Abstract
The GH3 family of acyl-acid-amido synthetases catalyze the ATP-dependent formation of amino acid conjugates to modulate levels of active plant hormones, including auxins and jasmonates. Initial biochemical studies of various GH3s show that these enzymes group into three families based on sequence relationships and acyl-acid substrate preference (I, jasmonate-conjugating; II, auxin- and salicylic acid-conjugating; III, benzoate-conjugating); however, little is known about the kinetic and chemical mechanisms of these enzymes. Here we use GH3-8 from Oryza sativa (rice; OsGH3-8), which functions as an indole-acetic acid (IAA)-amido synthetase, for detailed mechanistic studies. Steady-state kinetic analysis shows that the OsGH3-8 requires either Mg2+ or Mn2+ for maximal activity and is specific for aspartate but accepts asparagine as a substrate with a 45-fold decrease in catalytic efficiency and accepts other auxin analogs, including phenyl-acetic acid, indole butyric acid, and naphthalene-acetic acid, as acyl-acid substrates with 1.4–9-fold reductions in kcat/Km relative to IAA. Initial velocity and product inhibition studies indicate that the enzyme uses a Bi Uni Uni Bi Ping Pong reaction sequence. In the first half-reaction, ATP binds first followed by IAA. Next, formation of an adenylated IAA intermediate results in release of pyrophosphate. The second half-reaction begins with binding of aspartate, which reacts with the adenylated intermediate to release IAA-Asp and AMP. Formation of a catalytically competent adenylated-IAA reaction intermediate was confirmed by mass spectrometry. These mechanistic studies provide insight on the reaction catalyzed by the GH3 family of enzymes to modulate plant hormone action.
Keywords: Amino Acid, Enzyme Kinetics, Enzyme Mechanisms, Metabolic Regulation, Plant, Plant Hormone-Auxin
Introduction
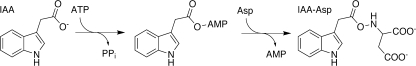
Auxins, such as indole-3-acetic acid (IAA),2 are a major class of plant hormones and control a range of cellular processes, including apical dominance, tropistic growth, lateral root formation, vascular tissue development, and regulation of plant senescence (1, 2). In Arabidopsis thaliana (thale cress), nearly 95% of auxins are found as amino acid and protein conjugates (3). Some of these molecules, such as IAA-aspartate (IAA-Asp) and IAA-glutamate, function as degradation intermediates, and others (IAA-alanine and IAA-leucine) provide a readily available metabolite pool for formation of free IAA (4–5). Biochemically, plants can modulate the levels of active auxin hormone by balancing the synthesis of the hormone with the formation of amide-linked conjugates (3). As part of this process, proteins of the GH3 family catalyze the ATP-dependent formation of auxin-amino acid conjugates (Fig. 1) (6).
FIGURE 1.
Overall reaction catalyzed by indole-acetic acid-amido synthetases.
Since the identification of the first GH3 transcript in soybean (Glycine max) by differential screening as an early auxin-responsive gene (6, 7), GH3-related genes have been found in various plant species. In the genome of Arabidopsis, the model plant of dicots, there are 19 GH3 genes (8–10). In addition, reports describe GH3 transcripts in Nicotiana tabacum (tobacco) (11), Citrus madurensis (calamondin) (12), Capsicum chinense (pungent pepper) (13), and Lycopersicon esculentum (tomato) (14). GH3 genes are also present in monocots. In the rice (Oryza sativa) genome, there are 13 GH3 genes (15–17). GH3 genes are also found in gymnosperms (Pinus pinaster) and moss (Physcomitrella patens) (18–20). Phylogenetic analysis of the GH3 family in Arabidopsis shows that the presence of three distinct groups (I, II, and III) (8–9). In rice, only groups I and II are present (15).
Biochemical analysis of members from each subgroup of the GH3 family demonstrates that these enzymes catalyze ligations of amino acids to jasmonic acid (JA) (group I), IAA (group II), salicylic acid (group II), and substituted benzoates (group III). Staswick et al. (8) were the first to examine the enzymatic function of a GH3 protein by analyzing GH3-11 (JAR1) from Arabidopsis. The Arabidopsis jar1 mutant was identified by screening mutants for reduced jasmonate hormone responses and the corresponding protein shown to catalyze the conjugation of isoleucine to JA to form isoleucyl-jasmonic acid, the active jasmonate hormone in plants (8). Subsequent analysis of Arabidopsis group II GH3 proteins (AtGH3-2, -3, -4, -5, -6, and -17) shows that these enzymes function as IAA-amido synthetases using ATP for the conjugation of various amino acids to IAA (14, 21–26). To date, only one group III member from Arabidopsis (AtGH3-12; PBS3) has been biochemically characterized and shown to accept 4-substituted benzoates as preferred substrates (25, 26). Analysis of multiple group II GH3 from rice (OsGH3-1, -8, and -13) also establish these proteins as IAA-amido synthetases involved in pathogen resistance and a range of auxin-dependent processes (27–30).
Mechanistic studies on the GH3 family of enzymes are limited to initial screening of acyl-acid substrate preference, amino acid use, and determination of basic kinetic parameters for only two members (OsGH3-8 and AtGH3-12) (8, 14, 21–30). The chemical mechanism of GH3 enzymes is postulated to consist of two half-reactions in which ATP is used to adenylate the carboxylate of an acyl molecule followed by nucleophilic attack of an amino acid to yield the conjugated amino acid product (Fig. 1) (8, 26). Here we evaluate the basic biochemical properties of OsGH3-8 and, as a representative GH3 family IAA-amido synthetase, determine its kinetic mechanism using initial velocity and product inhibition studies. In addition, we demonstrate the formation of a chemically competent adenylated-IAA reaction intermediate by mass spectrometry. This work provides insight on the molecular and biochemical basis for modulation of plant hormone action by enzymes of the GH3 family.
EXPERIMENTAL PROCEDURES
Materials
Generation of the pET28a-OsGH3.8 expression vector was previously described (28). Escherichia coli BL21-CodonPlus-RP cells were from Stratagene. Ni2+-nitrilotriacetic acid-agarose was bought from Qiagen. The HiLoad 26/60 Superdex-200 FPLC column was from Amersham Biosciences. All other reagents were purchased from Sigma.
Protein Expression and Purification
OsGH3-8 was heterologously expressed and purified as previously described (28). Briefly, E. coli BL21-CodonPlus-RP cells were transformed with pET28a-OsGH3-8 for protein expression. Cells were lysed by sonication, and the N-terminal His-tagged protein was purified by affinity chromatography and size-exclusion chromatography (28).
Enzymes Assay and Determination of Kinetic Parameters
IAA-amido synthetase activity of OsGH3-8 was assayed spectrophotometrically by coupling the formation of AMP to the reactions of myokinase, pyruvate kinase, and lactate dehydrogenase (31). The standard reaction performed at 25 °C contained 20 mm Tris-HCl (pH 8.0), 3 mm MgCl2, 1 mm ATP, 2 mm IAA, 3 mm aspartate, 1 mm dithiothreitol, 2 mm phosphoenolpyruvate, 200 μm NADH, 4 units of rabbit muscle myokinase, 4 units of rabbit muscle pyruvate kinase, and 4 units of rabbit muscle lactate dehydrogenase in a total of 200 μl. Initial velocities were determined using an Infinite 200 UV/visible microplate reader (Tecan) by monitoring the change in A340 nm (ϵ = 6220 m−1 cm−1). The ratio of product (i.e. IAA-Asp) formation is proportional to the rate of NADH conversion where two molecules of NADH are consumed for each molecule of IAA-Asp formed. Steady-state kinetic parameters were determined by initial velocity experiments. Measurements of the kcat and Km values for ATP (0.010–1 mm) were made at 2 mm IAA and 10 mm aspartate. Kinetic constants for IAA (0.025–2 mm) were measured at 1 mm ATP and 10 mm aspartate. Determination of the kinetic constants for aspartate (0.2–10 mm) used 1 mm ATP and 2 mm IAA. The aspartate concentration used for these assay was chosen to avoid substrate inhibition (28). The resulting data were fit to the Michaelis-Menten equation, v = kcat[S]/(Km + [S]), using Kaleidagraph (Synergy Software).
Characterization of Metal Ion Dependence and Substrate Specificity
Initial analysis of the metal ion dependence, amino acid specificity, and auxin-analog specificity of OsGH3-8 used the standard assay conditions with modifications. For screening of metal ion dependence, MgCl2 was replaced with 5 mm CaCl2, MnCl2, ZnCl2, or CuSO4. For analysis of Mg2+ and Mn2+ dependence, the metal concentration was varied (0.1–10 mm), and the resulting data were fit to the Michaelis-Menten equation. Screening of amino acid activity was performed by replacing aspartate with other amino acids (10 mm). Kinetic parameters for asparagine (2.5–75 mm) were determined at 1 mm ATP and 2 mm IAA. Analysis of OsGH3-8 activity using indole-3-butyric acid, phenyl-acetic acid (PAA), naphthalene-acetic acid (NAA), methyl-IAA, ethyl-IAA, 2,4-dichlorophenoxy-acetic acid, 3,6-dichloro-o-anisic acid (dicamba), and IAA hydrazide was performed by replacing IAA in the standard reaction with 1 mm concentrations of each compound. Kinetic parameters for indole-3-butyric acid (0.25–5 mm), PAA (0.1–5 mm), and NAA (0.1 - 5 mm) were determined at 1 mm ATP and 10 mm aspartate. Auxin-analog concentrations used in these assays were limited by solubility of the compound.
Kinetic Mechanism and Product Inhibition
Initial velocity experiments to analyze the kinetic mechanism were performed using standard assay conditions at varied concentrations of two substrates with the third substrate at a fixed concentration. Global curve-fitting in SigmaPlot (Systat Software) to the initial velocity data obtained from this matrix of substrate concentrations was used for modeling the data to the equations describing a Bi Uni Uni Bi Ping Pong mechanism, as follows: v = Vmax[A][B][C]/KiKmB[C] + KmC[A][B] + KmB[A][C] + KmA[B][C] + [A][B][C], where v is the initial velocity, Vmax is the maximum velocity, KA, KB, and KC are the Km values for substrates A, B, and C, respectively, and Ki is the substrate inhibition constant.
For product inhibition assays, enzymatic activity was measured in reactions under standard assay conditions containing either pyrophosphate (PPi) or IAA-Asp as the inhibitor and varied concentrations of one substrate with the other two at fixed concentrations. The PPi concentrations used in these assays did not inhibit the coupling system. The fitting analysis of data used SigmaPlot to fit data to the equations for competitive, v = Vmax/(1 + ((Km/[S])(1 + [I]/Ki))), noncompetitive, v = Vmax/((1 + [I]/Ki)(1 + Km[S])), or uncompetitive, v = Vmax/(1 + [I]/Ki + Km/[S]).
Mass Spectrometry and Analysis of the Adenylation Intermediate
To detect the adenylated intermediate, assays were performed at 25 °C with 50 mm Tris-HCl (pH 8.0), 3 mm MgCl2, 1 mm ATP, and 1 mm IAA (50 μl total volume) in the absence and presence of 1 mm aspartate. Reactions were initiated by the addition of 17 μg of OsGH3-8, and reactions without ATP, IAA, or OsGH3-8 were performed as controls. Additionally, the reaction was carried out with 1 mm PAA instead of IAA in both the absence and presence of aspartate to confirm that the peak detected was the predicted adenylated-acyl substrate intermediate. Assays were incubated for 1 min, and then 10 μl of each assay was added to 40 μl of 50% acetonitrile (v/v) and immediately analyzed by an ABI QSTAR XL (Applied Biosystems/MDS Sciex) hybrid quadrupole-time-of-flight MS/MS mass spectrometer equipped with a nanoelectrospray source (Protana XYZ manipulator). Negative mode nanoelectrospray was generated from borosilicate nanoelectrospray needles at −1.5 kV. TOF MS spectra were obtained using the Analyst QS software with an m/z range of 100–1100.
RESULTS
Steady-state Kinetic Analysis
Recombinant OsGH3-8 was overexpressed as a hexahistidine-tagged fusion protein in E. coli and purified to homogeneity by nickel-affinity and size-exclusion chromatography, as previously described (28). The steady-state kinetic parameters of OsGH3-8 for ATP, IAA, and aspartate were determined using a spectrophotometric assay (Table 1). The parameters obtained with this assay are similar to those previously determined using a mass spectrometry-based assay as follows: V/EtIAA = 20.3 min−1; KmIAA = 123 μm; V/EtATP = 14.1 min−1; KmATP = 50 μm; V/EtAsp = 28.8 min−1; KmAsp = 1580 μm (28). The kinetic values for OsGH3-8 are comparable to those reported for AtGH3-12 (26), the only other GH3 family member kinetically characterized.
TABLE 1.
Steady-state kinetic parameters for OsGH3–8
Reactions were performed as described under “Experimental Procedures.” All kinetic parameters are expressed as the mean ± S.E. for n = 3.
| Substrate | V/Et | Km | Kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1s−1 | |
| IAA | 22.1 ± 0.3 | 187 ± 18 | 1970 |
| ATP | 20.1 ± 1.3 | 36.0 ± 2.0 | 9305 |
| Aspartate | 19.6 ± 0.5 | 3910 ± 450 | 83.5 |
Metal Ion Dependence
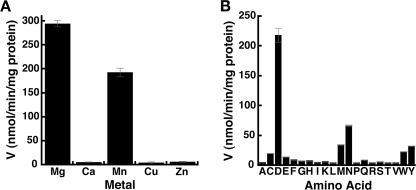
Because OsGH3-8 catalyzes an ATP-dependent reaction, we screened the effect of divalent metals (MgCl2, CaCl2, MnCl2, ZnCl2, and CuSO4) on enzymatic activity. The enzyme was active with Mg2+ and Mn2+ (Fig. 2A). The effect of metal ion concentration on OsGH3-8 activity was tested and yielded a V/Et = 25.3 ± 1.3 min−1 and Km = 515 ± 90 μm for MgCl2 and V/Et = 29.7 ± 4.6 min−1 and Km = 1990 ± 172 μm for MnCl2. These results indicate a 4-fold preference for Mg2+ over Mn2+ in the OsGH3-8 active site.
FIGURE 2.
Metal ion dependence and amino acid specificity of OsGH3-8. All assays were performed under standard reaction conditions as described under “Experimental Procedures.” Data shown are an average (n = 3) with S.E. indicated. A, activity of OsGH3-8 with divalent metals (5 mm) is shown. B, activity of OsGH3-8 with amino acids (10 mm) is shown. One-letter abbreviations for amino acids are used.
Amino Acid and Auxin-Analog Specificities
The Arabidopsis GH3 family members analyzed to date accept a range of amino acid substrates (8, 14, 26). To examine the amino acid specificity of OsGH3-8, aspartate was replaced with the other 19 amino acids in assays (Fig. 2B). The highest specific activity was observed with aspartate, asparagine was the next most active, and low activity was observed with methionine, tyrosine, and tryptophan. Velocity versus substrate analysis of OsGH3-8 for asparagine yielded the following kinetic values: V/Et = 7.6 ± 1.1 min−1; Km = 67.8 ± 17.6 mm; kcat/Km = 1.87 m−1 s−1. Overall, asparagine is 45-fold less efficient as a substrate for OsGH3-8 compared with aspartate. This suggests an important role for the aspartate side-chain carboxylate as a specificity determinant in the active site. Although methionine, tyrosine, and tryptophan also showed low activity (Fig. 2B), solubility of these amino acids prevented accurate kinetic analysis.
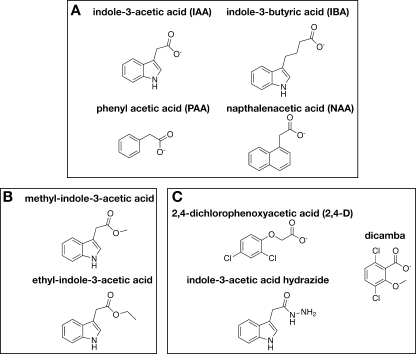
To probe the acyl group specificity of OsGH3-8, a range of auxin analogs were used (Fig. 3). Of these compounds, indole-3-butyric acid, PAA, and NAA (Fig. 3A) were accepted as substrates with 4.9-, 1.4-, and 8.9-fold reductions in catalytic efficiency, respectively, compared with IAA (Table 2). Although methyl- and ethyl-IAA were not substrates, these compounds inhibited OsGH3-8 with Ki values of 800 ± 283 and 560 ± 138 μm, respectively. Dicamba, 2,4-dichlorophenoxy-acetic acid, and IAA hydrazide (1 mm each) were neither substrates nor inhibitors of OsGH3-8.
FIGURE 3.
Chemical structures of auxin analogs. Structures of molecules that were substrates (A), inhibitors (B), or inactive with OsGH3-8 (C) are shown.
TABLE 2.
Kinetic analysis of OsGH3–8 with auxin analogs
Reactions were performed as described under “Experimental Procedures.” All kinetic parameters are expressed as the mean ± S.E. for n = 3.
| Substrate | V/Et | Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1s−1 | |
| Indole-3-butyric acid | 65.2 ± 4.7 | 2,700 ± 410 | 402 |
| PAA | 45.6 ± 1.7 | 540 ± 60 | 1410 |
| NAA | 13.3 ± 1.0 | 1000 ± 210 | 222 |
Initial Velocity Analysis of the Kinetic Mechanism
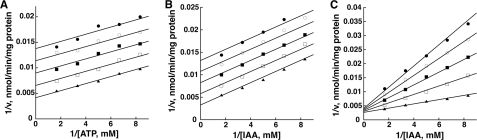
The kinetic mechanism of OsGH3-8 was determined by initial velocity experiments using a coupled enzyme assay in which two substrates were varied, whereas one substrate was maintained at a constant concentration (Fig. 4). The parallel lines observed in the double-reciprocal plots using either varied ATP and aspartate with fixed IAA (Fig. 4A) or varied IAA and aspartate with fixed ATP (Fig. 4B) indicate a Ping Pong type of mechanism for OsGH3-8. These two sets of parallel lines indicate that ATP and IAA are involved in the first half-reaction of the ping-pong mechanism. The intersecting lines observed in the double-reciprocal plot obtained with varied ATP and IAA at fixed aspartate (Fig. 4C) indicate that aspartate is the substrate that adds between the two product release steps in the kinetic mechanism and a substrate for the second half-reaction.
FIGURE 4.
Initial velocity analysis of the kinetic mechanism of OsGH3-8. Experimental data are indicated by the symbols in the double-reciprocal plots of the substrate variation experiments. The lines represent the global fit of all data to the equation for a Bi Uni Uni Bi Ping Pong mechanism. All assays were performed as described under “Experimental Procedures.” A, shown is a double-reciprocal plot for 1/v versus 1/(ATP, 0.12–0.6 mm) at a fixed IAA concentration (2 mm) with varied aspartate (1.2, 1.5, 2.0, 3.0, 6.0 mm; top to bottom). B, shown is a double-reciprocal plot for 1/v versus 1/(IAA, 0.12–0.6 mm) at a fixed ATP concentration (2 mm) with varied aspartate (1.2, 1.5, 2.0, 3.0, 6.0 mm; top to bottom). C, shown is a double-reciprocal plot for 1/v versus 1/(IAA, 0.12–0.6 mm) at a fixed aspartate concentration (10 mm) with varied ATP (0.12, 0.15, 0.2, 0.3, 0.6 mm; top to bottom) (C).
The initial velocity data were globally fit to the equation describing a Bi Uni Uni Bi Ping Pong mechanism with fitted kinetic parameters summarized in Table 3. Overall, the calculated kinetic parameters from the global fit (Table 3) are in good agreement with the experimentally determined kinetic parameters (Table 1).
TABLE 3.
Summary of fitted kinetic parameters
Parameters were derived by global fitting of initial velocity data to the equation for a Bi Uni Uni Bi Ping Pong kinetic mechanism as described under “Experimental Procedures.”
| Parameter | Value |
|---|---|
| Vmax (nmol min−1 mg protein−1) | 451 ± 77 |
| V/Et (min−1) | 30.1 ± 5.2 |
| KmA (IAA, μm) | 182 ± 27 |
| KmB (ATP, μm) | 115 ± 29 |
| KmC (Asp, μm) | 1060 ± 359 |
| Ki (Asp, μm) | 7350 ± 1006 |
Product Inhibition
Product inhibition studies were performed to determine the order of substrate addition and product release (supplemental Fig. 1). The resulting inhibition patterns and constants determined from best fits of the inhibition data are summarized in Table 4.
TABLE 4.
Product inhibition patterns
All assays were performed as described under “Experimental Procedures.” The inhibition patterns indicate the fit of the data (r2) to one of the three types of inhibition as follows: C, competitive; NC, non-competitive; UC, uncompetitive; –, no inhibition observed.
| Varied substrate | Inhibitor | Fixed substrates | Inhibition (r2) | Ki |
|---|---|---|---|---|
| mm | ||||
| ATP | PPi | IAA (0.15 mm), Asp (2 mm) | NC (0.997) | 3.52 ± 0.11 |
| ATP | PPi | IAA (1.0 mm), Asp (2 mm) | UC (0.959) | 0.16 ± 0.03 |
| ATP | PPi | Asp (10 mm), IAA (0.15 mm) | – | – |
| IAA | PPi | ATP (0.1 mm), Asp (2 mm) | NC (0.992) | 0.18 ± 0.01 |
| IAA | PPi | ATP (1.0 mm), Asp (2 mm) | NC (0.979) | 0.35 ± 0.03 |
| IAA | PPi | Asp (10 mm), ATP (0.15 mm) | – | – |
| Aspartate | PPi | IAA (0.15 mm), ATP (0.15 mm) | C (0.995) | 1.32 ± 0.07 |
| ATP | IAA-Asp | IAA (0.15 mm), Asp (2 mm) | UC (0.986) | 2.10 ± 0.37 |
| IAA | IAA-Asp | ATP (0.15 mm), Asp (2 mm) | UC (0.992) | 0.61 ± 0.18 |
| Aspartate | IAA-Asp | ATP (0.15 mm), IAA (0.15 mm) | NC (0.987) | 3.06 ± 0.42 |
The proposed product of the first half-reaction (i.e. PPi) was used as an inhibitor versus each substrate. Using PPi as an inhibitor versus varied ATP concentrations (and fixed aspartate), non-competitive inhibition was evident at an unsaturating IAA concentration, and uncompetitive inhibition was observed at a saturating ATP concentration. With varied IAA concentrations (and fixed aspartate), PPi acted as a non-competitive inhibitor at saturating and unsaturating concentrations of IAA. PPi did not inhibit OsGH3-8 under saturating aspartate conditions with either varied ATP or IAA concentrations. These data suggest that ATP binds to the enzyme before IAA in the first half-reaction (32). Competitive inhibition by PPi versus varied aspartate was observed and indicates that both molecules bind to the same form of the enzyme. This implies that PPi is released from an intermediate-bound form of the enzyme before aspartate binds.
To determine the order of product release, IAA-Asp was used as an inhibitor. Inhibition of OsGH3-8 by IAA-Asp displayed uncompetitive inhibition versus both ATP and IAA and non-competitive inhibition versus aspartate. These inhibition patterns are consistent with the release of IAA-Asp before release of the final product, which by process of elimination is AMP (32). Product inhibition by AMP could not be determined because of coupling system inhibition.
Detection of Adenylated Intermediate
Mass spectrometry was used to detect the adenylated intermediate formed by OsGH3-8 after incubation with IAA and ATP. For the IAA + ATP reaction in the absence of aspartate, a peak was detected at m/z = 504.326 that was not present in controls lacking either protein or substrates (supplemental Fig. 2A). The observed mass is consistent with a predicted m/z of an adenylated IAA molecule (IAA-AMP; Mr theoretical = 504.397). Analysis of the reaction after the addition of aspartate showed a loss in the IAA-AMP intermediate peak abundance with a concomitant appearance of an m/z = 291.201 peak corresponding to IAA-Asp (Mr theoretical = 291.284) (supplemental Fig. 2B) (28). A similar set of experiments was performed using PAA in place of IAA. Incubation of OsGH3-8 in the presence of PAA and ATP yielded a peak with m/z = 465.318, which corresponds with the predicted m/z of an adenylated PAA molecule (Mr theoretical = 465.360). After the addition of aspartate, mass spectrometry analysis showed a loss in signal of this peak. These experiments demonstrate the formation of the adenylated intermediate with the natural substrate and an auxin analog in the first half-reaction and verify the chemical competence of the observed intermediates for the second half-reaction.
DISCUSSION
In plants, the GH3 family of enzymes are involved in a range of hormone-dependent processes, including root growth, flowering, hypocotyl growth, and disease resistance (23, 25, 27, 29–30, 33–38). For example, the type I AtGH3-11 (JAR1) catalyzes the formation of the active jasmonate hormone isoleucyl-jasmonic acid, which promotes interaction of the F-box protein COI1 to degrade JAZ repressor proteins and activate jasmonate responses (39–41). Similarly, conjugation of IAA by type II GH3 to form IAA-Asp and IAA-Glu can modulate activity of the auxin receptor TIR1 by inactivating the auxin hormone and attenuating downstream events (42–43). Although the physiological roles of many GH3 proteins are being established, the molecular basis for how GH3 proteins recognize a range of acyl-acid substrates and different amino acids and catalyze a conjugation reaction remain to be established. Here we examine the catalytic properties of the IAA-amido synthetase OsGH3-8 and use this enzyme to determine the general kinetic and chemical mechanisms for the GH3 enzyme family.
Metal ions are essential for the activity of OsGH3-8. We compared the effects of various divalent cations and found the enzyme most active with Mg2+ and slightly less so with Mn2+ (Fig. 2A), as expected for an ATP-dependent enzyme.
Most of the group II GH3 proteins conjugate IAA to a range of amino acids (14). Six Arabidopsis GH3 (GH3-2, -3, -4, -5, -6, and -17) were previously shown to accept the acidic amino acids (aspartate and glutamate) along with methionine and tryptophan as substrates (14). Detailed analysis of OsGH3-8 shows that this enzyme accepts aspartate with a 45-fold preference over asparagine, the next most efficient amino acid substrate (Fig. 2B). This demonstrates that the carboxylate moiety of the aspartate side chain is critical for substrate recognition by OsGH3-8.
Previous analysis of OsGH3-8 established this enzyme as an IAA-amido synthetase (27–28). Here we examined the ability of the enzyme to accept other auxin analogs as substrates (Fig. 3 and Table 2). Of the substrates tested, the catalytic efficiency of PAA was comparable with that of IAA. Both substrates share an acetyl group with the phenyl ring of PAA approximating the position of the pyrrole ring of IAA. Kinetic analysis suggests that PAA readily fits within the active site region used for IAA binding. A longer acyl-carboxylate chain (i.e. butyl versus acetyl) is not preferred but can be accommodated with a 5-fold reduction in kcat/Km. If the acetate group of IAA and NAA were bound in the same orientation within the OsGH3-8 active site, then the naphthalene ring of NAA would be slightly skewed relative to the corresponding indole ring of IAA. This likely leads to steric clashes in the active site and the observed 9-fold decrease in catalytic efficiency with NAA versus IAA as a substrate. The methyl- and ethyl-IAA analogs, which cannot undergo catalysis, bind to the active site as inhibitors with Ki values 3–4-fold higher than the Km value for IAA. Given that ATP binds before IAA in the OsGH3-8 active site, it is likely that binding of the nucleotide slightly alters binding of these inhibitors relative to binding of an authentic substrate. Other auxin analogs (2,4-dichlorophenoxy-acetic acid, dicamba, and IAA hydrazide) did not bind to OsGH3-8 as either substrates or inhibitors at the concentrations used for these assays. Currently, the molecular determinants of acyl-acid specificity between the group I (JA), II (IAA and salicylic acid), and III (benzoate) GH3 proteins and of amino acid substrate specificity within each group remain unclear.
A detailed kinetic analysis of any GH3 enzyme has not been performed before. A number of possible three substrate-three product (i.e. Ter Ter) kinetic mechanisms are possible (32). Double-reciprocal plots of the initial velocity data clearly demonstrate that OsGH3-8 uses a Ping Pong mechanism for catalysis with ATP and IAA as substrates in the first half-reaction based on the observed parallel lines (Fig. 4, A and B), and aspartate as a substrate in the second half-reaction (Fig. 4C). The observed parallel lines eliminated a possible ordered Ter Ter mechanism and global fitting to other Ping Pong type kinetic mechanisms yielded the best fit of the data to a Bi Uni Uni Bi Ping Pong mechanism with fitted kinetic parameters that are similar to those observed experimentally (Tables 1 and 3). This is in agreement with the proposed chemical mechanism for GH3 proteins in which ATP and IAA are substrates for an adenylation reaction followed by the addition of aspartate for the formation of an amido-conjugate product (Fig. 1) (14). Next, we used product inhibition studies to determine whether substrate addition and product release occurred in either an ordered or random fashion.
Using PPi and IAA-Asp as inhibitors versus each substrate distinguishes the order of substrate binding and product release from OsGH3-8 (Table 4 and supplemental Fig. 1). The observed PPi inhibition patterns versus ATP, IAA, and aspartate are most consistent with an ordered addition of ATP followed by IAA in the first half-reaction (32). If ATP bound after IAA, then the PPi inhibition patterns for the substrates would be reversed. Competitive inhibition by PPi versus aspartate indicates that these ligands bind to the same form of the enzyme and, based on the proposed chemistry and the initial velocity experiments, this must be the adenylate-intermediate-bound form of OsGH3-8. IAA-Asp was used to determine the order of product release in the second half-reaction. The observed inhibitions patterns are most consistent with IAA-Asp leaving before AMP release. If IAA-Asp was the last product to leave the active site, then competitive inhibition would be observed versus ATP with non-competitive and competitive inhibition versus IAA and aspartate, respectively (32). Overall, experimental analysis of OsGH3-8 supports the proposed reaction sequence for the GH3 enzymes.
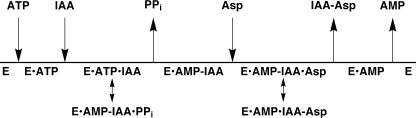
The initial velocity and product inhibition data lead to the Bi Uni Uni Bi Ping Pong mechanism shown in Fig. 5. In this mechanism ATP binds to the free enzyme followed by the addition of IAA to form an E·ATP·IAA ternary complex. The first half-reaction leads to release of PPi with formation of the proposed IAA-adenylate intermediate in the active site. In the second half-reaction, aspartate binds to the enzyme, and the amide bond between the amino acid and IAA is formed with cleavage of the adenylate anhydride. Last, IAA-Asp and AMP are sequentially released from the enzyme active site.
FIGURE 5.
Steady-state kinetic mechanism of OsGH3-8. The reaction sequence OsGH3-8 is diagrammed from left to right. Arrows show the addition or release of various substrates. The double-headed arrows represent the chemical reactions.
This work also provides the first direct evidence of an adenylated intermediate in a GH3-catalyzed reaction mechanism. Previously, ATP-[32P]PPi isotope exchange experiments with AtGH3-11 (JAR1) in the presence of JA showed an exchange of radiolabel into ATP, suggesting the presence of a reaction intermediate (8). Later, Okrent et al. (26) monitored the activity of AtGH3-12 (PBS3) using a PPi-dependent assay, which was consistent with an adenylation step in the first half-reaction. Using mass spectrometry, we now demonstrate that an adenylated intermediate is formed in an enzyme-specific reaction using ATP and either IAA or PAA as substrates. Moreover, the intermediate is catalytically competent as the addition of aspartate leads to loss of the intermediate and formation of the reaction product.
The metabolic utility of adenylation for the linking of two substrates plays a role in many enzyme reactions. Multiple other ATP-dependent enzymes, including Bradyrhizobium japonicum malonyl-CoA synthetase, E. coli fatty acyl-CoA synthetase, rat threonyl- and E. coli prolyl-aminoacyl-tRNA synthetases, Mycobacterium tuberculosis panthothenate synthetase, and Mycobacterium smegmatis cysteine ligase (44–50) use an acyl-adenylate intermediate in a Bi Uni Uni Bi Ping Pong mechanism for catalysis. This work demonstrates the biochemical role for formation of adenylated intermediates to processes associated with plant hormone regulation and further broadens the biological versatility of enzymes using this mechanistic strategy of chemical ligation.
Given the sequence similarity of GH3 family members in various plants (8–20), the kinetic mechanism of OsGH3-8 is likely shared across these enzymes. A significant gap in understanding how GH3 functions is a lack of structural information that defines the architecture of the catalytic, acyl-substrate binding, and amino acid substrate binding sites. Knowledge of the kinetic mechanism will aid in determining the combination of ligands to use for structural studies on these enzymes. In addition, quantification of kinetic parameters may better define the in vivo function the GH3 proteins. Earlier studies of GH3 enzymes employed qualitative assays and suggested promiscuous amino acid specificity (14). Similarly, assays of OsGH3-8 with a screen of amino acids suggests possible use of substrates other than aspartate; however, kinetic analysis demonstrates a preference for aspartate with the catalytic efficiencies of other possible amino acid substrates requiring non-physiological concentrations for effective turnover. Accurate definition of GH3 enzyme specificities will help to refine the biological roles of these enzymes and the extent of cross-talk in plant hormone responses and metabolism.
In summary, the GH3 enzymes are important regulators of plant hormone action but are poorly understood at the mechanistic level. Using OsGH3-8, we have investigated the catalytic properties of this enzyme, established the kinetic mechanism for the family of GH3 enzymes, and demonstrated the formation of an adenylated reaction intermediate. Additional structural and functional studies are needed to elucidate how members of this enzyme family recognize diverse acyl and amino acid substrate.
Supplementary Material
This work was supported by funds from the 111 Program (B07041) for the Huazhong Agricultural University-Donald Danforth Plant Science Center Joint Laboratory. This work was also supported by National Natural Science Foundation of China Grant 30930063 (to S. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- IAA
- indole-3-acetic acid (auxin)
- dicamba
- 3,6-dichloro-o-anisic acid
- IAA-Asp
- aspartyl indole-3-acetic acid
- JA
- jasmonic acid
- NAA
- napthalen-acetic acid
- OsGH3-8
- rice indole-3-acetic acid-amido synthetase GH3-8
- PAA
- phenyl-acetic acid.
REFERENCES
- 1.Woodward A. W., Bartel B. (2005) Ann. Bot. 95, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teale W. D., Paponov I. A., Palme K. (2006) Nat. Rev. Mol. Cell Biol. 7, 847–859 [DOI] [PubMed] [Google Scholar]
- 3.Ljung K., Hull A. K., Kowalczyk M., Marchant A., Celenza J., Cohen J. D., Sandberg G. (2002) Plant Mol. Biol. 49, 249–272 [PubMed] [Google Scholar]
- 4.LeClere S., Tellez R., Rampey R. A., Matsuda S. P., Bartel B. (2002) J. Biol. Chem. 277, 20446–20452 [DOI] [PubMed] [Google Scholar]
- 5.Rampey R. A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., Bartel B. (2004) Plant Physiol. 135, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagen G., Kleinschmidt A., Guilfoyle T. (1984) Planta 162, 147–153 [DOI] [PubMed] [Google Scholar]
- 7.Hagen G., Guilfoyle T. J. (1985) Mol. Cell. Biol. 5, 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staswick P. E., Tiryaki I., Rowe M. L. (2002) Plant Cell 14, 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Tian C.-e., Duan J., Wu K. (2008) Plant Growth Regul. 56, 225–232 [Google Scholar]
- 10.Hagen G., Guilfoyle T. (2002) Plant Mol. Biol. 49, 373–385 [PubMed] [Google Scholar]
- 11.Roux C., Perrot-Rechenmann C. (1997) FEBS Lett. 419, 131–136 [DOI] [PubMed] [Google Scholar]
- 12.Lahey K. A., Yuan R., Burns J. K., Ueng P. P., Timmer L. W., Kuang-Ren C. (2004) Mol. Plant Microbe Interact. 17, 1394–1401 [DOI] [PubMed] [Google Scholar]
- 13.Liu K., Kang B. C., Jiang H., Moore S. L., Li H., Watkins C. B., Setter T. L., Jahn M. M. (2005) Plant Mol. Biol. 58, 447–464 [DOI] [PubMed] [Google Scholar]
- 14.Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., Suza W. (2005) Plant Cell 17, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M., Kaur N., Tyagi A. K., Khurana J. P. (2006) Funct. Integr. Genomics 6, 36–46 [DOI] [PubMed] [Google Scholar]
- 16.Terol J., Domingo C., Talón M. (2006) Gene 371, 279–290 [DOI] [PubMed] [Google Scholar]
- 17.Jain M., Khurana J. P. (2009) FEBS J. 276, 3148–3162 [DOI] [PubMed] [Google Scholar]
- 18.Reddy S. M., Hitchin S., Melayah D., Pandey A. K., Raffier C., Henderson J., Marmeisse R., Gay G. (2006) New Phytol. 170, 391–400 [DOI] [PubMed] [Google Scholar]
- 19.Bierfreund N. M., Tintelnot S., Reski R., Decker E. L. (2004) J. Plant Physiol. 161, 823–835 [DOI] [PubMed] [Google Scholar]
- 20.Ludwig-Müller J., Jülke S., Bierfreund N. M., Decker E. L., Reski R. (2009) New Phytol. 181, 323–338 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Li Q., Li Z., Staswick P. E., Wang M., Zhu Y., He Z. (2007) Plant Physiol. 145, 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Wang M., Li Z., Li Q., He Z. (2008) Plant Signal. Behav. 3, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan S., Stone J. M. (2007) Planta 226, 21–34 [DOI] [PubMed] [Google Scholar]
- 24.Khan S., Stone J. M. (2007) Plant Signal. Behav. 2, 483–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobuta K., Okrent R. A., Stoutemyer M., Rodibaugh N., Kempema L., Wildermuth M. C., Innes R. W. (2007) Plant Physiol. 144, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okrent R. A., Brooks M. D., Wildermuth M. C. (2009) J. Biol. Chem. 284, 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. (2008) Plant Cell 20, 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q., Zhang B., Hicks L. M., Wang S., Jez J. M. (2009) Anal. Biochem. 390, 149–154 [DOI] [PubMed] [Google Scholar]
- 29.Domingo C., Andrés F., Tharreau D., Iglesias D. J., Talón M. (2009) Mol. Plant Microbe Interact. 22, 201–210 [DOI] [PubMed] [Google Scholar]
- 30.Zhang S. W., Li C. H., Cao J., Zhang Y. C., Zhang S. Q., Xia Y. F., Sun D. Y., Sun Y. (2009) Plant Physiol. 151, 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfleiderer G., Kreiling A., Wieland T. (1960) Biochem. Z. 333, 302–307 [PubMed] [Google Scholar]
- 32.Segel I. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Kinetic Systems, John Wiley and Sons, Inc., New York [Google Scholar]
- 33.Takase T., Nakazawa M., Ishikawa A., Kawashima M., Ichikawa T., Takahashi N., Shimada H., Manabe K., Matsui M. (2004) Plant J. 37, 471–483 [DOI] [PubMed] [Google Scholar]
- 34.Tanaka S., Mochizuki N., Nagatani A. (2002) Plant Cell Physiol. 43, 281–289 [DOI] [PubMed] [Google Scholar]
- 35.Park J. E., Park J. Y., Kim Y. S., Staswick P. E., Jeon J., Yun J., Kim S. Y., Kim J., Lee Y. H., Park C. M. (2007) J. Biol. Chem. 282, 10036–10046 [DOI] [PubMed] [Google Scholar]
- 36.Park J. E., Seo P. J., Lee A. K., Jung J. H., Kim Y. S., Park C. M. (2007) Plant Cell Physiol. 48, 1236–1241 [DOI] [PubMed] [Google Scholar]
- 37.Nakazawa M., Yabe N., Ichikawa T., Yamamoto Y. Y., Yoshizumi T., Hasunuma K., Matsui M. (2001) Plant J. 25, 213–221 [DOI] [PubMed] [Google Scholar]
- 38.Jagadeeswaran G., Raina S., Acharya B. R., Maqbool S. B., Mosher S. L., Appel H. M., Schultz J. C., Klessig D. F., Raina R. (2007) Plant J. 51, 234–246 [DOI] [PubMed] [Google Scholar]
- 39.Chini A., Fonseca S., Fernández G., Adie B., Chico J. M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F. M., Ponce M. R., Micol J. L., Solano R. (2007) Nature 448, 666–671 [DOI] [PubMed] [Google Scholar]
- 40.Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E. E. (2007) Plant Cell 19, 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H. S., Koo A. J., Gao X., Jayanty S., Thines B., Jones A. D., Howe G. A. (2008) Plant Physiol. 146, 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dharmasiri N., Dharmasiri S., Estelle M. (2005) Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- 43.Kepinski S., Leyser O. (2005) Nature 435, 446–451 [DOI] [PubMed] [Google Scholar]
- 44.Kim Y. S., Kang S. W. (1994) Biochem. J. 297, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Black P. N., DiRusso C. C., Sherin D., MacColl R., Knudsen J., Weimar J. D. (2000) J. Biol. Chem. 275, 38547–38553 [DOI] [PubMed] [Google Scholar]
- 46.Zheng R., Blanchard J. S. (2001) Biochemistry 40, 12904–12912 [DOI] [PubMed] [Google Scholar]
- 47.Allende C. C., Chaimovich H., Gatica M., Allende J. E. (1970) J. Biol. Chem. 245, 93–101 [PubMed] [Google Scholar]
- 48.Papas T. S., Mehler A. H. (1971) J. Biol. Chem. 246, 5924–5928 [PubMed] [Google Scholar]
- 49.Fan F., Luxenburger A., Painter G. F., Blanchard J. S. (2007) Biochemistry 46, 11421–11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bovee M. L., Pierce M. A., Francklyn C. S. (2003) Biochemistry 42, 15102–15113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.