the above results indicate that the inclusion of GalT2 in ERES depends on its interaction with Sar1 and that the integrity of site A is necessary for this selective inclusion at ERES.
FIGURE 5.
Sar1D198A selectively excludes GalT2 from COPII vesicles. Cells co-expressing Sar1 (A) or Sar1D198A (B) (blue) and GalT2 (A and B) or VSV-G (C) (green) were cultured in the presence or absence of the serine/threonine kinase inhibitor H89, as indicated at left, and immunostained for Sec23 (red). The fourth column is the blue-green-red merging, with insets corresponding to higher magnifications of the boxed areas. A, cells co-expressing GalT2 and Sar1-CFP. The arrowhead in each inset points to GalT2 not co-localizing with Sec23 in the −H89 condition and co-localizing with Sec23 (and with Sar1-CFP) in the +H89 condition. B, cells co-expressing Sar1D198A-CFP and GalT2. The arrowhead in each inset marks the lack of co-localization of GalT2 (arrowhead) and Sec23 (arrow) both in the −H89 and the +H89 condition. Note the co-localization of Sar1D198A-CFP and Sec23 in the +H89 condition. C, cells co-expressing Sar1D198A-CFP and VSV-G-YFP. The arrowhead points to VSV-G co-localizing with Sec23 and Sar1D198A-CFP in the +H89 condition.
Mutant Versions of Sar1 Do Not Affect VSV-G Traffic
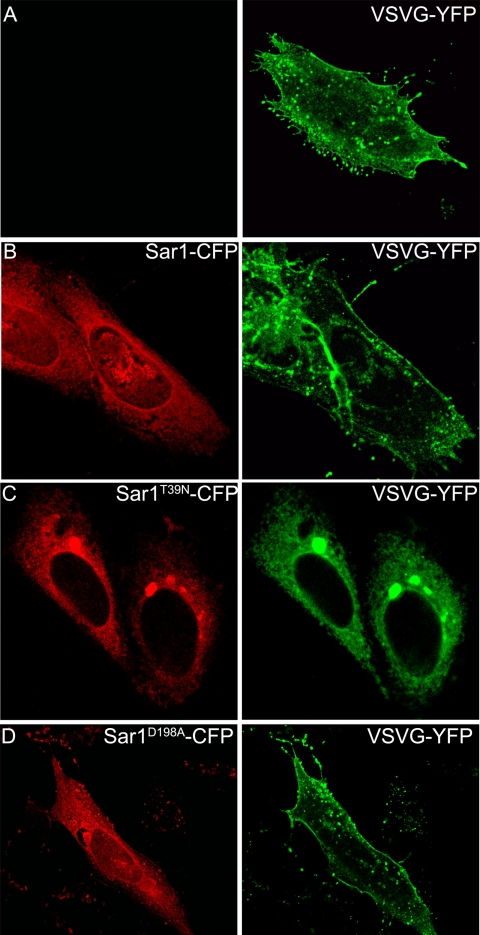
To investigate the functionality of the ERES of Sar1D198A-expressing cells for the concentration of other proteins not related to glycosyltransferases, VSV-G ts045 was co-expressed with Sar1D198A (Fig. 5C). In line with the results of Fig. 2 showing that the binding of VSV-G to Sar1 in vitro was not affected by the mutation in site A, in cells cultured in the presence of the inhibitor H89, VSV-G was found in ERES, as shown by the co-localization with Sec23. This result indicates that Sar1D198A is able to concentrate other proteins at ERES. Moreover, Sar1 mutants did not affect the ER exiting of VSV-G, because 18 h after transfection and when cells were incubated at the permissive temperature, VSV-G was found localized to the plasma membrane and to some extent to internal membranes in mock transfected cells (Fig. 6A) or in cells expressing either Sar1 (Fig. 6B) or Sar1D198A (Fig. 6D). As a control, co-transfection with Sar1T39N (GDP restricted) resulted in complete impairment of VSV-G from exiting the ER (Fig. 6C). Thus the results of experiments of Figs. 5 and 6 confirm that the impediment of Sar1 mutants in concentrating glycosyltransferases into ERES, and indirectly their transport to the Golgi, is a specific rather than a general effect on COPII cargo selection.
FIGURE 6.
Sar1D198A does not affect VSV-G traffic. CHO cells were transfected with VSV-G-YFP (A, green) or co-transfected with VSV-G-YFP (green) and Sar1-CFP (B, red) or with Sar1GDPT39N-CFP (GDP restricted) (C, red) or with Sar1D198A (D, red).
DISCUSSION
ER to Golgi transport of mammalian (16, 17) and plant (18, 19) glycosylating enzymes with type II membrane topology rely on the interaction of the basic amino acid motif (R/K)X(R/K) in the CT close to the transmembrane domain with Sar1. On the other hand, the ER to Golgi transport of type I membrane proteins is a Sec24-dependent process. Crystallographic analysis evidenced two independent binding sites in Sec24 for the ER/Golgi SNARE proteins Sed5 and Bet1p. Binding occurs via peptide sequences 202YNNSNPF208 and 237QLMLMEGQ245 in Sed5 or LXXLE in the context 46YSQSTLASLESSQ57 in Bet1p, which act as interacting signals with Sec24; an additional cargo binding site for the SNARE Sec22 was also identified in these studies (27). In vitro budding and cargo binding assays using Sec24 with alanine replacement in the B-site binding pocket identified specific amino acids in Sec24 that, although they do not interfere with COPII assembly, interfere with some (Bet1 and other ER/Golgi SNAREs) but not all (α-factor) cargo packaging (28). It was estimated that cargo binding sites in Sec24 reside ∼20 Å from the bilayer in membrane-bound COPII. A protein needs a CT with at least 20–25 amino acids (7–10 amino acids to span the distance and 10–15 amino acids to fit the binding sites) to reach to the Sec24 binding sites (27).
Because the CTs of glycosyltransferases are in general shorter than the 20 amino acids needed to reach binding sites in Sec24, additional cargo binding sites may exist in more than one element of the Sec23-Sec24-Sar1 COPII complex. We hypothesize that proteins with short CTs may interact directly with Sar1 and that those with longer CTs interact either directly or in a Sar1-dependent form (14) with other members of the ternary complex like Sec24. Computational docking delimited a region in the Sar1 molecule of best interaction possibilities with short peptides with the sequence of the CT of GalT2 (site A). Mutants of Sar1 with alanine replacements of amino acids in site A were generated, and their effects were tested in vitro and in cultured cells. In vitro, Sar1D198A showed a reduced ability to bind GalT2 while maintaining intact its ability to bind VSV-G (Fig. 2). In ex vivo experiments, when co-expressed in cells with the N-terminal domain of glycosyltransferases, Sar1 mutants produced a new steady state ER/Golgi distribution of the constructs that favors the ER compartment (Fig. 3). This effect was not observed for other Golgi residents, like the golgin GM130, which remains concentrated in the Golgi, essentially as in cells that did not receive the mutant Sar1 plasmid. The Sar1 mutants did not affect the traffic of other transmembrane cargo proteins that use the COPII/ER exiting mechanism, like the G protein of VSV, which concentrates at ERES (Fig. 5), exits the ER, and follows the route to the plasma membrane as in control cells (Fig. 6). It should be mentioned that Sar1 binds VSV-G and initiates cargo selection by recruiting it to prebudding complexes in a DXE sorting signal independent form and without participation of COPII components; the DXE sorting signal at the C terminus would became effective for ER exiting upon interaction with GTP-activated Sar1 and COPII components (14). The results of Figs. 2, 5, and 6 indicate that the COPII complex containing Sar1 with alanine substitutions in the glycosyltransferases CT binding site A is functional for other proteins but inefficient for packaging glycosyltransferases that use the (R/K)X(R/K) motif for interaction with Sar1 at the ER and, as a consequence, inefficient for their subsequent transport to the Golgi complex.
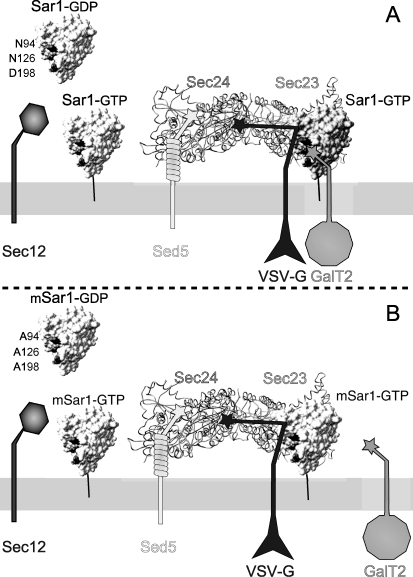
The fraction of glycosyltransferases localized to ER structures when co-expressed with Sar1D198A showed a reticular pattern, different from the punctate pattern shown by Sec23, characteristic of ERES. This result is indicative that Sar1 mutants were able to concentrate Sec23 but not GTs at those sites. Moreover, Sar1D198A-containing ERES were able to concentrate VSV-G, but GalT2 was selectively excluded from these sites. As indicated in Fig. 7, we hypothesize that the overexpressed mutant versions of Sar1 substitute the endogenous, normal version of Sar1 in the ternary complex Sec23-Sec24-Sar1. In doing so, the COPII complex would be completely functional for concentrating cargoes with long CTs but would fail to concentrate those cargoes with shorter CTs that use a direct interaction with Sar1 for loading, as is the case with glycosyltransferases and perhaps other type II membrane proteins with short CTs.
FIGURE 7.
Schematic representation of recruitment of membrane cargos by the ternary prebudding complex Sar1-Sec23-Sec24 (adapted from Ref. 15). A, cytosolic Sar1 converted to membrane-bound Sar1 by the GEF activity of Sec12 recruits the Sec23-Sec24 heterodimer. The external layer (not shown in the scheme) is completed with the binding of the heterotetrameric complex Sec13-Sec31 (15). Cargo proteins are recruited as the prebudding complex is being formed. Membrane proteins with long CTs, like Bet1 or Sed5 (27, 28), bind Sec24 through their respective LXXLE and YNNSNPF motifs (stars); VSV-G also binds Sec 24 through the DXE signal in a Sar1-dependent manner (14, 32), whereas membrane cargos with short CTs, like GalT2 and other glycosyltransferases, are recruited by direct interaction with Sar1 through the (R/K)X(R/K) motif. B, the prebudding complex formed with Sar1 with amino acids in site A replaced by alanines (mSar1) is still able to recruit VSV-G but has a reduced ability to bind the CT of GalT2 and consequently to load it in COPII vesicles, resulting in defective ER exit and in a steady state balance of ER-Golgi distribution favoring the ER.
Supplementary Material
Acknowledgments
We thank Javier Valdez-Taubas, Rodrigo Quiroga, and Mariana Ferrari for helpful discussions. The excellent technical assistance of Susana Deza and Gabriela Schachner with cell cultures and of Cecilia Sampedro and Carlos Mas for assistance with confocal microscopy is also acknowledged.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica del Ministerio de Ciencia, Tecnología e Innovación Productiva de la Nación Argentina, from the Secretaría de Ciencia y Tecnología de la Universidad Nacional de Córdoba, and from the Ministerio de Ciencia y Tecnología de la Provincia de Córdoba, Argentina. H. J. F. M., M. V., and G. M. are career members, and CQ fellow, of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) of Argentina.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- CT
- cytoplasmic tail
- GalT2
- GalT21–57-CFP
- GalNAcT
- GalNAcT1–27-CFP
- SialT2
- SialT21–57-CFP
- VSV-G
- vesicular stomatitis virus G protein ts045 fused to YFP
- CFP
- enhanced cyan fluorescent protein
- ERES
- ER exit site(s).
REFERENCES
- 1.Aridor M., Balch W. E. (2000) J. Biol. Chem. 275, 35673–35676 [DOI] [PubMed] [Google Scholar]
- 2.Barlowe C., Schekman R. (1993) Nature 365, 347–349 [DOI] [PubMed] [Google Scholar]
- 3.Bi X., Corpina R. A., Goldberg J. (2002) Nature 419, 271–277 [DOI] [PubMed] [Google Scholar]
- 4.Huang M., Weissman J. T., Beraud-Dufour S., Luan P., Wang C., Chen W., Aridor M., Wilson I. A., Balch W. E. (2001) J. Cell Biol. 155, 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aridor M., Weissman J., Bannykh S., Nuoffer C., Balch W. E. (1998) J. Cell Biol. 141, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balch W. E., McCaffery J. M., Plutner H., Farquhar M. G. (1994) Cell 76, 841–852 [DOI] [PubMed] [Google Scholar]
- 7.Kuehn M. J., Herrmann J. M., Schekman R. (1998) Nature 391, 187–190 [DOI] [PubMed] [Google Scholar]
- 8.Springer S., Schekman R. (1998) Science 281, 698–700 [DOI] [PubMed] [Google Scholar]
- 9.Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 10.Lederkremer G. Z., Cheng Y., Petre B. M., Vogan E., Springer S., Schekman R., Walz T., Kirchhausen T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10704–10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salama N. R., Chuang J. S., Schekman R. W. (1997) Mol. Biol. Cell 8, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonny B., Schekman R. (2001) Curr. Opin. Cell Biol. 13, 438–443 [DOI] [PubMed] [Google Scholar]
- 13.Bannykh S. I., Nishimura N., Balch W. E. (1998) Trends Cell Biol. 8, 21–25 [DOI] [PubMed] [Google Scholar]
- 14.Aridor M., Fish K. N., Bannykh S., Weissman J., Roberts T. H., Lippincott-Schwartz J., Balch W. E. (2001) J. Cell Biol. 152, 213–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonifacino J. S., Glick B. S. (2004) Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 16.Giraudo C. G., Maccioni H. J. (2003) Mol. Biol. Cell 14, 3753–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y., Linstedt A. D. (2006) J. Cell Biol. 174, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005) Plant J. 41, 81–94 [DOI] [PubMed] [Google Scholar]
- 19.Schoberer J., Vavra U., Stadlmann J., Hawes C., Mach L., Steinkellner H., Strasser R. (2009) Traffic 10, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 21.Lavigne P., Bagu J. R., Boyko R., Willard L., Holmes C. F., Sykes B. D. (2000) Protein Sci. 9, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo F., Feytmans E. (1998) J. Mol. Biol. 277, 1141–1152 [DOI] [PubMed] [Google Scholar]
- 23.Seemann J., Jokitalo E., Pypaert M., Warren G. (2000) Nature 407, 1022–1026 [DOI] [PubMed] [Google Scholar]
- 24.Storrie B., Pepperkok R., Nilsson T. (2000) Trends Cell Biol. 10, 385–391 [DOI] [PubMed] [Google Scholar]
- 25.Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K., Lippincott-Schwartz J. (2001) J. Cell Biol. 155, 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T. H., Linstedt A. D. (2000) Mol. Biol. Cell 11, 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossessova E., Bickford L. C., Goldberg J. (2003) Cell 114, 483–495 [DOI] [PubMed] [Google Scholar]
- 28.Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. (2003) Cell 114, 497–509 [DOI] [PubMed] [Google Scholar]
- 29.Xiang Z., Soto C. S., Honig B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraudo C. G., Daniotti J. L., Maccioni H. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller P., Toomre D., Díaz E., White J., Simons K. (2001) Nat. Cell Biol. 3, 140–149 [DOI] [PubMed] [Google Scholar]
- 32.Nishimura N., Balch W. E. (1997) Science 277, 556–558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.