Abstract
Copines are highly conserved proteins with lipid-binding activities found in animals, plants, and protists. They contain two calcium-dependent phospholipid binding C2 domains at the amino terminus and a VWA domain at the carboxyl terminus. The biological roles of most copines are not understood and the biochemical properties required for their functions are largely unknown. The Arabidopsis copine gene BON1/CPN1 is a negative regulator of cell death and defense responses. Here we probed the potential biochemical activities of BON1 through mutagenic studies. We found that mutations of aspartates in the C2 domains did not alter plasma membrane localization but compromised BON1 activity. Mutation at putative myristoylation residue glycine 2 altered plasma membrane localization of BON1 and rendered BON1 inactive. Mass spectrometry analysis of BON1 further suggests that the N-peptide of BON1 is modified. Furthermore, mutations that affect the interaction between BON1 and its functional partner BAP1 abolished BON1 function. This analysis reveals an unanticipated regulation of copine protein localization and function by calcium and lipid modification and suggests an important role in protein-protein interaction for the VWA domain of copines.
Keywords: Arabidopsis, Calcium, Calcium/Binding Proteins, Membrane, Protein/Myristoylation, Signal Transduction, Signal Transduction/Calcium, Signal Transduction/Protein Kinases, BON1, Copine
Introduction
Copines are evolutionarily conserved proteins present in protozoa, plants, nematodes, and mammals (1). They were first identified in preparations of Ca2+-dependent, phospholipid-binding proteins in Paramecium (1). Genome information has revealed their existence in many organisms, and there is often more than one member in each organism. Their ubiquitous presence in different organisms and their high sequence conservation suggest important roles of these proteins in conserved cellular processes. The biological roles of several copine genes are starting to be revealed. The BON1/CPN1 gene from Arabidopsis thaliana is the first copine gene being genetically characterized (2, 3). The loss of BON1 function leads to de-repression of defense responses and consequently a reduction in plant growth (4). There are three copine genes (BON1, BON2, and BON3) in Arabidopsis, and they have overlapping functions in repressing programmed cell death and defense responses (5). In Dictyostelium, one copine gene CPNA is required for cytokinesis, contractile vacuole function, and development (6, 7). In human embryonic kidney 293 (HEK293) cells, a dominant-negative copine construct interferes with signaling from the tumor necrosis factor-α (TNF-α) receptor by repressing NF-κB transcription (8). In Caenorhabditis elegans, one copine protein is associated with nicotinic acetylcholine receptors (9) and another regulates the activity of a channel protein TRPM (10). All copine genes studied so far appear to have signaling and regulatory roles although no common processes can be readily seen.
Copine proteins have distinct domains with well studied structure homology although the biochemical activities utilized for their biological roles are not well studied. The copine proteins are characterized by two C2 domains at the amino terminus and a von Willebrand A domain (VWA)4 at the C terminus. The C2 domain, named after the second conserved domain in protein kinase C (PKC), comprises a characteristic 8-stranded antiparallel β-sandwich of about 130 amino acids, with 3 key inter-strand loops that are responsible for binding both Ca2+ and membranes. Based on this homology, the C2 domain was suggested to contain the calcium switch required for localization to the membrane (11). BON1, BON1 association protein 1 (BAP1), and many other C2 domain containing proteins bind to phospholipids in a calcium-dependent manner (1, 2) and this lipid binding property is thought to contribute to membrane localization. Although many C2 proteins are associated with membranes, some may not localize to membranes by themselves. For instance, activated PKC isozymes are each localized to unique intracellular sites and this localization is mediated by their selective interactions with specific anchoring proteins (12).
The VWA domain consists of alternating α-helices and β-strands that adopt a classic α/β Rossmann-fold (13). Similar to many VWA-containing proteins, copines contain a metal ion-dependent adhesion site, which is critical for manganese and magnesium binding (1, 14). This domain is likely involved in protein-protein interactions. BAP1, a functional partner of BON1, interacts with the VWA domain of BON1 (2, 15). A number of proteins interacting with the human copine VWA domains have been isolated through the yeast two-hybrid screen and they are mostly signaling molecules often with coiled-coil domains (16). Sequence alignment has suggested homology of the VWA domain to protein kinases. Conserved motifs for kinases are preserved in copines and the VWA domain of human copine III is suggested to possess an intrinsic protein kinase activity (17). It is yet to be determined if the copines do indeed function as kinases.
In addition, a potential myristoylation site was predicted for the Arabidopsis BON1 protein. Myristoylation is the linkage of myristic acid by an amide bond to a N-terminal amino acid residue of the protein, and this post-translational lipid modification confers the protein a tendency to associate with membranes (18). Myristoylation is found to target some plant proteins to membranes. For instance, a hydrophilic cation-binding protein AtPCaP1, and a calcineurin B-like (CBL1) could localize to the plasma membrane through this lipid modification (15, 19, 20). The role of this potential modification in BON1 is unknown.
To further understand how copine proteins function, we carried out mutagenesis on the Arabidopsis BON1 protein to investigate the biochemical properties required for its biological functions. In this study we examined three potential biochemical activities of BON1: calcium binding, myristoylation, and kinase activity. Residues potentially essential for these activities were mutated and the function of these mutant proteins were analyzed in the bon1-1 (referred as bon1) mutant with a dwarf phenotype. This study reveals unanticipated findings on the function of these domains/motifs and has implications in understanding the regulation and activities of copine proteins in general.
EXPERIMENTAL PROCEDURES
Plant Growth Conditions
Arabidopsis plants were grown at 22 °C under continuous fluorescent light (100 μmol m−2 s−1) and 50–70% relative humidity. Arabidopsis seeds were either directly sown on soil or grown on Petri dishes containing half-strength Murashige-Skoog medium (21) (Sigma) with 2% sucrose and 0.7% agar.
Plasmid Construction and Plant Transformation
A shorter version of the previously described pBON1::BON1:HA (2) was utilized for mutagenesis. This is a 5.5-kb PstI-BamHI fragment of the previous genomic fragment of BON1 with three copies of the HA epitope tag (sequence: YPYDVPDYA). It contains 2.2 kb of the BON1 native promoter and the full-length genomic coding region. Site-directed mutagenesis was carried out with the QuikChange® II XL Site-directed Mutagenesis Kit (Stratagene). A total of 13 constructs were made. Two complementary primers were used as instructed by the manufacturer and the forward primers are shown as follows (from 5′ to 3′): D63A-1, CGAGACCGCGCCGTGCTCTCC; D69A-1, TTGCAGAGTGCTCCTATGGTTG; D122A-1, CGTGTGTATGCTGTTGACACC; D122AD124A-1, CGTGTGTATGCTGTTGCCACCAAATTTC; D209A-1: AATCTAAGGCTCTTTTTTCA; D215A-1, GCAACTTTAGGCCCCCTTTTTGG; D269A-1, GAATGCTCAGCCTTTAACTCC; A350VG353A-1, GATTTCACAGTTTCAAATGTAAATCCCCGC; K391A-1, TGACTCAGACGCACGTTTCCCT; G2A-1, CAAAAATTATGGCGAATTGTTGC; LinkerK-1, GTTGCAGTTTTATAACGCAGCCATACGTTCCCCTGCCTGGG.
Sequential mutagenesis was carried out to introduce multiple mutations. The mutagenized BON1 and wild-type control were subcloned into a binary vector as described previously (2).
For yeast two-hybrid analysis, the BON1/pGAD construct described previously (2) was used as a template for mutagenesis to generate A350V/G353A and LinkerK mutations. For BON1:HA purification, the BON1:HA fusion gene (2) was expressed under a 35S promoter in the pGreen0229 vector.
For protoplast analysis, a construct p35S::BON1:GFP was generated using a binary vector as previously described (22). Both wild-type and mutant constructs were transformed into Col-0 protoplasts according to a protocol previously described (23). GFP signals were visualized under a confocal microscope (Leica SP2). Agrobacterium tumefaciens strains GV3101 (24) carrying different constructs were used to transform the wild-type or the bon1 mutant plants via floral dip transformation (25).
Protein Extraction and Detection
Membrane fractionation and immunoprecipitation (IP) were carried out as described previously (2) with minor modifications. Total proteins were spun at 60,000 × g with a TLA100 rotor for 20 min for membrane fractionation. IP was carried out by incubating the total protein extracts or fractions with anti-HA antibody (COVANCE, Emeryville, CA) followed by incubating with Protein G-agarose beads. The anti-HA antibody was used to detect BON1-HA. Gel-based liquid chromatography-mass spectrometry-mass spectrometry (GeLC-MS-MS) is used to detect proteins on gel slices.
For differential solubilization assay, total proteins were centrifuged at 10,000 × g and the supernatants were spun at 100,000 × g. The resulting microsomal pellet was solubilized with 0.5% sodium deoxycholate for 30 min at 4 °C followed by centrifugation at 100,000 × g. The supernatant is the 0.5% soluble fraction. The pellet was further solubilized with 1.0% sodium deoxycholate and centrifuged at 100,000 × g. This supernatant is the 1.0% soluble fraction and the pellet is the insoluble fraction.
Yeast Two-hybrid Analysis
The BON1/pGAD and BAP1/pGBD plasmids and the vector controls were transformed into the yeast pJG69-4a strain (26) as a pair. Double transformants were grown on synthetic complete (SC) medium without tryptophan and leucine (SC-Leu-Trp) plates to select both plasmids as described (14). Single colonies were inoculated into SC-Leu-Trp liquid medium for overnight growth. Cells were resuspended in SC-Leu-Trp liquid medium at A600 = 1 and an equal amount of serial dilutions were spotted on both SC-Leu-Trp and SC medium without leucine, tryptophan, histidine, and adenine (SC-Leu-Trp-His-Ade) plates. Plates were left at 28 °C for 3 and 5 days for growth analysis.
RESULTS
Mutations in Calcium-binding Aspartate Residues Compromised BON1 Function
The C2 domains of a few copine proteins including BON1 can promote lipid vesicle aggregation in a calcium-dependent manner, suggesting their calcium- and lipid-binding activities (1, 2). Conserved aspartates found in the C2 domain are thought to be essential for calcium binding (11, 27). To determine whether these calcium- and lipid-binding activities of copines are important for their biological roles, we mutated the conserved aspartate residues in BON1 and asked if they would affect the biological functions of BON1. In BON1, these aspartates are Asp63, Asp69, Asp122, and Asp124 in the C2A domain, and Asp209, Asp215, and Asp269 in the C2B domain. They were changed into alanines singly or in combination in an HA-tagged BON1 construct driven by its native promoter (Fig. 1). Both wild-type and mutant BON1 genes were transformed into bon1 mutants and growth phenotypes of the transgenic plants were analyzed at the T2 stage.
FIGURE 1.
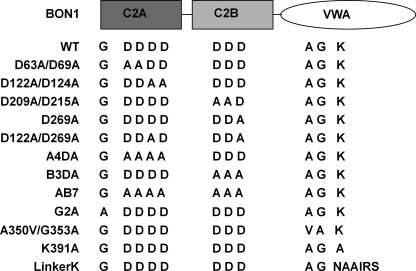
Diagram of the wild-type (WT) and mutant BON1 proteins. For WT, the first G represents glycine 2. The first four Ds are the four conserved aspartates 63, 69, 122, and 124 in the C2A domain. The following three Ds are the three conserved aspartates 209, 215, and 269 in the C2B domain. In the VWA domain, A, G, and K represent Ala350, Gly353, and Lys391, respectively. In the LinkerK construct, six amino acids (DSDKRF) around Lys391 were mutated into NAAIRS.
All of the 52 bon1 lines with the wild-type BON1 transgene showed rescued (wild-type) phenotype (Table 1, Fig. 2A), indicating that this pBON1::BON1:HA is a functional fusion. Mutant BON1 genes with either single or double aspartate mutations showed good rescue of the bon1 mutant phenotype. Fourteen of 16 lines of BON1(D63A/D69A) with the first two conserved aspartates in C2A mutated had the wild-type phenotype. All 26 lines of BON1 (D122A/D124A) where the last two aspartates in C2A were mutated exhibited wild-type phenotype. Twenty-nine of 31 lines of BON1 (D122A/D269A) had wild-type growth phenotype. All of the 30 lines with BON1(D209A/D215A) and 106 of 109 lines with BON1(D269A) showed fully rescued growth phenotype.
TABLE 1.
Summary of growth phenotype of transgenic plants in bon1
The wild-type constructed pBON1::BON1:HA was mutagenized to generate specific mutations described in the legend to Fig. 1. All constructs were transformed into bon1 mutants, and T2 transgenic plants grown at 22 °C for 3 weeks were analyzed for growth phenotype.
| BON1 transgenic | Total | Rescued | Not rescued |
|---|---|---|---|
| WT | 52 | 52 | 0 |
| D63A/D69A | 16 | 14 | 2 |
| D122A/D124A | 26 | 26 | 0 |
| D122A/D269A | 31 | 29 | 2 |
| D209A/D215A | 30 | 30 | 0 |
| D269A | 109 | 106 | 3 |
| A4DA | 16 | 0 | 16 |
| B3DA | 35 | 3 | 32 |
| AB7 | 8 | 0 | 8 |
| G2A | 16 | 0 | 16 |
| A350V/G353A | 32 | 0 | 32 |
| K391A | 22 | 22 | 0 |
| LinkerK | 15 | 0 | 15 |
FIGURE 2.
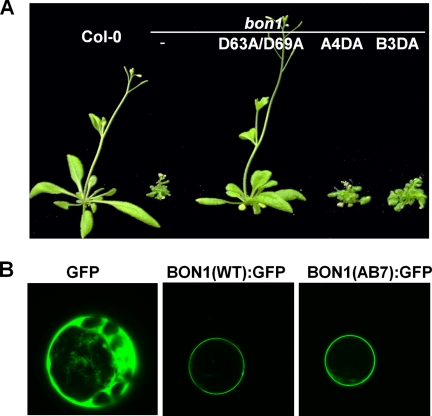
Characterization of BON1 proteins mutated at the designated aspartates. A, growth phenotype of transgenic bon1 plants grown at 22 °C. Shown is the rescued phenotype displayed by a transgenic plant with the BON1(D63A/D69A) mutant gene and non-rescued phenotype displayed by two transgenic plants with BON1(A4DA) and BON1(B3DA). B, localization of wild-type and mutant BON1:GFP proteins in protoplasts. Constructs of p35S::BON1:GFP were transformed into Col-0 protoplasts and GFP signals were visualized with confocal microscopy. The GFP alone control is expressed ubiquitously, and the wild-type BON1 is localized to plasma membrane. The BON1(AB7) mutant protein is also found on the plasma membrane.
In contrast, mutating all conserved aspartates in either of the C2 domains abolished or dramatically reduced the BON1 activity. Of 16 transgenic lines with BON1(A4DA) containing alanine substitution at all four aspartates in C2A, all exhibited a bon1-like mutant phenotype. Of 35 transgenic lines with BON1(B3DA) containing an alanine substitution at all three aspartates in C2B, only 3 exhibited wild-type phenotype. For BON1(AB7) with all seven Asp to Ala substitutions in the two C2 domains, no line of 8 exhibited rescued phenotype (Fig. 2A and Table 1). Therefore these conserved aspartates play essential roles in BON1 function.
Mutations in Aspartate Residues do Not Alter the Subcellular Localization of BON1
We asked if these mutations in aspartates affect the function of BON1 by altering its subcellular localization. To this end, a BON1:GFP fusion was expressed under the strong 35S promoter in Arabidopsis protoplasts to assist protein localization. The wild-type BON1:GFP was localized to the plasma membrane, similar to the BON1 localization pattern observed in stable transgenic plants (2) (Fig. 2B). The AB7 mutation was introduced to this p35S::BON1:GFP construct and its localization was analyzed in Arabidopsis protoplasts. Surprisingly, BON1(AB7):GFP was found to be on the plasma membrane similarly to the wild-type BON1:GFP (Fig. 2B), indicating that these aspartates are not essential for subcellular localization of BON1.
Mutation in a Predicted Myristoylation Site of Glycine 2 Abolishes BON1 Function and Alters Its Localization
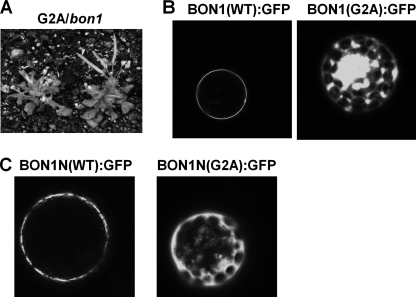
A putative myristoylation site was found at the second residue glycine in BON1. Because myristoylation has been shown to mediate membrane localization, we investigated if this is the case for BON1. A glycine to alanine mutation was introduced in pBON1::BON1:HA, and the construct was transformed into bon1. Of 16 BON1(G2A) lines generated, none of them could rescue the bon1 mutant phenotype (Table 1 and Fig. 3A), suggesting that N-terminal myristoylation of BON1 is required for its proper function.
FIGURE 3.
Characterization of BON1 proteins with a G2A mutation. A, growth phenotype of transgenic bon1 plants with the BON1(G2A) construct grown at 22 °C. The plants exhibited a bon1-like dwarf phenotype. B, localization of BON1(G2A):GFP in protoplasts. The p35S::BON1:GFP constructs were expressed transiently in Col-0 protoplasts. Although the wild-type protein is localized to the plasma membrane, the G2A mutant protein lost its specific plasma membrane localization. C, localization of BON1N:GFP and BON1N(G2A):GFP in protoplasts. BON1N is a truncated BON1 with the first 116 amino acids of BON1. The p35S::BON1N:GFP constructs were expressed transiently in Col-0 protoplasts. The wild-type truncated BON1 protein is localized to the plasma membrane, but the G2A version lost the specific plasma membrane localization.
The same G2A mutation was introduced into p35S:: BON1:GFP and expressed in Arabidopsis protoplasts to determine its localization. Although the wild-type BON1 is found exclusively at the plasma membrane as previously reported (2), the G2A mutant is found partially on the plasma membrane and mostly inside the plasma membrane (Fig. 3B). Thus, glycine 2 and likely its myristoylation are critical for both BON1 function and its plasma membrane localization.
The N-terminal Fragment of BON1 Is Sufficient for Plasma Membrane Localization
We further tested if the N-terminal part of BON1 is sufficient to confer plasma membrane localization. We fused the first 116 amino acids of the BON1 protein with GFP and transiently expressed the fusion in protoplasts. This BON1N:GFP fusion is specifically localized to the plasma membrane similarly to the full-length BON1:GFP (Fig. 3C), indicating that this short fragment of BON1 without the intact C2 domain is sufficient for targeting BON1 localization.
We mutated glycine 2 to alanine in BON1N:GFP to make BON1N(G2A):GFP. This mutated N-terminal BON1 lost its specific plasma membrane localization and was distributed among plasma membranes and some inner membranes (Fig. 3C), further supporting an important role of myristoylation in protein localization.
The N-terminal Peptide of BON1 Is Modified
We used mass spectrometry to determine whether the N-terminal of BON1 is indeed lipid modified. A BON1:HA fusion was expressed in bon1-1 under the strong 35S promoter of tobacco mosaic virus and it rescued the bon1-1 phenotype (data not shown). We purified the BON1:HA protein from this transgenic line by IP with antibodies for the HA epitope. IPed products were separated on SDS-PAGE gels and the BON1:HA protein band was sliced out and subject to in-gel digestion with chymotrypsin or trypsin. Peptides were then extracted from the gel and subjected to GeLC-MS/MS analysis. Using the Mascot program, BON1 peptides were detected in all BON1:HA samples, three digested with chymotrypsin and three digested with trypsin. The coverage of BON1 peptides identified were 21, 44, 45, 50, 32, and 37%, respectively, in these six samples.
Almost all regions of BON1 were identified when data from the six samples were combined (Fig. 4). Sequences not detected by mass spectrometry (except for the N-terminal peptides) were either too small or had too many charges that they are not predicted to be detected by the instruments used. The failure of identifying only the N-peptide of BON1 indicates that the N terminus of BON11 is modified.
FIGURE 4.
Mass spectrometry identification of BON1 peptides. Shown is the BON1 amino acid sequences. Six BON1:HA samples digested with chymotrypsin or trypsin were analyzed by mass spectrometry. Matched chymotrypsin peptides are shown in gray, and matched trypsin peptides are underlined. All other regions not matched by any peptides were predicted not to be detectable except for the N-terminal 34-amino acid peptide (chemotrypsin) and the 44-amino acid peptide (trypsin).
We further used a high concentration of organic solvent to extract peptides from the digested gel slices to prevent the loss of N-peptides due to their high hydrophobicity when modified. The extracted samples were analyzed on MALDI and SYNAP. Although we could extract and detect a control peptide that is synthetically myristoylated, no N-peptides of BON1 either non-modified or modified could be detected by these means. Therefore, the N-peptides of BON1 must be modified and there might be additional modifications other than myristoylation.
Myristoylation Might Be a Specific Feature for Plant Copines
We asked if myristoylation might be a general feature of copine proteins. To this end, we identified 124 proteins in the pfam data base with a canonical copine structure of two C2 domains and a VWA domain. These include proteins from various plant, animal, and protozoa species. No glycine is seen in the second residue in any of the non-plant copines. In contrast, Gly2 is found in all except for one plant copines. Using the NCBI blast, we pulled out 19 non-redundant plant copines with high similarity to the BON1 protein (Table 2) and all of them have glycine at the second residue. Interestingly, all except two were predicted to be myristoylated according to the program. BON1 is predicted with low confidence and BON3 is predicted with high confidence. Surprisingly, BON2 is not predicted to be myristoylated although it has been genetically shown to have a similar function to BON1.
TABLE 2.
Prediction of N-myristyolation in 19 plant copines
Ninteen proteins were identified as plant copines by querying the whole protein database with BON1 with the NCBI Blast program. Shown are their protein IDs, sequences of their N terminus, the species, and the predicted probability of N-myristoylation. The abbreviations used are: At: Arabidopsis thaliana; Vv: Vitis vinifera, Rc: Ricinus communis; Pt: Populus trichocarpa; Ps: Picea sitchensis; Mt: Medicago truncatula; Os: Oryza sativa; Zm: Zea mays; Sb: Sorghum bicolor. No: no modification predicted. Low, medium, and high: modification predicted with low, medium, and high confidence.
| Protein ID | Sequences | Species | Prediction |
|---|---|---|---|
| AAK98797 | MGNCCS- - -DVASGAGATAGVGGS | At (BON1) | Low |
| NP_568180 | MGSCWS- - -DGSYAGGGMVGVGGG | At (BON2) | No |
| XP_002280913 | MGNCCS- - -DDGGGRSAVGGTSAS | Vv | Low |
| XP_002511086 | MGNCCS- - -DEAGGRAAVGGTLAA | Rc | Low |
| XP_002318175 | MGNCFS- - -DVAGGRAAVGGSAAA | Pt | High |
| XP_002321753 | MGNCCS- - -DVAGGRAAVGGSGAA | Pt | Low |
| XP_002267420 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
| XP_002268132 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
| XP_002266991 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
| ABR16493 | MGCCFS- - -DIAGGQQAVGGTGVN | Ps | Medium |
| XP_002512304 | MGGCVSG- -DVKGGKQAIGGAQQR | Rc | Medium |
| ABN08161 | MGSCFS- - -DLKGAKQAVGGVNAQ | Mt | Medium |
| XP_002319577 | MGLCFS- - -DVRGGKQAIGGTSQQ | Pt | No |
| NP_172362 | MGGCLSG- -DVKGGKQAIGGVQQR | At (BON3) | High |
| NP_001046976 | MGNCCS- - -DEMGGGGGHAGRHSV | Os | High |
| CAO49680 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
| NP_001147038 | MGNCCS- - -DEVSHAGAHPVGSAT | Zm | High |
| XP_002439702 | MGGCFSLDGDVRGGMEAVGGGVTA | Sb | High |
| CAN68494 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
| CAN74634 | MGGCFS- - -DVRGGKQAVGVGLTG | Vv | Low |
Mutations in the VWA Domain Affected BON1 Function
The VWA domain of BON1 has a weak sequence homology to protein kinases and contains conserved kinase motifs including the glycine loop (Ala350–Gly353) for ATP binding and lysine site (Lys391) for catalytic activity. To determine whether the putative kinase activity is required for BON1 function, we introduced A350V/G353A mutations in the glycine loop and a K391A mutation at the conserved lysine residue in the pBON1::BON1:HA construct (Fig. 1). When transformed into bon1, the mutant gene BON1(A350V/G353A) failed to rescue the bon1 dwarf phenotype. All of the 32 transgenic lines showed bon1-like dwarf phenotype (Table 1). In contrast, the BON1(K391A) mutant gene appeared to be functional. All of the 22 transgenic lines exhibited a wild-type phenotype (Table 1).
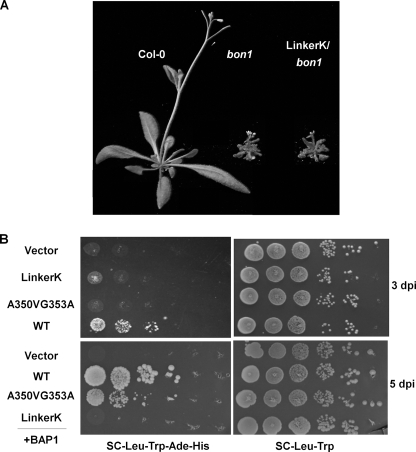
We asked if Lys391 may play a structural role rather than a catalytic role for BON1 by introducing a 6-amino acid stretch, NAAIRS, around this residue. This Asp-Ala-Ala-Ile-Arg-Ser hexapeptide is capable of forming either α-helices and β-sheets depending on its local context (28) and therefore is suitable for mutagenesis studies (29). When this mutant pBON1::BON1(LinkerK):HA gene was introduced into bon1, none of the 15 transgenic lines exhibited a wild-type phenotype (Table 1 and Fig. 5A), suggesting this segment is essential for BON1 function.
FIGURE 5.
Functional analysis of the BON1 VWA domain. A, growth phenotype of transgenic bon1 plants with the BON1(LinkerK) construct. Transgenic plants with the LinkerK mutation exhibited a bon1-like dwarf phenotype. B, yeast two-hybrid analysis of BON1 and BAP1 interaction. Yeast cells containing both BON1 and BAP1 were grown on SC-Leu-Trp plates for growth control and on SC-Leu-Trp-His-Ade plates for interaction assays. The wild-type BON1 has a strong interaction with BAP1. The A350V/G353A mutation greatly weakens BON1-BAP1 interaction because the growth on selection medium was only visible at 5 days after inoculation (5 dpi) but not at 3 dpi. No interaction was observed between BON1(LinkerK) and BAP1 even at 5 dpi.
Mutations in the VWA Domain Affected BON1-BAP1 Interaction
As the LinkerK mutation rather than the K391A mutation disrupts the BON1 function, we asked if the LinkerK mutation abolishes its interaction with its functional partner BAP1 protein. In the yeast two-hybrid assay, the BON1 full-length protein was expressed as the GAL4-AD fusion and the BAP1 was co-expressed as a GAL4-BD fusion. The wild-type BON1 interacts with BAP1 and reconstitutes the functional GAL4 protein to drive the expression of reporter genes essential for yeast growth (2) (Fig. 5B). In contrast, the BON1(LinkerK) mutant protein together with BAP1 could not confer yeast growth on the selection medium (Fig. 5B). Interestingly, the A350V/G353A mutation is found to have a much weaker interaction with BAP1. At 3 days after inoculation, no growth could be observed for yeasts co-expressing BAP1 and BON1(A350V/G353A), whereas those expressing BAP1 and the wild-type BON1 had already fully grown (Fig. 5B). At 5 days after inoculation, some growth was observed for BON1(A350V/G353A) (Fig. 5B), suggesting a much reduced interaction.
Protein Expression Analysis in Transgenic Plants
To determine whether or not the lack of bon1 rescuing activity is merely due to a reduced level of the mutant proteins, the BON1:HA transgenes were subjected to expression analysis. Northern blot analysis detected BON1:HA transcripts with a predicted size and comparable level from all the transgenic bon1 lines tested (data not shown), indicating the transgenes were not silenced. However, Western blot analysis could not detect BON1:HA from total protein extracts, suggesting that this pBON1:BON1:HA construct has a lower expression level than the previous BON1:HA construct possibly due to a shorter promoter (2). We therefore used IP with an anti-HA antibody to detect BON1:HA on three representative transgenic lines of each BON1:HA construct. Total proteins were separated into soluble and microsome fractions that were subsequently IPed with anti-HA antibodies. BON1:HA was detected in one or two of the three lines for the following BON1 constructs: wild-type, D63A/D69A, D122A/D124A, D209A/D215A, D122A/D215A, D269A, G2A, A350V/G353A, K391A, and LinkerK, but no signal was detected for any lines of BON1:HA with A4DA, B3DA, or AB7 (Fig. 6A and data not shown). There is no strict correlation between detection and activity, as no BON1:HA protein could be detected in many rescuing transgenic line, suggesting that a low expression of BON1:HA below the detection level could still provide enough BON1 activity. The AB7 mutant does not appear to have intrinsic protein instability, as the BON1(AB7) mutant protein has a similar GFP signal level as the wild-type BON1 in protoplasts. Therefore, the lack of activity of BON1(A4DA) and BON1(AB7) is likely due to a loss of protein activities rather than merely a low expression level.
FIGURE 6.
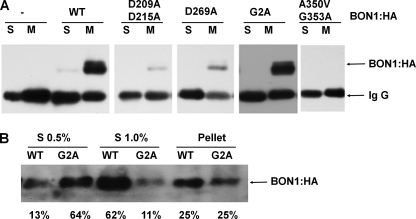
Expression of wild-type and mutant BON1 proteins in transgenic plants. A, expression of BON1:HA in bon1 transgenic plants. Soluble and membrane fractions of total proteins were IPed with anti-HA antibodies. BON1:HA can be detected from IPed products by Western blot in some lines (such as WT, D209A/D215A, D269A, and G2A) but not others (such as A350V/G353A); however, there is no correlation of its detection and its rescuing activity. All detected BON1:HA were mostly in the membrane fractions. B, solubilization of WT and G2A BON1:HA proteins from membrane fractions. Microsome pellets were step solubilized by 0.5 and 1.0% sodium deoxycholate. Shown are the two soluble fractions (S 0.5% and S 1.0%) and the pellet fraction analyzed by Western blot with the percentage of BON1 protein in each fraction shown at the bottom. The majority (62%) of the WT BON1 was in the soluble fraction of 1.0% detergent, whereas the majority (64%) of the G2A BON1 was in the soluble fraction of 0.5% detergent.
All detected mutant BON1:HA proteins were found in membrane fractions, indicating that they are mainly membrane associated similar to the wild-type BON1 (Fig. 6A, and data not shown). As the G2A mutant BON1 expressed in protoplasts was distributed not only in plasma membrane but also on inner membranes (Fig. 3), we determined if G2A and WT BON1 proteins were associated to different domains or subdomains of the membranes in transgenic plants. To avoid differential localization due to differential expression levels, a WT transgenic line and a G2A transgenic line with comparable total BON1 protein levels were chosen for this analysis. Because the two-phase partitioning experiment did not give consistent partitioning of the proteins, we used ionic detergent sodium deoxycholate to step solubilize proteins from the membrane fractions. At a concentration of 0.5% detergent, 13% of the WT and 61% of the G2A BON1 proteins were distributed to the soluble fraction. At a concentration of 1.0% detergent, 62% of the WT and 11% of the G2A was further solubilized (Fig. 6B). Thus the G2A BON1 is less associated with membrane fractions resistant to higher concentrations of detergents, indicating a location alteration in the subdomains of the membrane.
DISCUSSION
Putative Calcium Binding Activity Is Required for BON1 Function but Not for Its plasma Membrane Localization
Copine proteins have two calcium-dependent lipid binding C2 domains and some copine proteins, including BON1, were shown to indeed have calcium-dependent lipid binding activity in vitro. However, the biological roles of this activity have not been demonstrated. Using a mutagenesis approach, we showed for the first time that conserved aspartates mediating calcium binding are required for the biological roles of copine. Although mutating one or two aspartates in one C2 domain does not significantly compromise BON1 activity, mutating all conserved aspartates in the C2A domain or the C2B domain greatly reduced the BON1 activity. We did obtain very few wild-type among the much more mutant looking lines with BON1(B3DA) where three aspartates in the C2B domain were mutated. These lines might have a higher expression level to compensate for the reduced activity of the BON1(B3DA) mutant protein. Therefore both C2A and C2B are required for BON1 function, and the two C2 domains are not functionally redundant.
It has been thought that the lipid-binding activity of C2 is responsible for membrane localization of BON1. In our study, we found that mutating all seven aspartate residues in the C2 domains of BON1 did not significantly alter its plasma membrane localization. In contrast, a single G2A mutation abolished the exclusive plasma membrane localization of BON1, indicating that myristoylation at glycine 2 is required for BON1 plasma membrane localization. The BON1 protein without the myristoylation site glycine 2 is still membrane associated in both transient and stable expression systems. When overexpressed in protoplasts, it is found on the plasma membrane as well as inner membranes whose exact nature is yet to be determined. In transgenic plants, BON1 G2A is more readily solubilized from membranes by a lower concentration of detergent, suggesting that myristoylation is critical for targeting BON1 to detergent-resistant domains of the plasma membrane. This explains the much tighter association of BON1 to the membranes in plants than in vitro calcium-dependent binding of BON1 to lipids (2). Recently, some signaling proteins such as receptor kinases and calcium signaling molecules were found to reside in detergent-resistant domains of the plasma membranes (30), suggesting an intriguing possibility that BON1 is in physical proximity to some of these molecules and perhaps have signaling interaction with them.
Our study reveals that calcium binding activity and possibly the calcium-dependent lipid-binding activity are required for BON1 function. They likely play a regulatory role on the copine function rather than only contribute to the localization of copines. Calcium is a universal signal for diverse biological process, and is critical for the cells to interpret calcium signals varying in concentration and dynamics. Copines might have been evolved to act as calcium sensors to facilitate signaling events for a variety of pathways.
Myristoylation Is Critical for the Function of Plant Copines
N-Myristoylation of copines might be a new feature acquired by higher plants. Sequence analysis revealed that glycine 2 exists in almost all copines found in higher plants; however, it does not exist in any copines from animals or protists. Most of the plant copines are predicted to have N-myristoylation, but some, including BON2, are not predicted to be so. Genetic studies indicate that BON2 has a similar but not identical function as BON1 (5). It will be interesting to analyze the subcellular localization of BON2 to determine whether it is associated with other parts of the cell and if it has more dynamic localization than BON1. It remains possible that BON2 is myristoylated in plants although the prediction score is very low. In any case, myristoylation appears to be unique to plant copines and it confers a stronger association of BON1 to perhaps special subdomains of the plasma membrane.
Very little is known about subcellular localization of other copine proteins with the exception of CPNA from Dictyostelium and NRA-1 from C. elegans. CPNA was typically found in the cytoplasm without specific membrane localization but was observed to bind transiently to the plasma membrane and intracellular vacuoles in some starved cells (6). NRA-1 is involved in regulating nicotinic receptor and it is found to largely localize to the plasma membrane (9). Thus the localization of copines could be dynamic and probably responsive to calcium signals that are regulated by diverse signaling events.
The G2A mutation disrupting potential myristoylation not only alters the location of the BON1 protein but also abolishes its biological activity. This suggests that specific association with the plasma membrane of BON1 is essential for its proper function. Consistent with this, overexpression of the BON1 VWA domain in wild-type Arabidopsis caused a bon1-like phenotype (31), presumably resulting from mislocalization of the VWA domain to a different location. Association with the plasma membrane or its subdomain may enable BON1 to interact with its proper binding proteins. Alternatively but not mutually exclusively, myristoylation may confer additional properties to BON1 other than localization, similar to the tomato disease resistance protein Pto. Pto is a protein kinase, and its N-myristoylation is critical for its signaling but not its localization (32). The effect of myristoylation on signaling is not known yet. It is possible that BON1 has additional modification other than myristoylation at its N terminus. This is suggested by mass spectrometry analysis that detected all predicted peptides of BON1 except for the N-terminal peptide of 34 amino acids either un-modified or myristoylated. Dual fatty acyl modification was observed in the CBL protein in Arabidopsis where myristoylation at glycine 2 is required for acylation of cysteine 3 (20). BON1 has a cysteine at residue 4, which could potentially be modified after glycine 2 is myristoylated. This intriguing possibility will be further tested and the implication of lipid modification on calcium signaling molecules awaits further exploring.
Role of the VWA Domain in Mediating BAP1 Interaction
The VWA domains of copines including BON1 have features of canonical kinase motifs. Although a kinase activity was associated with a purified human copine III, it is unknown if copines indeed possess kinase activity and if catalytic activity is critical for copine function. This study suggests that the Arabidopsis copine protein BON1 may not use kinase activity for its function as mutating the essential catalytic residue Lys391 did not abolish BON1 activity. Rather, this VWA domain may take a structural role particularly in mediating interaction with BAP1 as a correlation was observed between BON1 activity and the interaction between BON1 and BAP1. The LinkerK mutation abolishes the interaction of BON1 with BAP1 and also disrupts the BON1 function. In addition, the A350V/G353A mutation, although potentially affecting ATP binding, greatly reduces the BON1-BAP1 interaction. This correlation also further supports the idea that BAP1 is a functional partner of BON1 and it could mediate BON1 function. A structural role rather than a catalytic activity has been proposed for other proteins with kinase motifs. For instance, the Arabidopsis SSP gene is involved in embryogenesis and encodes an interleukin-1 receptor-associated kinase. The SSP protein possesses canonical kinase motifs but the kinase catalytic activity is dispensable for its function (33). Mammalian interleukin-1 receptor-associated kinase proteins often lack kinase activity but rather contribute to the assembly of receptor complexes (34). The protein-protein interaction function of BON1 may apply generally to other copines as multiple interacting proteins have been isolated for human copines from yeast two-hybrid screens.
The VWA domain of BON1 may nevertheless possess an ATP-binding activity as altering the glycine loop disrupted BON1 function. This activity could provide a second level of regulation by ATP in addition to calcium, and therefore BON1 might be highly regulated by the cellular state. An alternative but not mutually exclusive function for the glycine loop is to provide a structure for BON1 to interact with other proteins as the mutations in this motif weakens its interaction with BAP1.
In summary, our structure-function analysis of an Arabidopsis copine protein BON1 reveals an unexpected regulation of copine protein localization and function. Future studies of BON1 and other copines should generate further insights in the signaling mode of copines by the C2 domains and the VWA domains.
Acknowledgments
We thank Dr. Z. Sheng for discussion on mass spectrometry analyses and the Cornell Proteomic Facility for support. We also thank W. Qian, Y. Zhu, Z. Bao, W. Underwood, and J. Vadassery for discussion and comments on the manuscript.
This work was supported, in whole or in part, by a grant from the National Science Foundation (to J. H.).
- VWA
- von Willebrand A
- SC
- synthetic complete
- IP
- immunoprecipitation.
REFERENCES
- 1.Creutz C. E., Tomsig J. L., Snyder S. L., Gautier M. C., Skouri F., Beisson J., Cohen J. (1998) J. Biol. Chem. 273, 1393–1402 [DOI] [PubMed] [Google Scholar]
- 2.Hua J., Grisafi P., Cheng S. H., Fink G. R. (2001) Genes Dev. 15, 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jambunathan N., Siani J. M., McNellis T. W. (2001) Plant Cell 13, 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S., Hua J. (2004) Plant Cell 16, 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S., Yang H., Grisafi P., Sanchatjate S., Fink G. R., Sun Q., Hua J. (2006) Plant J. 45, 166–179 [DOI] [PubMed] [Google Scholar]
- 6.Damer C. K., Bayeva M., Hahn E. S., Rivera J., Socec C. I. (2005) BMC Cell Biol. 6, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damer C. K., Bayeva M., Kim P. S., Ho L. K., Eberhardt E. S., Socec C. I., Lee J. S., Bruce E. A., Goldman-Yassen A. E., Naliboff L. C. (2007) Eukaryot. Cell 6, 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomsig J. L., Sohma H., Creutz C. E. (2004) Biochem. J. 378, 1089–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottschalk A., Almedom R. B., Schedletzky T., Anderson S. D., Yates J. R., 3rd, Schafer W. R. (2005) EMBO J. 24, 2566–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Church D. L., Lambie E. J. (2003) Genetics 165, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizo J., Südhof T. C. (1998) J. Biol. Chem. 273, 15879–15882 [DOI] [PubMed] [Google Scholar]
- 12.Banci L., Cavallaro G., Kheifets V., Mochly-Rosen D. (2002) J. Biol. Chem. 277, 12988–12997 [DOI] [PubMed] [Google Scholar]
- 13.Whittaker C. A., Hynes R. O. (2002) Mol. Biol. Cell 13, 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomsig J. L., Creutz C. E. (2000) Biochemistry 39, 16163–16175 [DOI] [PubMed] [Google Scholar]
- 15.Yang H., Li Y., Hua J. (2006) Plant J. 48, 238–248 [DOI] [PubMed] [Google Scholar]
- 16.Tomsig J. L., Snyder S. L., Creutz C. E. (2003) J. Biol. Chem. 278, 10048–10054 [DOI] [PubMed] [Google Scholar]
- 17.Caudell E. G., Caudell J. J., Tang C. H., Yu T. K., Frederick M. J., Grimm E. A. (2000) Biochemistry 39, 13034–13043 [DOI] [PubMed] [Google Scholar]
- 18.Thompson G. A., Jr., Okuyama H. (2000) Prog. Lipid Res. 39, 19–39 [DOI] [PubMed] [Google Scholar]
- 19.Nagasaki N., Tomioka R., Maeshima M. (2008) FEBS J. 275, 2267–2282 [DOI] [PubMed] [Google Scholar]
- 20.Batistic O., Sorek N., Schültke S., Yalovsky S., Kudla J. (2008) Plant Cell 20, 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murashige T., Skoog F. (1962) Physiol. Plant 15, 473–497 [Google Scholar]
- 22.Tzfira T., Tian G. W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005) Plant Mol. Biol. 57, 503–516 [DOI] [PubMed] [Google Scholar]
- 23.Zhai Z., Sooksa-nguan T., Vatamaniuk O. K. (2009) Plant Physiol. 149, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koncz C., Schell J. (1986) Mol. Gen. Genet. 204, 383–396 [Google Scholar]
- 25.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 26.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davletov B., Perisic O., Williams R. L. (1998) J. Biol. Chem. 273, 19093–19096 [DOI] [PubMed] [Google Scholar]
- 28.Wilson I. A., Haft D. H., Getzoff E. D., Tainer J. A., Lerner R. A., Brenner S. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 5255–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosher R. A., Durrant W. E., Wang D., Song J., Dong X. (2006) Plant Cell 18, 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kierszniowska S., Seiwert B., Schulze W. X. (2009) Mol. Cell. Proteomics 8, 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Jambunathan N., McNellis T. W. (2005) Planta 221, 85–94 [DOI] [PubMed] [Google Scholar]
- 32.de Vries J. S., Andriotis V. M., Wu A. J., Rathjen J. P. (2006) Plant J. 45, 31–45 [DOI] [PubMed] [Google Scholar]
- 33.Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009) Science 323, 1485–1488 [DOI] [PubMed] [Google Scholar]
- 34.Janssens S., Beyaert R. (2003) Mol. Cell 11, 293–302 [DOI] [PubMed] [Google Scholar]