Abstract
In Saccharomyces cerevisiae, aminopeptidase I (Ape1p) and α-mannosidase (Ams1p) are known cargoes of selective autophagy. Atg19p has been identified as an Ape1p receptor and targets Ape1p to the preautophagosomal structure (PAS). Under nutrient-rich conditions, transport of Ams1p to the vacuole largely depends on Atg19p. Here, we show that Atg34p (Yol083wp), a homolog of Atg19p, is a receptor for Ams1p transport during autophagy. Atg34p interacted with Ams1p, Atg11p, and Atg8p using distinct domains. Homo-oligomerized Ams1p bound to the Ams1-binding domain of Atg34p; this binding was important for the formation of a higher order complex named the Ams1 complex. In the absence of the interaction of Atg34p with Atg8p, the Ams1 complex was targeted to the preautophagosomal structure but failed to transit to the vacuole, indicating that the interaction of Atg34p with Atg8p is crucial for the Ams1 complex to be enclosed by autophagosomes. Atg34p and Atg19p have similar domain structures and are important for Ams1p transport during autophagy.
Keywords: Autophagy, Protein Degradation, Trafficking, Yeast, Yeast Genetics, Autophagy-related Genes, Cytoplasm-to-vacuole Targeting
Introduction
Macroautophagy (hereafter simply called autophagy) has been described as a non-selective degradation system of cytoplasmic constituents. Recently, selective degradation of proteins and organelles by autophagy has been shown to be important for the homeostasis of eukaryotic cells by eliminating unnecessary or harmful proteins and organelles such as mitochondria and peroxisomes.
Thirty-three autophagy-related (ATG)2 genes have been identified as requisites for several types of autophagy in yeast. Among them, 18 genes are essential for autophagosome formation under starvation conditions (1). Atg proteins encoded by these genes are organized into the preautophagosomal structure (PAS), which plays a central role in autophagosome formation near the vacuole (2). During autophagy, cytoplasmic constituents are sequestered by the double membrane of an autophagosome. The outer membrane of the autophagosome fuses with the vacuole membrane. The inner structure, an autophagic body, is released into the vacuolar lumen and degraded by the action of vacuolar hydrolases.
Previous analysis showed that the resident vacuolar hydrolases, α-mannosidase (Ams1p) and aminopeptidase I (Ape1p), are transported to the vacuole via selective autophagy (3, 4). Ams1p oligomerizes after synthesis and associates with a precursor Ape1p (prApe1p) by the action of Atg19p (4, 5). Ams1p, prApe1p, and Atg19p assemble into a large complex called the cytoplasm-to-vacuole targeting (Cvt) complex, which was identified as an electron-dense structure localized close to the vacuole by electron microscopy (3). Atg19p mediates the association of the Cvt complex with the PAS by interacting with Atg8p and Atg11p (5, 6). The Cvt complex is selectively enclosed in a Cvt vesicle (about 150 nm in diameter) under nutrient-rich conditions and in an autophagosome (about 500 nm in diameter) under starvation conditions. After transport to the vacuole, prApe1p is processed into mature Ape1p (mApe1p).
Under nutrient-rich conditions, Atg19p is essential for Ams1p transport (4, 5). In this study, we show that Ams1p is transported to the vacuole under starvation conditions in an Atg19p-independent manner. We found that Atg34p, a homolog of Atg19p, functions as a receptor for Ams1p. Ams1p bound to the Ams1-binding domain of Atg34p and formed a higher order complex called the Ams1 complex. The interaction of Atg34p with Atg8p was essential for sequestration of the Ams1 complex into an autophagosome.
EXPERIMENTAL PROCEDURES
Strains and Media
Standard methods were used for yeast manipulation (7). Cells were grown in either YEPD (1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose) or SD + casamino acid medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acid, and 2% glucose) with appropriate supplements. Autophagy was induced by addition of 400 ng/ml rapamycin (Sigma) or by shifting to a nitrogen starvation medium (SD(−N); 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose).
The yeast strains used in this study are listed in supplemental Table S1. GFP-tagged Atg strains were generated using a PCR-based gene modification method (8). For Atg34ΔC4p-GFP and Atg34AEEAp-GFP strains, mutations were introduced in primers used to amplify GFP fragments. To disrupt ATG genes, disruption cassettes were amplified from disruptants in laboratory stocks and used for transformation. Tetrad dissection was used to create several disruptants. mRFP-prApe1 strains were generated by transforming cells with pPS129 digested with AvrII (the plasmid was a gift from Dr. Daniel J. Klionsky, University of Michigan).
ATG34 was cloned into the pRS316 plasmid (9). Plasmids expressing Atg34HA,EA, Atg34HA, and Atg34EA were generated using a QuikChange site-directed mutagenesis kit (Stratagene). The GFP sequence of the pRS316 GFP-ATG8 plasmid was substituted for the 2× mCherry sequence to create the pRS316 2× mCherry-ATG8 plasmid.
Microscopy
Fluorescence microscopy was performed using a total internal reflection fluorescence microscopy system (Olympus) equipped with a UPlanSApo 100× oil objective (numerical aperture, 1.40) and a CoolSNAP HQ charge-coupled device camera (Nippon Roper). Blue (Sapphire 488-20, Coherent) and yellow lasers (85-YCA-010, Melles Griot) were used for excitation of GFP and mRFP/mCherry, respectively. A U-MNIBA2, mirror unit from which the excitation filter was removed was used for GFP visualization, and an FF593-Di02-25 × 36 dichroic mirror and an FF593-Em02-25 bandpass filter (Semrock) were used to visualize mRFP/mCherry. For simultaneous observation of GFP and mRFP/mCherry, both lasers were combined to obtain a fluorescent image. Fluorescence was filtered with a DM488–561 dichroic mirror and an EM505–540&572IF bandpass filter (Olympus) and was split into two channels using a U-SIP splitter (Olympus) equipped with a 565DCLP dichroic mirror (Chroma). For each channel, the fluorescent light was further filtered with an FF495-Em02-25 bandpass filter (Semrock) for the GFP channel and an FF593-Em02-25 bandpass filter (Semrock) for the mRFP/mCherry channel. Both channels were imaged on different parts of the charge-coupled device camera. Images were acquired using MetaMorph software (Molecular Devices).
Immunoblotting Analysis
Cell lysates were prepared by NaOH/2-mercaptoethanol extraction with a slight modification (10). An anti-Atg34p antiserum was raised against bacterially expressed Atg34p (a gift from Dr. Inagaki, Hokkaido University, Sapporo, Japan) and affinity-purified using the antigen. To detect GFP and Ape1p, anti-GFP antiserum (Invitrogen) and anti-Ape1p antiserum (6) were used, respectively.
Yeast Two-hybrid Analysis
Yeast two-hybrid assays were carried out as described previously (11). Briefly, a series of truncated ATG34 constructs were cloned into a pGAD-based plasmid. Full-length ATG34, AMS1, ATG11, ATG8, and APE1 were cloned into a pGBD-based plasmid. pGAD- and pGBD-based plasmids were cotransformed into the PJ69-4A host strain. Cells harboring the indicated plasmids were cultured in synthetic complete medium lacking leucine and tryptophan (SC(−LW)). Five microliters of 0.1-A600 unit aliquots of cells were spotted onto SC(−LW) or SC(−LW) lacking adenine (SC(−ALW)) plates. These plates were incubated for 3–4 days at 30 °C.
RESULTS
Yol083wp Is Involved in Ams1p Transport to Vacuole
α-Mannosidase (Ams1p) is a vacuolar hydrolase that is selectively transported to the vacuole via the Cvt pathway under nutrient-rich conditions and via autophagy under starvation conditions (4). Under nutrient-rich conditions, Ams1p transport largely depends on Atg19p, a receptor for Ams1p transport (5).
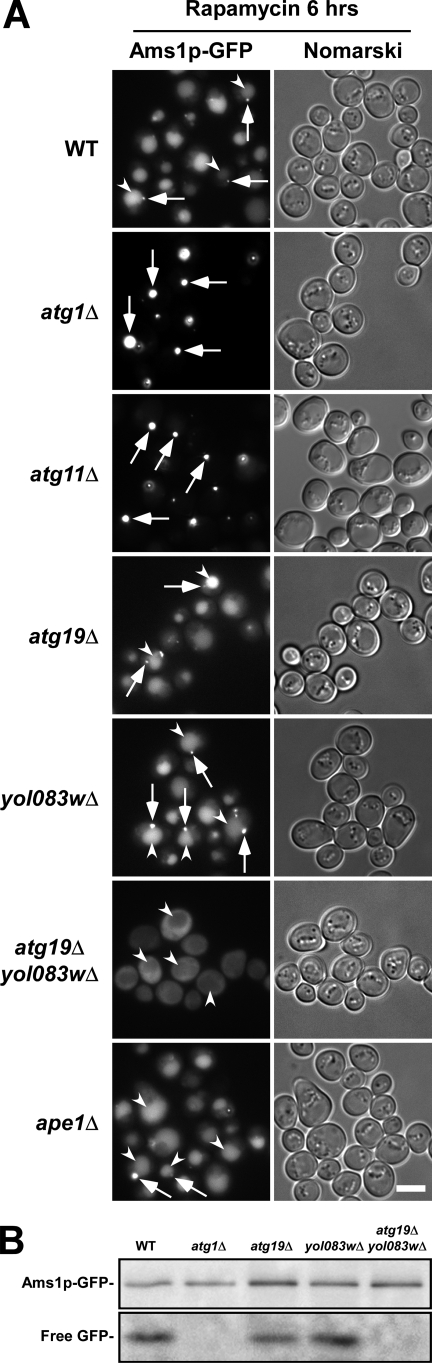
The dynamics of Ams1p during autophagy was analyzed by fluorescence microscopy using Ams1p-GFP expressed from the chromosome. We induced autophagy by adding rapamycin to growth medium unless otherwise noted. In this study, we refer to the rapamycin-treated conditions as “starvation conditions.” In wild-type cells under starvation conditions, Ams1p-GFP localized to a dot proximal to the vacuole (Fig. 1A, arrows) as well as the vacuolar lumen (Fig. 1A, arrowheads). In atg1Δ cells, GFP staining was detected as a bright dot(s) exclusively outside the vacuole (Fig. 1A). These results show that vacuolar staining with GFP enables the monitoring of Ams1p-GFP transport by autophagosomes. Vacuolar staining was hardly detected in atg11Δ cells (Fig. 1A), indicating that Atg11p is involved in selective transport of Ams1p by autophagy.
FIGURE 1.
Yol083wp is involved in Ams1p transport to vacuole. A, cells chromosomally expressing Ams1p-GFP were cultured in SD + casamino acid medium to a density of approximately A600 = 1.5, and rapamycin was added to induce autophagy. After 6 h of incubation, cells were observed by fluorescence microscopy. Arrows and arrowheads indicate Ams1p dot structures (Ams1 complex) and vacuoles, respectively. All strains were in the same genetic background (GYS337). The scale bar represents 5 μm. B, cells used in A were treated with rapamycin for 3 h, and cell lysates were prepared by the alkaline lysis method. The equivalent of 0.1 A600 units was subjected to immunoblot with an anti-GFP antiserum.
Ams1p transport depends on Atg19p under nutrient-replete conditions (5, 12). Under starvation conditions, however, Ams1p was effectively transported even in the atg19Δ strain (Fig. 1A). We assumed that another receptor protein would play a role in Ams1p transport under starvation conditions. Genome-wide two-hybrid analyses showed that Yol083wp interacts with Ams1p (13, 14). Moreover, bioinformatics analysis using a BLAST search suggests that Yol083wp is a homolog of Atg19p. Thus, YOL083W is additionally disrupted in atg19Δ cells. The transport of Ams1p-GFP was abolished in the atg19Δyol083wΔ double disruptant, showing that Yol083wp acts for Ams1p transport in the absence of Atg19p (Fig. 1A). In contrast, the transport of Ams1p was normal in the yol083wΔ strain (Fig. 1A). This result indicates that either Yol083wp or Atg19p is sufficient for Ams1p transport during autophagy. The Ams1p transport is defective in ape1Δ cells during vegetative growth (5) but seemed normal under starvation conditions (Fig. 1A), suggesting that the prApe1p-Atg19p complex is dispensable for Ams1p transport.
Ams1p-GFP did not stain the vacuolar lumen in the atg1Δ, atg11Δ, or atg19Δyol083wΔ strains but exhibited different staining patterns: Ams1p-GFP formed a dot structure in the atg1Δ and atg11Δ strains, whereas it dispersed throughout the cytoplasm in the atg19Δyol083wΔ strain (Fig. 1A), indicating that either Yol083wp or Atg19p serves to assemble Ams1p to the dot structure and subsequently to the PAS, the center of autophagosome formation (2). In atg19Δ cells, the dot structure marked by Ams1p must be localized to the PAS since Ams1p was normally transported to the vacuole, but this Ams1p dot is distinct from the Cvt complex because in these cells the Ape1 complex is apart from the PAS (5, 6). Hence, the dot structure marked by Ams1p is referred to as the “Ams1 complex.”
In addition, we monitored Ams1p transport to the vacuole by immunoblot. When Ams1p-GFP is delivered to the vacuole, the GFP moiety is cleaved off by vacuolar hydrolases. As the GFP moiety is relatively stable in the vacuole, the level of Ams1p-GFP transport could be estimated by the amount of free GFP (6). The immunoblot showed that Ams1-GFP was normally expressed in all strains (Fig. 1B). The GFP moiety was detected in wild-type cells but not in atg1Δ cells (Fig. 1B). Double disruption of ATG19 and YOL083W prevented the generation of free GFP, but disruption of either ATG19 or YOL083W did not (Fig. 1B). Taken together, we conclude that Yol083wp is involved in Ams1p transport via autophagosomes in a manner similar to Atg19p. Thus, we name YOL083W ATG34. Atg34p is dispensable for Ape1p transport, for Ams1p transport under nutrient-rich conditions, and for bulk autophagy (supplemental Fig. S1).
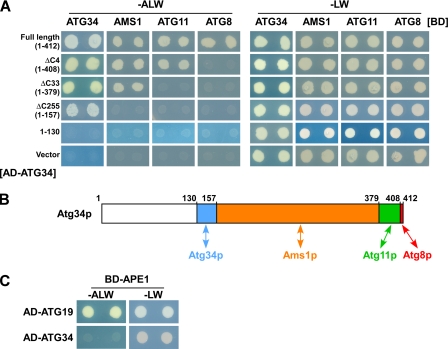
Atg34p Interacts with Ams1p, Atg8p, and Atg11p
Yeast two-hybrid analysis showed that full-length Atg34p interacts with Ams1p, Atg8p, Atg11p, and itself (Fig. 2A). We generated four truncated forms of Atg34p: Atg34p lacking the C-terminal four residues (Atg34ΔC4p, residues 1–408), Atg34p lacking the 33 C-terminal residues (Atg34ΔC33p, residues 1–379), Atg34p lacking residues 158–412 (Atg34ΔC255p, residues 1–157), and the first 130 N-terminal residues of Atg34p (Atg34(1–130), residues 1–130).
FIGURE 2.
Atg34p interacts with Ams1p, Atg8p, and Atg11p. A, Atg34p interacts with Ams1p, Atg8p, Atg11p, and itself using distinct domains. PJ69-4A cells harboring pGAD-ATG34, pGAD-ATG34ΔC4, pGAD-ATG34ΔC33, pGAD-ATG34ΔC255, pGAD or an empty vector were transformed with pGBD-ATG34, pGBD-AMS1, pGBD-ATG11, pGBD-ATG8, or an empty vector. Cells were cultured in SC(−LW) medium overnight. Five microliters of 0.1-A600 unit aliquots of cells were spotted onto SC(−LW) or SC(−ALW) plates. These plates were incubated for 3–4 days at 30 °C. B, schematic diagram of binding domains in Atg34p. Atg34p interacts with at least four proteins. Blue, orange, green, and red regions represent domains required for Atg34p, Ams1p, Atg11p, and Atg8p interactions, respectively. C, Atg34p does not interact with prApe1p, whereas Atg19p does. PJ69-4A cells harboring pGAD-ATG19 and pGBD-APE1 or pGAD-ATG34 and pGBD-APE1 were assayed as in A.
Atg34ΔC4p did not bind to Atg8p but did bind to Ams1p, Atg11p, and itself (Fig. 2A). Atg34ΔC33p did not interact with Atg8p or Atg11p (Fig. 2A). Further truncation just before the putative coiled coil domain (Atg34ΔC255p) abrogated the interaction with Ams1p (Fig. 2A). When the putative coiled coil domain was deleted, Atg34p no longer bound any interacting protein identified here (Atg34(1–130); Fig. 2A). These results show that Atg34p associates with Ams1p, Atg8p, Atg11p, and itself using distinct domains (Fig. 2B). Atg34p is not involved in Ape1p transport, whereas Atg19p is (supplemental Fig. S1). Consistently, Atg34p was not capable of interacting with prApe1p, but Atg19p was (Fig. 2C).
Association of Atg34p with Ams1p Is Essential for Ams1 Complex Formation
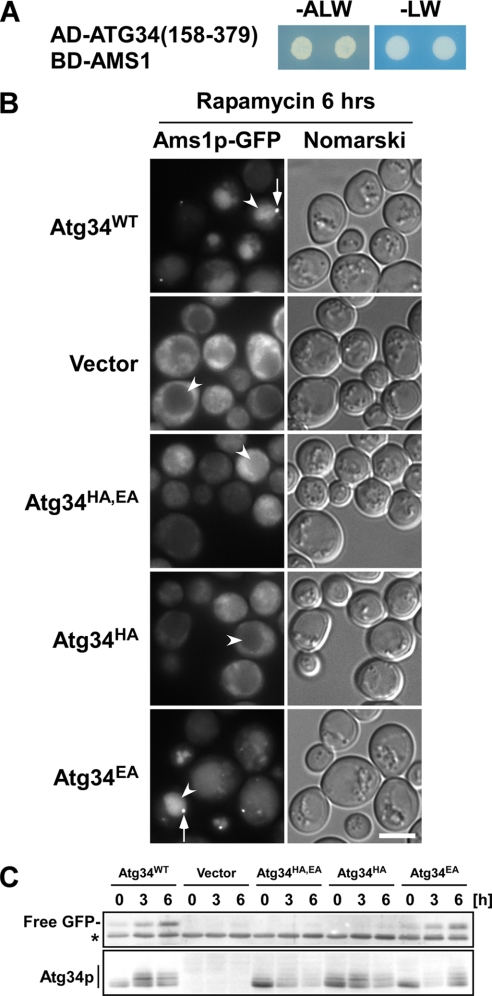
Our two-hybrid analysis of Atg34p indicates that residues 158–379 are essential for the interaction with Ams1p (Fig. 2B). We confirmed that this domain was sufficient for the interaction with Ams1p (Fig. 3A).
FIGURE 3.
Association of Atg34p with Ams1p is essential for Ams1 complex formation. A, Atg34(158–379) is sufficient to interact with Ams1p. Cells harboring the indicated plasmids were assayed as in Fig. 2A. B, Ams1p-GFP cells lacking both ATG19 and ATG34 (GYS360) and expressing Atg34p mutants from pRS316-based plasmids were used. Cells harboring the indicated plasmids were cultured in SD + casamino acid medium to a density of approximately A600 = 1.5, and rapamycin was added to induce autophagy. After 6 h of incubation, cells were observed by fluorescence microscopy. Arrows and arrowheads indicate Ams1 complexes and vacuoles, respectively. The scale bar represents 5 μm. C, cells used in B were collected at the times indicated after rapamycin addition, and lysates were prepared by the alkaline lysis method. Cell lysates equivalent to 0.1 A600 units were subjected to immunoblot with an anti-GFP antiserum and anti-Atg34p antibodies. Transport of Ams1p-GFP was monitored by emergence of free GFP by proteolysis of Ams1p-GFP in the vacuole. An asterisk indicates nonspecific bands.
Recently, a structure of the Ams1-binding domains of Atg34p and Atg19p has been solved by nuclear magnetic resonance analysis (see the accompanying paper by Watanabe et al. (22)). An in vitro pulldown assay showed that two conserved His and Glu residues in the domain are important for the interaction with Ams1p (see the accompanying paper by Watanabe et al. (22)). Based upon these data, we generated three Atg34p mutants, which harbor a change of His296 to Ala (Atg34HAp), Glu297 to Ala (Atg34EAp), or both (Atg34HA,EAp). The in vivo activity of these proteins was monitored by fluorescence microscopy and GFP cleavage assay using an Ams1p-GFP strain lacking both ATG19 and ATG34. In cells carrying an empty vector, Ams1p-GFP exhibited a cytoplasmic pattern, whereas wild-type Atg34p enabled Ams1p to be assembled into the Ams1 complex (Fig. 3B, arrows) and to be transported to the vacuole (Fig. 3B, arrowheads). Atg34EAp acted as a wild-type Atg34p, but Atg34HAp and Atg34HA,EAp neither formed the Ams1 complex nor transported Ams1p to the vacuole (Fig. 3B). Immunoblot analysis showed that cells expressing Atg34HAp or Atg34HA,EAp are defective in Ams1p transport (Fig. 3C). These results led to the conclusion that the His296 residue is critical for the formation of the Ams1 complex by binding to Ams1p in vivo.
Interaction with Atg8p via Atg8 Family-interacting Motif (AIM) Is Important for Atg34p to Target Ams1 Complex to Autophagosomes
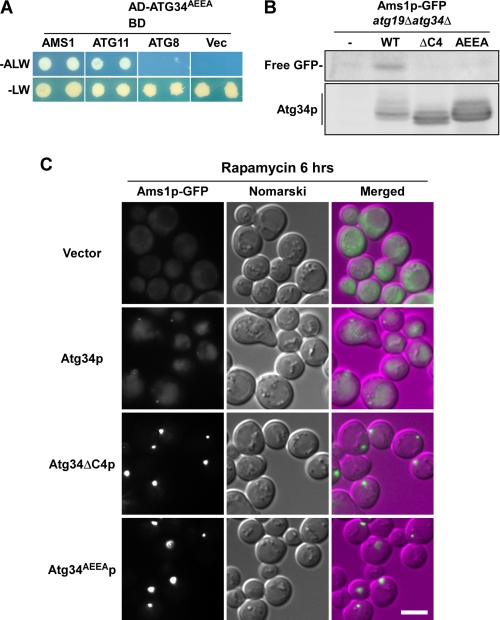
We next examined whether the interaction of Atg34p with Atg8p is essential for its activity. Recently, our groups have found an AIM at the C terminus of Atg19p (Trp412-Glu-Glu-Leu415) and shown that the Atg19p mutants harboring changes of either Trp412, Leu415, or both residues to Ala fail to interact with Atg8p (15, 16). Atg34p also has an AIM at the C terminus (Trp409-Glu-Glu-Ile412). We replaced the Trp409 and Ile412 residues with Ala to make Atg34AEEAp. Yeast two-hybrid analysis showed that Atg34AEEAp interacted normally with Ams1p and Atg11p but did not interact with Atg8p (Fig. 4A). This result was the same as that of Atg34ΔC4p (Fig. 2A). Next, Atg34ΔC4p and Atg34AEEAp were expressed in the atg19Δatg34Δ strain expressing Ams1p-GFP, and their activities were analyzed by monitoring Ams1p-GFP cleavage. Free GFP was generated only in the cells expressing wild-type Atg34p but not in the cells expressing Atg34ΔC4p or Atg34AEEAp (Fig. 4B), showing that the interaction between Atg34p and Atg8p is essential for Ams1p transport.
FIGURE 4.
Interaction with Atg8p via AIM is important for Atg34p to target Ams1 complex to autophagosome. A, Atg34AEEAp does not interact with Atg8p. The interaction was monitored by yeast two-hybrid assay as described in Fig. 2A. B, Atg34ΔC4p and Atg34AEEAp are defective in Ams1p transport. atg19Δatg34Δ cells expressing Ams1p-GFP (GYS360) and carrying an empty vector (−) or expressing wild-type Atg34p (WT), Atg34ΔC4p (ΔC4), or Atg34AEEAp (AEEA) were cultured in SD + casamino acid medium, and rapamycin was added to induce autophagy. After incubation for 6 h, cells were collected, and cell lysates were prepared by the alkaline lysis method. Cell lysates equivalent to 0.1 A600 units were subjected to immunoblot with anti-GFP and anti-Atg34p antibodies. C, localization of Ams1p-GFP in the Atg34p mutants lacking an interaction with Atg8p. Cells used in B were observed by fluorescence microscopy. For comparison of the fluorescence intensity, Ams1p-GFP images are shown using the same brightness and contrast. The scale bar represents 5 μm.
Ams1p-GFP dispersed throughout the cytoplasm in atg19Δatg34Δ cells (Figs. 1A and 4C). This pattern reverted to normal when wild-type Atg34p was expressed in these cells (Fig. 4C). In cells expressing Atg34ΔC4p or Atg34AEEAp, Ams1p-GFP was not transported to the vacuole but accumulated outside the vacuole as a brighter dot relative to that observed in cells expressing wild-type Atg34p (Fig. 4C). This suggests that Atg34p mutants lacking the interaction with Atg8p retain the ability to form the Ams1 complex.
Atg34p Is Selectively Transported to Vacuole as Component of Cvt Complex
In wild-type cells, Atg34p-GFP was transported to the vacuole (Fig. 5), whereas the vacuole was not labeled by GFP in atg1Δ cells (supplemental Fig. S2A), indicating that the Atg34p transport depends on autophagy. Moreover, in atg1Δ cells, Atg34p-GFP accumulated as a dot outside the vacuole and was colocalized with prApe1p, which is a marker of the Cvt complex (supplemental Fig. S2B). Shintani et al. (5) showed that the Cvt complex is targeted to the PAS in the absence of Atg1p. Together with these results, we conclude that Atg34p is first targeted to the PAS and then delivered to the vacuole by autophagy as is Atg19p.
FIGURE 5.
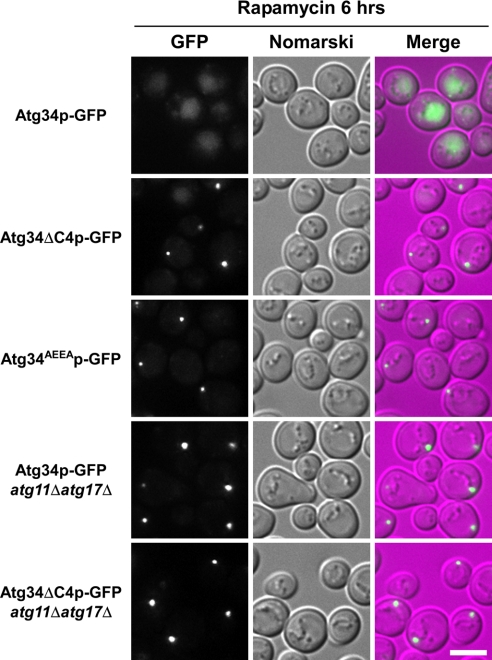
Atg34p mutants defective in interaction with Atg8p are not enclosed in autophagosomes. Cells chromosomally expressing each GFP fusion protein were cultured in SD + casamino acid medium to a density of approximately A600 = 1.5 and treated with rapamycin for 6 h before fluorescence microscopy. Atg34p-GFP (GYS390), Atg34ΔC4p-GFP (GYS805), Atg34AEEAp-GFP (GYS822), Atg34p-GFP atg11Δatg17Δ (GYS812), and Atg34ΔC4p-GFP atg11Δatg17Δ (GYS820) strains were used. For comparison of the fluorescence intensity, Atg34p-GFP images are shown using the same brightness and contrast. The scale bar represents 5 μm.
We next created Atg34ΔC4p-GFP and Atg34AEEAp-GFP strains and examined the localization of these proteins by fluorescence microscopy. During autophagy, they were detected as a dot in the cytoplasm but were not transported to the vacuole (Fig. 5). The fluorescence intensity of the dots was much brighter than observed in wild-type cells (Fig. 5), indicating that these Atg34p mutants are not able to be enclosed in autophagosomes and consequently accumulate outside the vacuole. Together, it is likely that Ams1p and Atg34ΔC4p/Atg34AEEAp form the Ams1 complex but accumulate outside the vacuole because of a defect in sequestration into autophagosomes.
Under starvation conditions, both Atg11p and Atg17p act as scaffold proteins and are required for organization of the PAS (17). In atg11Δatg17Δ cells, both Atg34p-GFP and Atg34ΔC4p-GFP were still detected as dots (Fig. 5), indicating that PAS organization is not required for dot formation. Thus, we conclude that Atg34p is associated with the Cvt complex. Immunoelectron microscopy with anti-Atg34 antibodies demonstrated that Atg34p was localized on the periphery of the Cvt complex enclosed in autophagic bodies (supplemental Fig. S3).
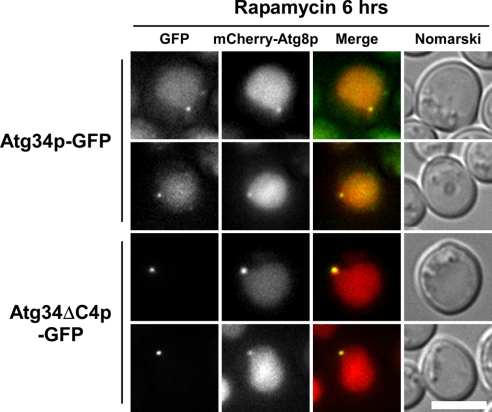
Next, we examined PAS localization of Atg34ΔC4p by using 2× mCherry-Atg8p. Vacuoles were stained with mCherry fluorescence, indicating that autophagy is normal in this strain (Fig. 6). Nevertheless, Atg34ΔC4p-GFP was not transported to the vacuole (Fig. 6). We evaluated the extent of colocalization by counting cells with dots labeled by both GFP (Atg34p) and mCherry (PAS). Atg34p-GFP and mCherry-Atg8p were colocalized in 74% of the cells (n = 43), and Atg34ΔC4p-GFP and mCherry-Atg8p were colocalized in 62% of the cells (n = 34). These results indicate that Atg34ΔC4p-GFP was targeted to the PAS in a normal manner but that it was not engulfed by autophagosomes. It seems likely that Ams1p accumulates at the PAS in Atg34ΔC4p- and Atg34AEEAp-expressing cells (Fig. 4C), showing that the interaction with Atg8p is important for the Ams1 complex to be delivered to autophagosomes.
FIGURE 6.
Atg34pΔC4p-GFP is targeted to PAS. Cells harboring the 2× mCherry ATG8 plasmid were cultured and treated with rapamycin for 6 h. Atg34p-GFP was localized to the PAS and was successively delivered to the vacuole. Atg34ΔC4p was targeted to the PAS but failed to transit to the vacuole. Note that 2× mCherry-Atg8p labels the vacuolar lumen, indicating that autophagy is normal in the Atg34ΔC4p-GFP strain. The scale bar represents 5 μm.
Atg34p Is Normally Delivered to Vacuole in Absence of Known Cargoes
prApe1p is important for recruitment of Atg19p to a dot structure (5). Thus, we examined whether known cargoes are responsible for recruitment of Atg34p to a dot structure. In ams1Δape1Δ cells, Atg34p formed a dot and was normally transported to the vacuole, whereas Atg19p was defective in dot formation (supplemental Fig. S4). The delivery of Atg34p to the vacuole was not affected in the ape1Δams1Δ cells, raising the possibility that other cargoes exist.
We showed that Atg34p acts as a receptor for Ams1p under starvation conditions. Selective autophagy may have important roles in adaptation to various nutrient environments. The physiological significance of selective autophagy will be elucidated by examining the activities of receptors for selective autophagy under diverse culture conditions.
DISCUSSION
In this study, we showed that Atg34p acts as a receptor for the selective transport of Ams1p to the vacuole during autophagy. Ams1p assembles into a homo-oligomer in the cytoplasm after synthesis (4). Formation of an Ams1p-receptor complex by interaction with Ams1p-binding domains in Atg34p and Atg19p is the first step for Ams1p transport (Fig. 3B). Next, the Ams1p-receptor complex is assembled into a higher order complex, called the Ams1 complex, which can be recognized as an Ams1p-GFP dot by fluorescence microscopy (Figs. 1A and 3B). This complex is targeted to the PAS and enclosed in an autophagosome via the interaction of the four residues at the C terminus of Atg34p with Atg8p (Figs. 4–6).
The Ams1p transport under nutrient-rich conditions was defective in atg19Δ cells (supplemental Fig. S1), but this defect was not observed during autophagy (Fig. 1A). Under starvation conditions, Atg34p was detected in a ladder pattern by immunoblot (Fig. 3C). The upper bands disappeared following λ-phosphatase treatment (data not shown). This result suggests that Atg34p is phosphorylated under starvation conditions. The activity of Atg34p might be regulated by its phosphorylation.
We examined the involvement of Atg34p in transport of other selective cargoes of autophagosomes. Pexophagy was normal in atg34Δatg19Δ cells (supplemental Fig. S5A). We also examined peroxisome degradation in nitrogen-starved cells by assessing the cleavage of Pex14p-GFP to free GFP. Peroxisome degradation was normal in atg34Δatg19Δ cells (supplemental Fig. S5B). It is thus unlikely that Atg34p and Atg19p act as receptors for the degradation of peroxisomes. In contrast, atg11Δ cells showed a partial defect (supplemental Fig. S5B), suggesting that Atg11p can recognize peroxisomes without Atg34p and Atg19p.
Recently, we have found that leucine aminopeptidase III (Lap3p) is a selective cargo of the autophagosome. Overexpressed Lap3p concentrated around the Cvt complex is degraded in the vacuole by autophagy in an Atg11p-independent manner (18). Ald6p, a cytosolic acetaldehyde dehydrogenase, is known to be a cargo preferentially degraded by autophagy. This degradation does not depend on Atg11p either (19). Thus, there may be Atg11p-independent mechanisms of selective autophagy.
In mammalian cells, p62 and NBR1 are proteins that interact with LC3, a mammalian homolog of yeast Atg8p, and are thought to interact with polyubiquitinated protein aggregates and participate in their elimination (20, 21). Interestingly, p62 and NBR1 also contain AIMs, which are important for binding to LC3.
We believe that there are more receptors other than Atg34p and Atg19p and more cargoes for selective autophagy. Identification of new receptors and new cargoes will lead to a fundamental understanding of the physiological roles of selective autophagy in eukaryotic cells.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of recombinant Atg34p from Dr. Fuyuhiko Inagaki (Hokkaido University, Sapporo, Japan). We thank Dr. Nobuo N. Noda and Yasunori Watanabe for helpful discussion and Dr. Hayashi Yamamoto for plasmid construction. We also thank the National Institute for Basic Biology Center for Analytical Instruments for technical assistance.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- ATG
- autophagy-related
- PAS
- preautophagosomal structure
- Cvt
- cytoplasm-to-vacuole targeting
- AIM
- Atg8 family-interacting motif
- prApe1p
- precursor Ape1p
- SD
- synthetic dextrose
- SC
- synthetic complete medium.
REFERENCES
- 1.Suzuki K., Ohsumi Y. (2010) FEBS Lett. 584, 1280–1286 [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. (1997) J. Cell Biol. 139, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchins M. U., Klionsky D. J. (2001) J. Biol. Chem. 276, 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki K., Kamada Y., Ohsumi Y. (2002) Dev. Cell 3, 815–824 [DOI] [PubMed] [Google Scholar]
- 7.Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (eds) (1998) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 8.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 9.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath A., Riezman H. (1994) Yeast 10, 1305–1310 [DOI] [PubMed] [Google Scholar]
- 11.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. (2001) Mol. Cell 7, 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 14.Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M., Yamamoto K., Kuhara S., Sakaki Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 16.Noda N. N., Ohsumi Y., Inagaki F. (2010) FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 18.Kageyama T., Suzuki K., Ohsumi Y. (2009) Biochem. Biophys. Res. Commun. 378, 551–557 [DOI] [PubMed] [Google Scholar]
- 19.Onodera J., Ohsumi Y. (2004) J. Biol. Chem. 279, 16071–16076 [DOI] [PubMed] [Google Scholar]
- 20.Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 21.Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 22.Watanabe Y., Noda N. N., Kumeta H., Suzuki K., Ohsumi Y., Inagaki F. (2010) J. Biol. Chem. 285, [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.