Abstract
Most cells produce ATP in the mitochondria by oxidative phosphorylation. However, macrophages, which are major players in the innate immune system, use aerobic glycolysis to produce ATP. HIF-1 (hypoxia-inducible factor-1) regulates expression of glycolysis-related genes and maintains macrophage glycolytic activity. However, it is unclear how HIF-1 activity is maintained in macrophages during normoxia. In this study, we found that macrophages lacking membrane type 1 matrix metalloproteinase (MT1-MMP/MMP-14), a potent invasion-promoting protease, exhibited considerably lower ATP levels than wild-type cells. HIF-1 was activated by an unanticipated function of MT1-MMP, which led to the stimulation of ATP production via glycolysis. The cytoplasmic tail of MT1-MMP bound to FIH-1 (factor inhibiting HIF-1), which led to the inhibition of the latter by its recently identified inhibitor, Mint3/APBA3. We have thus identified a new function of MT1-MMP to mediate production of ATP so as to support energy-dependent macrophage functions by a previously unknown non-proteolytic mechanism.
Keywords: Glycolysis, Hypoxia, Macrophage, Matrix Metalloproteinase, Transcription Factors
Introduction
Macrophages are major players in the innate immune system and regulate early inflammatory responses against invading pathogens (1, 2). During inflammation, macrophages produce cytokines, growth factors, and reactive oxygen molecules, thereby contributing to the pathology of various diseases (1–3). Inflammation often leads to hypoxic conditions, and macrophages must be able to survive and function following migration into hypoxic inflamed tissues. Most cellular functions are dependent upon energy production, and macrophages are characterized by a unique system to maintain production of ATP following movement from a normoxic to a hypoxic environment.
Most cells produce ATP by oxidative phosphorylation under normoxic conditions. However, oxidative phosphorylation is inefficient under hypoxic conditions, and cells must rely on anaerobic glycolysis for energy production as a result of adaptation to hypoxic stress. Macrophages, however, are unique in using glycolysis for ATP production constitutively rather than oxidative phosphorylation even during normoxia. This characteristic is advantageous for macrophages, which must move into and function in a hypoxic environment during an inflammatory response. Glycolytic activity is regulated by HIF-1, whose activity is stimulated by hypoxia and leads to increased expression of glycolysis-related genes (4, 5). HIF-1 is a heterodimeric transcription factor composed of regulatory (α) and non-regulatory (β) subunits, which binds to hypoxia response elements (HREs)2 localized within the promoter regions of target genes. HIF-1α activity is regulated in response to cellular oxygen tension. Loss of HIF-1α activity in macrophages by targeted disruption of its gene has been reported to reduce ATP production to 20% of that observed in wild-type cells (6). The resulting reduction in ATP concentration causes a severe defect in energy-dependent macrophage functions, including the recruitment of cells to acutely inflamed tissues, cytokine production, migration, and invasion of tissue (6). However, it is unclear how HIF-1 activity is maintained in macrophages during normoxic conditions, which would otherwise suppress HIF-1 activity in other types of cells.
Macrophages must traverse various tissue barriers by degrading components of the extracellular matrix so as to move to sites of inflammation. Degradation of the extracellular matrix is mediated by the concerted action of multiple proteases expressed by macrophages (7, 8). MT1-MMP is one such invasion-promoting protease expressed in macrophages (9) and other types of invasive cells, such as malignant tumor cells (7, 10–13). Substrates of MT1-MMP include not only extracellular matrix proteins but multiple other proteins localized to the pericellular milieu. As such, the proteolytic action of MT1-MMP regulates a variety of cellular functions (7, 10–13). Indeed, MT1-MMP knock-out (MT1−/−) mice display defects in multiple cell types and do not survive beyond weaning (14–17).
We have recently reported that infiltration of macrophages into sites of TPA-induced acute inflammation is impaired in MT1−/− mice (9), although infiltration of such macrophages into sites of chronic inflammation has been reported to be unaffected in a separate study (18). Macrophages derived from monocytes isolated from the bone marrow of MT1−/− mice exhibit reduced invasive activity due to the lack of proinvasive MT1-MMP. MT1−/− macrophages also exhibit reduced migratory activity, and this defect is overcome by the expression of recombinant exogenous MT1-MMP in the cells. However, the migration-promoting activity of MT1-MMP is not associated with its protease activity; rather, it is linked to its cytoplasmic tail (CPT) (9). Thus, MT1-MMP promotes macrophage migration in a non-proteolytic manner. In this study, we show that the concentration of ATP in MT1−/− macrophages is 60% lower than that in WT cells due to reduced glycolytic activity in the knock-out cells. This unanticipated role for MT1-MMP in regulating aerobic glycolysis in macrophages is substantiated further by the identification of a signaling cascade that links MT1-MMP to HIF-1α.
EXPERIMENTAL PROCEDURES
Plasmids
The mouse FIH-1 (factor inhibiting HIF-1) cDNA was amplified from macrophages by RT-PCR. Human FIH-1 and Mint3 cDNAs were amplified from HEK293 cells by RT-PCR. Expression constructs encoding Mint3-NT (encoding amino acids 1–214) or mutant MT1-MMPs were prepared as described previously (9, 19). These proteins were expressed in cells using lentivirus (pLenti6) vectors (Invitrogen). The mammalian expression vector pcDNA3.1 (Invitrogen) was used to express proteins for immunoprecipitation and reporter assays.
Mouse Strains
Mt1-mmp+/− mice (9) were backcrossed against C57BL/6 mice 12 times. Mice were maintained under specific pathogen-free conditions. Experiments were performed on 10–18-day-old littermates of WT and MT1−/− mice.
Cell Culture
Bone marrow-derived macrophages were obtained as described previously (9, 19). Mouse embryonic fibroblasts (MEFs) were prepared as described previously (20) and cultured in DMEM supplemented with 10% FBS. HEK293 cells were purchased from ATCC and cultured in DMEM high glucose (Sigma) supplemented with 10% FBS. For the experiments under normoxia, cells were cultured with 21% O2 and 5% CO2. For the experiments under hypoxia, cells were cultured with 1% O2 and 5% CO2 in a model 9200 incubator (Wakenyaku) for 24 h.
Measurement of Lactic Acid
Cells were seeded onto 6-well plates (1 × 106/well) in triplicate. Conditioned medium was collected after 24 h, and lactic acid was measured using an l-lactic acid kit (R-Biopharm).
Measurement of ATP Concentrations
ATP levels in cultured cells were determined using the ATP Bioluminescence Assay Kit CLS II (Roche Applied Science). ATP levels were normalized according to the total protein concentration, determined using a Bradford assay kit (Bio-Rad).
Detection of HIF-1α and p300/CBP
Nuclear proteins were prepared from cells using a nuclear extract kit (Active Motif), and 10 μg of nuclear lysate per reaction was analyzed with the TransAM HIF-1 kit (Active Motif), according to the manufacturer's protocol. This kit comprises assay plates coated with an HRE oligonucleotide that binds HIF-1. Binding of HIF-1 was then detected using an anti-HIF-1α antibody. To detect p300/CBP, anti-p300/CBP antibodies (clones RW109, RW105, and RW128 mixed antibodies, Upstate) were used instead of the anti-HIF-1α antibody.
RNA Isolation, Reverse Transcription, and Real-time PCR
Total RNA isolation, reverse transcription, and real-time PCR were performed as described previously (19). The following specific primers were used: β-actin sense (5′-gccaacacagtgctgtctgg-3′) and antisense (5′-atctgctggaaggtggacag-3′), GLUT-1 sense (5′-gggcatgtgcttccagtatgt-3′) and antisense (5′-acgaggagcaccgtgaagat-3′), and PGK-1 sense (5′-tgtcgctttccaacaagctg-3′) and antisense (5′-ggtggctcataaggacaacg-3′).
Preparation of Lentiviral Vectors
Lentiviral vectors were prepared as described previously (9, 19). Lentiviral vectors were transduced into macrophages at a multiplicity of infection of 3, and MT1-MMP expression was detected by RT-PCR using the following specific primers: MT1-MMP sense (5′-atgtctcccgcccctcgacc-3′) and antisense (5′-acattggccttgatctcagt-3′) and β-actin sense (5′-gccaacacagtgctgtctgg-3′) and antisense (5′-atctgctggaaggtggacag-3′).
Reporter Assay
The reporter assay monitoring the HIF-1α C-terminal activation domain (CAD) activity was performed as described previously (19).
Knockdown Experiments Using shRNAs
shRNA sequences used for knockdown of target proteins were as follows: mouse FIH-1 1 (5′-caccggacctcgaatacctgcaagacgaatcttgcaggtattcgaggtcctttt-3′ and 2 (5′-caccggaagattgtcatggacttctcgaaagaagtccatgacaatcttcc-3′), human FIH-1 1 (5′-caccgctgaccgacacaaatcttgtcgaaacaagatttgtgtcggtcagctttt-3′) and 2 (5′-caccggaagattgtcatggacttctcgaaagaagtccatgacaatcttcctttt-3′), and mouse Mint3 (5′-caccgccagttcctacaggagaacacgaatgttctcctgtaggaactggc-3′).These sequences were subcloned into the pENTR/U6 TOPO vector (Invitrogen) and then transferred by recombination into the lentivirus vector pLenti6 BLOCKiT. shRNA-expressing lentiviral vectors were generated and used according to the manufacturer's instructions.
Knockdown Experiments Using siRNA
Target sequences for siRNA were designed and synthesized (B-bridge). siRNAs were provided as a mixture containing three different siRNA target sequences. The following sequences were used for human Mint3: 5′-gauggaacuugaugaguca-3′, 5′-gggaggugcaccucgagaa-3′, and 5′-gguucuugguccuguauga-3′. Control siRNAs were 5′-auccgcgcgauaguacgua-3′, 5′-uuacgcguagcguaauacg-3′, and 5′-uauucgcgcguauagcggu-3′. Cells were seeded (1 × 104 cells/well onto 24-well plates) and transfected with a 10 nm siRNA mixture (containing the three target sequences) using LipofectamineTM RNAiMAX (Invitrogen), according to the manufacturer's instructions.
Immunoprecipitation
The immunoprecipitation assay to detect an interaction between FIH-1 and Mint3 in macrophages was performed as previously described (19).
GST Pull-down Assay
Recombinant proteins, such as GST, GST-CPT, GST-CPT(R/A), GST-FIH-1, His6-FIH-1, and His6-Mint3 were prepared as previously described (19). Glutathione-Sepharose 4B (GE Healthcare)-conjugated GST fusion proteins (10 μg) were preincubated with 0.5 mg/ml BSA in lysis buffer for 30 min at 4 °C. Then 2 μg of His6-tagged FIH-1 or Mint3 was resuspended in lysis buffer containing 0.5 mg/ml BSA and added to the beads along with synthesized competitor peptides (CPT (RRHGTPRRLLYCQRSLLDKV) or control peptide (KTLRYSDLVRQCRGLRHPLR); Invitrogen). After mixing on a rotator for 2 h, the beads were washed four times with lysis buffer. The proteins were eluted using Laemmli sample buffer and analyzed by SDS-PAGE followed by immunoblotting using an anti-His6 antibody (Roche Applied Science) or an anti-GST antibody (GeneTex).
Immunoblotting
Cells were lysed with lysis buffer and centrifuged at 20,000 × g for 15 min at 4 °C. The supernatants were collected, and total protein content was measured using the Bradford assay (Bio-Rad). To fractionate subcellular proteins, the subcellular proteome extraction kit (Merck) was used. Lysates were separated by SDS-PAGE, transferred to membrane filters, and subjected to immunoblotting using an anti-MT1-MMP mouse antibody (Daiichi Fine Chemical), anti-p300/CBP mouse antibody (Upstate), anti-lamin A/C mouse antibody (BD Biosciences), anti-HIF-1α mouse antibody (EXALPHA), anti-Mint3 mouse antibody (BD Biosciences), anti-FIH-1 goat antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-transferrin receptor mouse antibody (Invitrogen), anti-lactate dehydrogenase A (LDHA) rabbit antibody (Novus Biologicals), rabbit anti-PHD1 to -3 antibodies (Novus Biologicals), anti-actin mouse antibody (CHEMICON), or anti-FLAG epitope M2 antibody (Sigma).
Immunostaining
Cells were fixed with 4% paraformaldehyde and permeabilized using 0.01% Triton X-100 for 10 min. After blocking in PBS containing 5% goat serum and 3% BSA, cells were incubated with mouse anti-Mint3 antibody (BD Biosciences) or rabbit anti-FIH-1 antibody (Novus Biologicals) for 1 h and then washed three times and incubated for 1 h with anti-mouse IgG Alexa488 conjugate or anti-rabbit Alexa594 (Invitrogen). Cells were counterstained with Hoechst33342, washed five times with PBS, mounted, and observed by CCD microscopy.
To analyze the co-localization of FIH-1 with Mint3 in the Golgi body, FIH-1 signal intensities from whole cells were quantified using ImageJ software (National Institutes of Health) as described previously (19). FIH-1 signals exceeding the average signal intensity in each cell by at least 2-fold were extracted and merged with Mint3 signals.
Statistical Analysis
Data represent the mean ± S.D. Unpaired Student's t test was used for analyzing differences between experimental groups.
RESULTS
Reduced ATP Content in MT1-MMP-deficient Macrophages
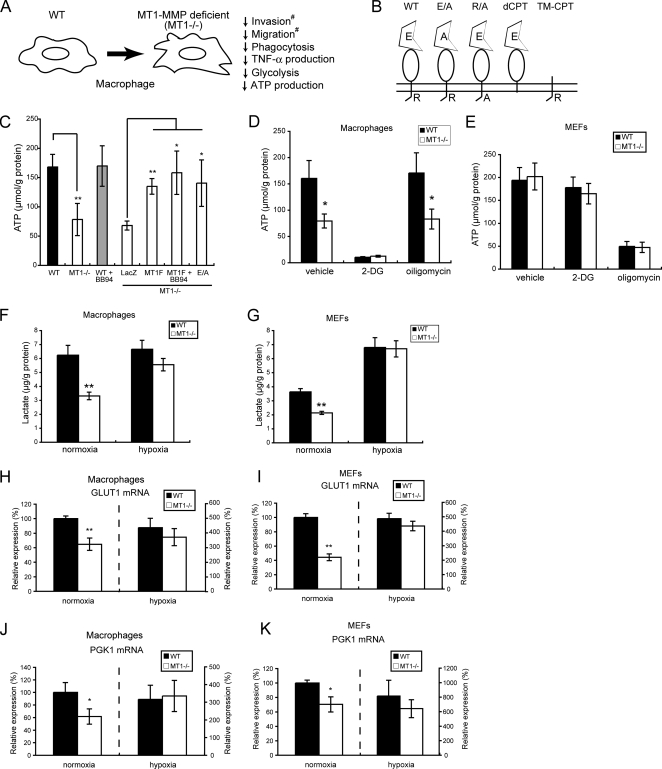
During a follow-up study of our previous observation that MT1−/− macrophages exhibit impaired migratory activity (9), we found that the mutant cells also exhibited defects in phagocytosis (supplemental Fig. S1) and TNF-α production (supplemental Fig. S2), and these phenotypes are summarized in Fig. 1A. Fig. 1B summarizes the various MT1-MMP expression constructs used to identify important domains for its function. To explore the underlining mechanism through which MT1-MMP deficiency affects multiple macrophage functions, we analyzed possible alterations in cytokine signaling and the regulation of actin polymerization in the mutant macrophages. Although no differences were observed in these analyses, we eventually found that the level of ATP in MT1−/− macrophages is 60% lower than that in WT cells (Fig. 1C, WT and MT1−/−). Forced expression of a FLAG-tagged MT1-MMP (MT1F) in MT1−/− macrophages mitigated this reduction in ATP level. Co-treatment of MT1F-transfected cells with BB94, a synthetic inhibitor of MMP protease activity, failed to abolish the effect of MT1F expression on ATP levels (Fig. 1C, MT1F + BB94), and expression of a protease-defective mutant of MT1-MMP containing a Glu240 → Ala substitution (E/A) in the catalytic site also restored ATP levels in MT1−/− macrophages (Fig. 1C, E/A). Thus, forced expression of MT1-MMP can boost ATP production in MT1−/− macrophages, but this effect is not linked to its protease activity. Consistent with these results, ATP levels in WT cells were not inhibited by exposure of the cells to BB94 (Fig. 1C, WT + BB94). The previously reported restoration of migratory activity of MT1−/− macrophages through the expression of MT1-MMP constructs exactly mirrors this restoration of ATP levels in the cells (9).
FIGURE 1.
MT1-MMP augments ATP production in macrophages but not in fibroblasts. A, illustration showing defects in cellular functions of MT1−/− macrophages. The functions labeled with a number sign were reported in a previous study (9). B, MT1-MMP constructs containing a mutation in either the protease domain or the cytoplasmic tail. C, ATP levels in WT and MT1−/− macrophages. Values were normalized relative to total protein. MT1F, FLAG-tagged MT1-MMP; E/A, protease-deficient mutant MT1F; LacZ, a control; BB94, a synthetic MMP inhibitor. D and E, lack of MT1-MMP affects ATP level in macrophages (D) but not in MEFs (E). 2-Deoxyglucose (2-DG; 100 μg/ml) and oligomycin (5 μg/ml) were used to inhibit glycolysis- and oxidative phosphorylation-derived ATP production, respectively. Black bar, WT; white bar, MT1−/−. F and G, MT1-MMP augments glycolytic activity in both macrophages (F) and MEFs (G). Cells were cultured under normoxic conditions, and lactate that accumulated in the culture media was measured. Hypoxia augmented glycolytic activity in both types of cell. H–K, MT1−/− cells express lower levels of glycolytic enzymes. Expression of mRNAs encoding GLUT-1 (H and I) and PGK-1 (J and K) were measured by RT-PCR in both WT and MT1−/− macrophages (H and J) and MEFs (I and K). Note that mRNA levels are low in both types of MT1−/− cells cultured during normoxia. Hypoxia induced expression of these genes so as to override the effect of MT1-MMP deficiency. Values are presented relative to the levels in WT cells under normoxic conditions, which were set at 100%. Black bar, WT; white bar, MT1−/−. The data in C–K were analyzed using Student's t test. *, p < 0.05; **, p < 0.01.
MT1-MMP Augments ATP Production Specifically in Macrophages
Most cells produce ATP in the mitochondria via oxidative phosphorylation, whereas macrophages are unique in that they generate ATP via glycolysis. Indeed, the addition of 2-deoxyglucose, an inhibitor of glycolysis, to the culture medium inhibited ATP production in macrophages (Fig. 1D, 2-DG) but not in mouse embryonic fibroblasts (MEFs) (Fig. 1E, 2-DG). Conversely, the addition of oligomycin, an inhibitor of oxidative phosphorylation, to the culture medium inhibited ATP production in MEFs (Fig. 1E, oligomycin) but not in macrophages (Fig. 1D, oligomycin). Because MT1-MMP deficiency does not affect ATP levels in MEFs, MT1-MMP may modulate glycolysis specifically in macrophages.
We next analyzed the glycolytic activity of the cells by measuring production of lactate, which is an end product of glycolysis. The level of lactate was lower in MT1−/− macrophages than in WT macrophages (Fig. 1F, normoxia), and this reduction was roughly proportional to the reduction in ATP levels in MT1−/− macrophages. In contrast, the level of lactate in WT MEFs was ∼40% lower than that in WT macrophages (Fig. 1, F and G, normoxia), and the level in MT1−/− MEFs was reduced even further (Fig. 1G, normoxia). Consistent with this reduction of glycolytic activity in MT1−/− cells, we observed reduced expression of the mRNAs encoding the glycolysis-related enzymes, PGK-1 (phosphoglycerate kinase) and GLUT-1 (hypoxia-regulated glucose transporter), in both MT1−/− macrophages (Fig. 1, H and J, normoxia) and MT1−/− MEFs (Fig. 1, I and K, normoxia) compared with the levels in their WT counterparts. Thus, a lack of MT1-MMP reduces glycolytic activity in both macrophages and MEFs, although there is little impact upon ATP production in MEFs due to the activity of the oxidative phosphorylation cycle.
Hypoxia enhances the glycolytic activity of most types of cell (21). However, whereas exposure to hypoxia augments the glycolytic activity of MT1−/− macrophages, it does not increase the glycolytic activity of WT cells (Fig. 1F, hypoxia). Thus, glycolytic activity in WT macrophages appears to be operating at maximal levels regardless of the oxygen tension. In contrast, hypoxia enhances the glycolytic activity in both WT and MT1−/− MEFs (Fig. 1G, hypoxia), and the effect of hypoxia upon glycolytic activity overrides the effect of MT1-MMP expression in WT cells.
We also observed that expression of the mRNAs encoding GLUT-1 (Fig. 1, H and J) and PGK-1 (Fig. 1, I and K) was enhanced following exposure of macrophages or MEFs to hypoxia. In particular, although the glycolytic activity of WT macrophages was unaffected by hypoxia (Fig. 1F), the expression of mRNAs encoding GLUT-1 and PGK-1 was enhanced 4.3- and 3.0-fold, respectively. This apparent discrepancy may be explained by a scenario whereby the glycolytic activity of MT1-MMP-expressing macrophages (WT) is already functioning at a maximal rate that cannot be further enhanced by increased expression of GLUT-1 or PGK-1 (Fig. 1F). Taken together, the results show that WT macrophages express higher levels of GLUT-1 and PGK-1 mRNAs and greater glycolytic activity than is observed in MT1−/− cells during normoxia but that these differences can be overridden following exposure of cells to hypoxia. Thus, the effect of MT1-MMP expression on glycolytic activity of macrophages is only apparent during normoxia.
MT1-MMP Regulates the Activity of HIF-1α during Normoxia
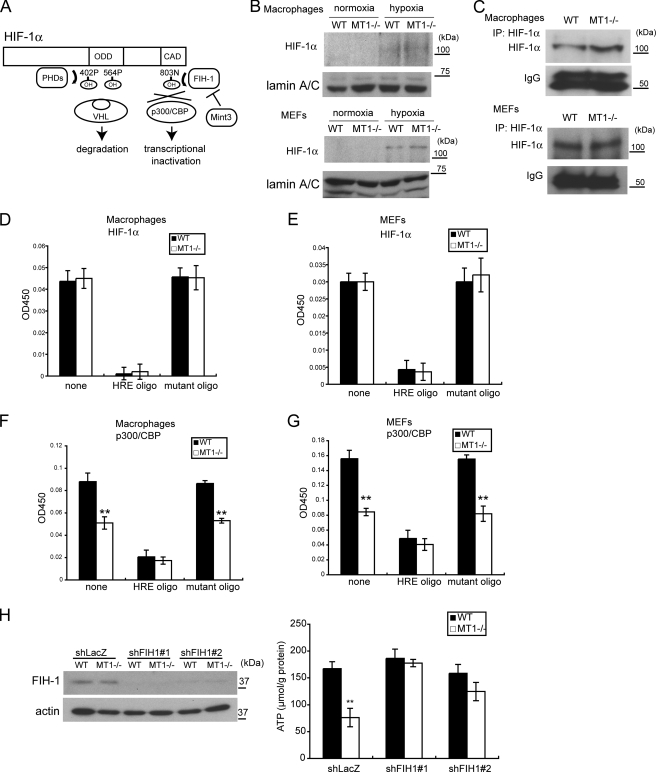
HIF-1 is the master transcription factor regulating expression of glycolysis-related genes, and its HIF-1α subunit is subject to regulatory mechanisms that respond to oxygen tension in the cell (Fig. 2A). Under oxygen-rich conditions, HIF prolyl hydroxylases (PHDs) mediate hydroxylation of proline residues in the oxygen-dependent degradation domain of HIF-1α, leading to degradation of the protein in a proteasome-dependent manner (22, 23). In addition, FIH-1 mediates hydroxylation of an asparagine residue (Asn803) located in the CAD of HIF-1α in an oxygen-dependent manner (24, 25) and prevents HIF-1α from binding the transcriptional co-activator p300/CBP (26). Mint3 inhibits FIH-1 activity by competing with HIF-1α for binding to FIH-1 (19). Given the effect of MT1-MMP deficiency on the expression of HIF-1 target genes, we next compared both the expression and activity of HIF-1α protein in MT1−/− and WT cells.
FIGURE 2.
MT1-MMP regulates ATP production through an effect upon HIF-1α. A, schematic depiction of how HIF-1α is regulated by PHDs and FIH-1. ODD, oxygen-dependent degradation domain. B, the amount of nuclear HIF-1α protein is low during normoxia in WT and MT1−/− macrophages (top) and MEFs (bottom). Nuclear extracts were subjected to Western blot for detection of HIF-1α and lamin A/C. Hypoxia increased the amount of HIF-1α but not lamin A/C. C, detection of nuclear HIF-1α protein in cells cultured during normoxia. 40-fold more nuclear extract than that used in the former experiment (B) was subjected to immunoprecipitation (IP: HIF-1α) to concentrate the protein, followed by Western blot for detection. D and E, the presence or absence of MT1-MMP does not affect the amount of HIF-1α that can bind HRE. Nuclear extracts from WT and MT1−/− macrophages (D) and MEFs (E) were subjected to the plate assay. Free HRE or mutant oligonucleotides were used as competitors. F and G, the amount of p300/CBP associated with HIF-1α is affected by MT1-MMP. Nuclear extracts from WT and MT1−/− macrophages (F) and MEFs (G) were subjected to the plate assay. H, FIH-1 activity is reduced in WT macrophages compared with that observed in MT1−/− cells. FIH-1 protein was detected by Western blot as in the left panel. Expression of FIH-1 was knocked down using two shRNA sequences (#1 and #2). Down-regulation of FIH-1 results in greater ATP production (right). Note that the level of ATP in WT cells is comparable with that observed in the FIH-1 knockdown cells. shRNA targeted to the LacZ mRNA (shLacZ) served as a negative control. The data in D–H were analyzed using Student's t test. *, p < 0.05; **, p < 0.01.
The expression of HIF-1α protein was sufficiently low under normoxic conditions that we could not detect it by conventional immunoblot analysis of nuclear extracts prepared from macrophages and MEFs (Fig. 2B, normoxia). Concentration of the protein from the extracts by immunoprecipitation prior to immunoblot analysis allowed us to detect it in both cell types at a comparable level (Fig. 2C). Exposure of the cells to hypoxia increased the amount of HIF-1α protein (Fig. 2B, hypoxia), and the PHDs responsible for this regulation were detected in both macrophages and MEFs at a similar level (supplemental Fig. S3, A and B). We next captured the heterodimeric form of HIF-1 by exposing the nuclear extracts to an HRE oligonucleotide-coated plate. A similar amount of HIF-1α in extracts derived from either macrophages or MEFs bound to HRE (Fig. 2, D and E, none), and this binding was specific to the HRE sequence. Measurement of the amount of p300/CBP associated with HIF-1α on the plate using an anti-p300/CBP antibody demonstrated a lower level of association between p300/CBP and HIF-1α in MT1−/− macrophages and MEFs compared with that observed in the corresponding WT cells (Fig. 2, F and G). Because the overall level of expression of CBP and p300 did not differ between MT1−/− and WT cells (supplemental Fig. S3, C and D), we hypothesized that the lack of MT1-MMP leads to inhibition of the binding of p300/CBP to HIF-1α.
Because FIH-1 regulates the ability of HIF-1α to bind p300/CBP, MT1-MMP may affect FIH-1 activity. FIH-1 was expressed at a similar level in WT and MT1−/− macrophages, as demonstrated by immunoblot analysis (Fig. 2H, shLacZ). To examine whether FIH-1 plays a role in ATP production in macrophages, we knocked down FIH-1 expression in macrophages using a lentivirus vector expressing shRNA sequences targeting its mRNA (Fig. 2H, shFIH-1#1 and shFIH-1#2). Knockdown of FIH-1 in MT1−/− macrophages led to an increase in ATP production, showing that FIH-1 suppresses ATP production in the absence of MT1-MMP (Fig. 2H). However, this suppressor activity of FIH-1 was abrogated in WT cells (Fig. 2H).
MT1-MMP Activates HIF-1α by Abrogating FIH-1 Activity
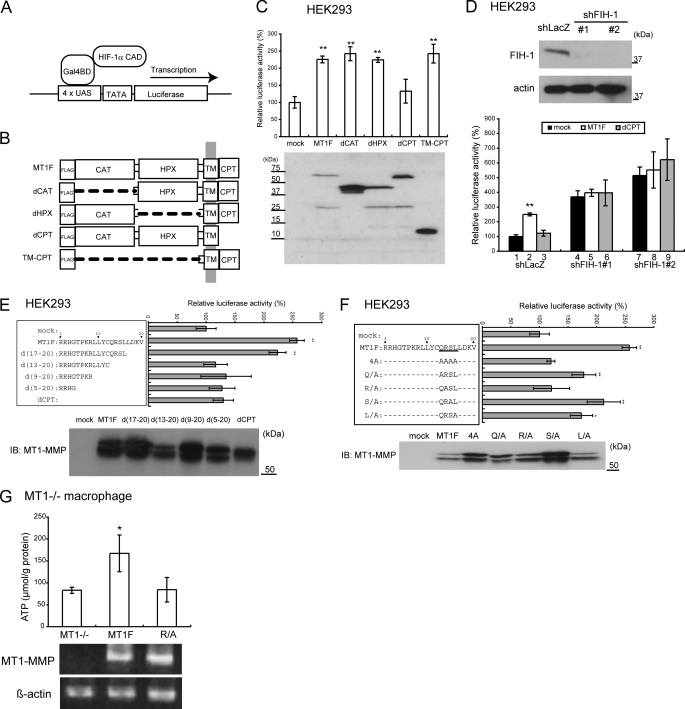
To examine whether MT1-MMP regulates FIH-1 activity against HIF-1α, we made use of a luciferase reporter assay to monitor the transcriptional activity of the HIF-1α CAD fragment fused to the Gal4 DNA-binding domain (19) (Fig. 3A). The Gal4-CAD protein cannot be modified by PHDs because it lacks the oxygen-dependent degradation domain, but it does retain Asn803 from the HIF-1α CAD, which is modified by FIH-1. Thus, this construct allows us to assess the effect of MT1-MMP on the activity of FIH-1 through its effect on the Gal4-CAD surrogate transcriptional reporter. The human embryonic kidney cell line HEK293 was used for this assay because it lacks expression of endogenous MT1-MMP.
FIGURE 3.
The cytoplasmic tail of MT1-MMP regulates HIF-1α. A, schematic illustration of the assay to monitor transcriptional activity of HIF-1α. Gal4BD, Gal4 DNA-binding domain; UAS, Gal4 binding site; TATA, thymidine kinase minimal promoter. B, deletion constructs of MT1-MMP. CAT, catalytic domain; HPX, hemopexin-like domain; TM, transmembrane domain; CPT, cytoplasmic tail. A FLAG tag was fused to the N terminus of each construct. C, the CPT of MT1-MMP is important for activation of the HIF-1α CAD in HEK293 cells. Expression of MT1-MMP constructs was confirmed by Western blot (bottom). D, MT1-MMP regulates HIF-1α CAD activity in the presence of FIH-1 in HEK293 cells. Knockdown of FIH-1 in HEK293 cells and detection of the protein by Western blot are presented (top). E, delineation of the MT1-MMP CPT sequence required for activation of HIF-1α CAD. The CPT mutants containing sequential deletions (left) and their effects on the reporter assay (right) are presented. Expression of the MT1-MMP derivatives in the cells was confirmed by Western blot (lower panels). IB, immunoblot. F, identification of amino acid residues in the CPT essential for activation of HIF-1α CAD activity. Expression of the MT1-MMP protein constructs was confirmed by Western blot. G, FLAG-tagged MT1-MMP (MT1F) or its mutant containing an R/A substitution in the CP tail was expressed in MT1−/− macrophages, and ATP was measured (top). Expression of the genes was detected by RT-PCR (bottom). The data in C–G were analyzed using Student's t test. *, p < 0.05; **, p < 0.01.
Various FLAG-tagged deletion mutants of MT1-MMP were prepared (Fig. 3B) and expressed in the cells to examine their effect on reporter gene expression. Expression of full-length MT1-MMP (MT1F) enhanced reporter expression up to 2.3-fold over the mock-transfected control (Fig. 3C, mock versus MT1F). Expression of mutants lacking the catalytic domain (dCAT), the hemopexin-like domain (dHPX), or the entire extracellular domain (TM-CPT) could enhance reporter gene expression (Fig. 3C). However, a construct encoding MT1-MMP lacking the CPT (dCPT) failed to enhance reporter gene expression (Fig. 3C, dCPT). The expression level of each mutant was greater than that of MT1F. However, this does not affect the above mentioned interpretation because the level of expression of MT1F was already greater than the level needed to saturate reporter gene expression (data not shown).
Endogenous FIH-1 appears to exert a suppressor activity against the HIF-1α CAD in HEK293 cells because knockdown of the expression of the former enhanced reporter gene expression by 4–5-fold (Fig. 3D, shFIH#1 and shFIH#2). Forced expression of MT1-MMP failed to boost reporter expression any further in FIH-1 knockdown cells (Fig. 3D, MT1F + shFIH). Based on these results, we conclude that MT1-MMP inhibits FIH-1 activity in a CPT-dependent manner.
Critical Amino Acid Residues in the CPT
The CPT of MT1-MMP comprises 20 amino acids (Fig. 3E, left). We generated a series of deletion expression constructs lacking 4, 8, 12, or 16 amino acids from the C-terminal end. Deletion of the first four amino acids (d(17–20)) did not affect the CAD-activating activity of MT1F (Fig. 3E, right). However, further deletion of the preceding four amino acids (d(13–20)) reduced the activity of the construct to a level similar to that of dCPT. Substitution of four amino acids (CPT 13–16) to alanine also abolished this activity (Fig. 3F). Individual substitutions of these amino acids with alanine reduced the CAD-activating activity to varying degrees, but individual substitution of the arginine residue at position 14 to alanine (R/A) had the greatest inhibitory effect (Fig. 3F).
The dependence of ATP production in macrophages upon this specific sequence within the CPT was confirmed by demonstrating that expression in MT1−/− macrophages of a variant MT1F containing the same (R/A) mutation failed to stimulate ATP production (Fig. 3G).
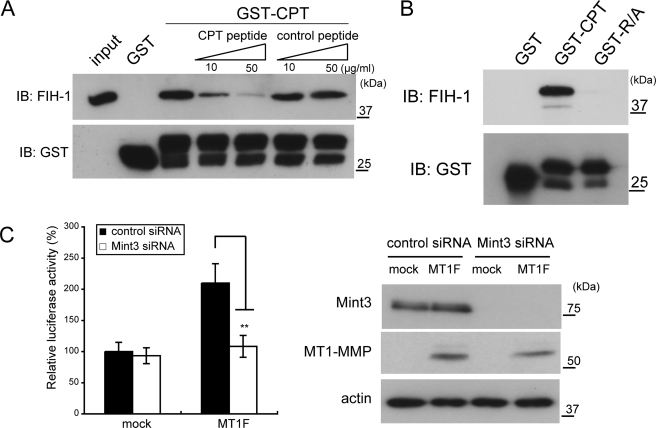
The CPT of MT1-MMP Binds FIH-1
Because MT1-MMP abrogated FIH-1 activity in a CPT-dependent manner, we next examined whether the CPT interacts with FIH-1. A CPT peptide fused to glutathione S-transferase (GST-CPT) was used in a pull-down analysis to test binding of His6-tagged FIH-1. Indeed, GST-CPT bound to FIH-1, but GST alone did not (Fig. 4A). The presence of free CPT peptide in the binding reaction inhibited the binding of GST-CPT to FIH-1 in a dose-dependent manner (GST-CPT + CPT peptide), whereas competition using a control peptide did not (GST-CPT + control peptide). Furthermore, we found that a GST-CPT fusion protein harboring the R/A substitution (GST-R/A) failed to bind FIH-1 (Fig. 4B).
FIGURE 4.
The cytoplasmic tail of MT1-MMP binds FIH-1. A, FIH-1 binds the CPT of MT1-MMP. A pull-down assay was performed using the GST-CPT fusion and His6-tagged FIH-1 proteins. FIH-1-bound GST-CPT was detected by Western blot (IB). CPT and control peptides were used for the competition assay. B, the CPT mutant peptide (R/A), which cannot activate HIF-1α, failed to bind FIH-1. C, activation of HIF-1α CAD by MT1-MMP requires Mint3. Expression of MT1F enhances CAD activity. The effect of MT1F was diminished by knockdown of Mint3 using siRNA (left). Protein expression was confirmed by Western blot (right). The data in C were analyzed using Student's t test. **, p < 0.01.
MT1-MMP Inhibits FIH-1 Using Mint3
We recently identified Mint3/APBA3 as a new FIH-1 inhibitor (19). Mint3 competes with HIF-1α for binding to FIH-1 and thereby inhibits the ability of FIH-1 to modify HIF-1α. We also observed that Mint3 inhibits FIH-1 constitutively in macrophages by forming a stable complex that localizes predominantly to the Golgi area, whereas Mint3 expressed in HEK293 cells does not inhibit FIH-1 constitutively (19). However, because expression of MT1-MMP in HEK293 cells abrogates FIH-1 activity, MT1-MMP may make use of Mint3 to inhibit FIH-1. To test this idea, we examined whether MT1-MMP requires the presence of Mint3 to activate HIF-1α CAD activity using the luciferase reporter assay. We knocked down Mint3 expression in HEK293 cells prior to performing the reporter gene assay. Subsequent transfection of these cells with MT1F revealed that they had lost the ability to stimulate reporter gene expression (Fig. 4C). The association of p300/CBP with HIF-1α in HEK293 cells was also enhanced by the expression of MT1-MMP in a Mint3-dependent manner (supplemental Fig. S4). These results suggest that MT1-MMP mediates inhibition of FIH-1 via an effect upon Mint3 activity in HEK293 cells.
MT1-MMP Uses Mint3 to Stimulate ATP Production in Macrophages
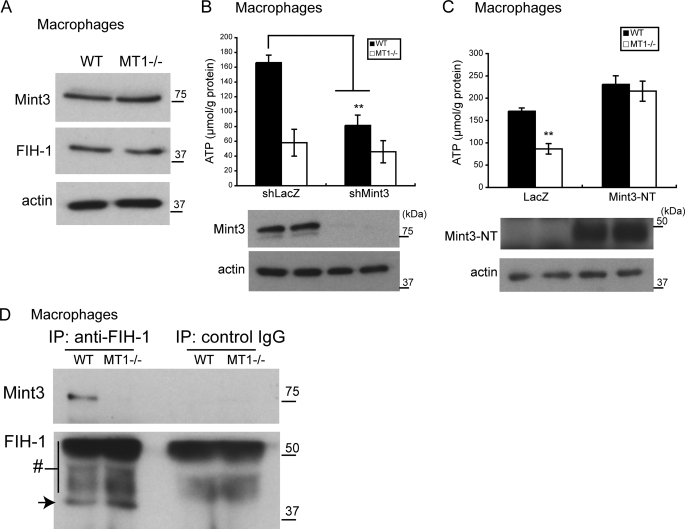
We next examined whether MT1-MMP stimulates ATP production in macrophages through an effect upon Mint3. Mint3 and FIH-1 are expressed in both MT1−/− and WT macrophages at a similar level (Fig. 5A). Knockdown of Mint3 in WT macrophages led to an ∼50% reduction in ATP content (Fig. 5B, shLacZ versus shMint3 in WT cells), suggesting that Mint3 contributes to ATP production in WT cells. In contrast, the ATP content of MT1−/− macrophages was already ∼40% of that observed in WT macrophages, and was not reduced any further by Mint3 knockdown (Fig. 5B, shLacZ and shMint3 in MT1−/− cells). Forced expression in MT1−/− macrophages of Mint3-NT, a minimal inhibitory fragment of Mint3 (19) (supplemental Fig. S5), enhanced ATP production (Fig. 5C) to a level comparable with that observed following knockdown of the expression of FIH-1 (Fig. 2H). Therefore, we conclude that, as is observed in HEK293 cells, MT1-MMP uses Mint3 to inhibit FIH-1 in WT macrophages.
FIGURE 5.
MT1-MMP mediates inhibition of FIH-1 by Mint3. A, Mint3 and FIH-1 are expressed at a similar level in MT1−/− and WT macrophages as detected by Western blot using specific antibodies. B, Mint3 plays a role in ATP production in WT macrophages. Knockdown of Mint3 in WT but not in MT1−/− macrophages diminishes ATP production (top). Mint3 protein expression was assessed by Western blot (bottom). C, forced expression of Mint3NT stimulates ATP production in MT1−/− macrophages to a level similar to that observed in WT cells. Protein expression was confirmed by Western blot (bottom). D, FIH-1 binds Mint3 in WT but not in MT1−/− macrophages. Binding of FIH-1 to Mint3 was demonstrated by immunoprecipitation (IP) of FIH-1 followed by detection of Mint3 by Western blot (top). The arrow indicates FIH-1, and the number symbol indicates rabbit immunoglobulin (bottom). The data in B and C were analyzed using Student's t test. **, p < 0.01; n = 3/group.
MT1-MMP Mediates Binding of Mint3 to FIH-1
Mint3 binds FIH-1 and abrogates the function of the latter to suppress HIF-1α in WT macrophages. Indeed, we showed binding of Mint3 to FIH-1 by immunoprecipitation of FIH-1, followed by immunoblot analysis to detect Mint3 (Fig. 5D, WT). However, such a complex was not detected in MT1−/− macrophages despite the expression of both proteins (Fig. 5D, MT1−/−). The latter result is consistent with the fact that Mint3 failed to inhibit FIH-1 in MT1−/− macrophages (Fig. 5B). As was observed in WT macrophages, overexpression of MT1-MMP in HEK293 cells induced association of FIH-1 with Mint3, whereas expression of a mutant MT1-MMP lacking the CTP (dCTP) failed to do so (supplemental Fig. S6). The CPT of MT1-MMP could bind FIH-1, but MT1-MMP was not detected in immunoprecipitates of FIH-1 (not shown). Therefore, it was still not clear how MT1-MMP mediates inhibition of FIH-1 using Mint3.
MT1-MMP Changes the Subcellular Localization of FIH-1
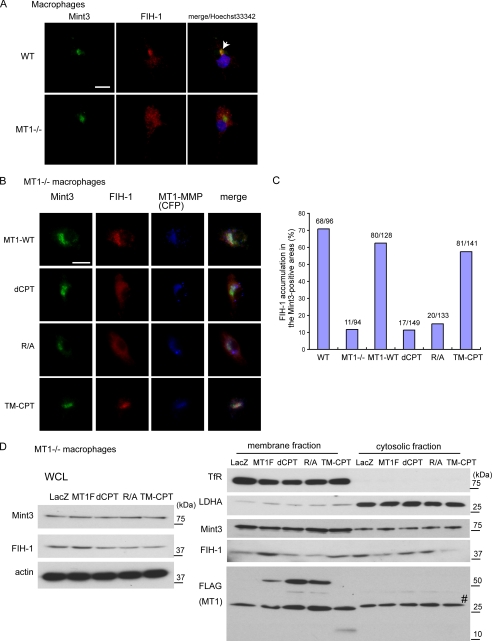
To visualize MT1-MMP-mediated inhibition of FIH-1 by Mint3, we next analyzed localization of these proteins by immunohistochemistry (Fig. 6A). In a previous study, we observed that FIH-1 formed a complex with Mint3, which co-localized with a Golgi marker within the perinuclear region of WT macrophages, and that knockdown of Mint3 expression in these cells liberated FIH-1 from the Golgi to the cytoplasm (19). Thus, FIH-1 localized to the perinuclear region represents FIH-1 that is inhibited by Mint3. We first confirmed co-localization of FIH-1 and Mint3 within the perinuclear region of WT macrophages by immunohistochemistry (Fig. 6A, WT). In contrast, FIH-1 was distributed diffusely throughout the cytoplasm of MT1−/− macrophages, whereas Mint3 remained perinuclear (Fig. 6A, MT1−/−). The results are consistent with the idea that MT1-MMP inhibits FIH-1 by promoting an interaction of the latter with Mint3.
FIGURE 6.
MT1-MMP promotes the association of FIH-1 with Mint3. A, FIH-1 and Mint3 co-localize in WT but not in MT1−/− macrophages. Complex formation between FIH-1 and Mint3 was visualized by immunohistochemistry. Hoechst33342 is a marker for the nucleus. The arrow indicates a representative pattern of co-localization between FIH-1 and Mint3 in WT macrophages. Bar, 25 μm. B, co-localization of FIH-1 and Mint3 observed in WT macrophages was reconstituted in MT1−/− cells by expressing cyan fluorescent protein-fused MT1-MMP. Various fusions of MT1-MMP mutants were expressed in MT1−/− macrophages. Bar, 25 μm. Note that translocation of FIH-1 to sites of co-localization was induced by expression of MT1-MMP fusions only when an intact CPT was present. C, FIH-1 signals exceeding twice the average signal intensity in each cell were digitally extracted and merged with Mint3 signals. The numbers of cells in which FIH-1 signals were clearly enriched in the Mint3-positive areas were counted. D, various MT1-MMP mutants were expressed in MT1−/− macrophages, and subcellular fractionated proteins from cells were analyzed by Western blot. The transferrin receptor (TfR) serves as a representative of membrane proteins, and lactate dehydrogenase A (LDHA) serves as a representative of cytosolic proteins. The number symbol indicates a nonspecific band detected by anti-FLAG antibodies. WCL, whole cell lysate.
MT1-MMP is localized to the Golgi until it is exported to the cell surface (27). Mint3 is presumably localized in close proximity to MT1-MMP in the Golgi, because Mint3 is reported to bind furin (28) (supplemental Fig. S5), which binds pro-MT1-MMP (29) and cleaves off the propeptide sequence of the latter, leading to its activation (10). Therefore, FIH-1 bound to the CPT of MT1-MMP may be localized in sufficiently close proximity to Mint3 to interact with the latter. To test whether MT1-MMP can mediate translocation of FIH-1 to the perinuclear region, we first generated constructs expressing cyan fluorescent protein fused to either MT1-MMP (MT1-WT) or variants of the latter that contained mutations in the CPT (Fig. 1B). Following expression of the constructs in MT1−/− macrophages, we analyzed the localization of FIH-1 and Mint3 by immunohistochemistry. A significant fraction of the MT1-WT-associated signal was found to co-localize with Mint3 in the perinuclear region (Fig. 6B, MT1-WT). The fusions of the various MT1-MMP derivatives exhibited a localization similar to that of the MT-WT fusion (Fig. 6B, dCPT, R/A, and TM-CPT). In addition to the signal from the MT1-WT fusion detected in the perinuclear region, we observed an additional podosome-like punctuate signal within the cells (supplemental Fig. S7). The expression of either the MT1-WT or the TM-CPT fusions induced co-localization of FIH-1 with Mint3 (Fig. 6B, MT1-WT and TM-CPT). However, the expression of the mutant MT1-MMP fusions characterized by a deletion of the CPT (dCPT) or an R/A substitution failed to recruit the cytoplasmic FIH-1 to co-localize with Mint3 (Fig. 6B, dCPT, R/A). To observe areas of accumulation of the FIH-1 signal with that of Mint3 more clearly, we extracted digital FIH-1 signals that exceeded twice the average of the FIH-1 signals of each cell, as described previously (19). We then counted the number of cells that exhibited clear co-localization of FIH-1 with Mint3, and the results confirmed that the MT1-MMP CPT mediates co-localization of FIH-1 with Mint3 (Fig. 6C). To confirm these results further, we examined the localization of FIH-1 following subcellular fractionation of cell proteins. In MT1−/− macrophages, FIH-1 proteins were mainly detected in the cytoplasmic fraction (Fig. 6D, LacZ). Expression of FLAG-tagged MT1-MMP (MT1F) or TM-CPT decreased the amount of FIH-1 protein in the cytoplasmic fraction and increased the protein in the membrane fraction, where both MT1-MMP and Mint3 exist (Fig. 6D, MT1F and TM-CPT). However, expression of dCPT or R/A mutants could not alter the FIH-1 localization (Fig. 6D, dCPT and R/A). These results support the conclusion obtained from the immunohistochemical analysis (Fig. 6, B and C).
Taken together, the CPT of MT1-MMP alleviates the repressive effect of FIH-1 on HIF-1α by mediating an interaction of FIH-1 with Mint3, thereby freeing HIF-1α to stimulate glycolysis and the resulting ATP production (Fig. 7).
FIGURE 7.
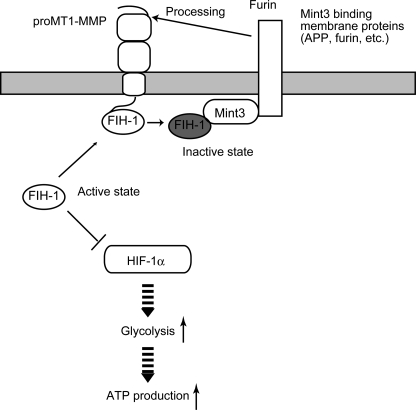
The MT1-MMP CPT enhances the transcriptional activity of HIF-1 via suppression of FIH-1. MT1-MMP and Mint3 localize to the plasma membrane compartment, particularly within the Golgi. FIH-1 localizes predominantly to the cytoplasm, where it inhibits the transcriptional activity of HIF-1α. The CPT of MT1-MMP is able to bind FIH-1. Although this association is not stable, it recruits the cytoplasmic FIH-1 to the Golgi. Mint3 lines the plasma membrane within the Golgi by binding to membrane proteins, such as furin and APP, via its C-terminal domain, which is separated from its FIH-1 binding domain located at the N terminus (supplemental Fig. S4). Furin is a protease that binds MT1-MMP, cleaves the propeptide sequence of MT1-MMP, and converts pro-MT1-MMP into the mature enzyme. Thus, if FIH-1 binds transiently to the CPT of MT1-MMP, it may increase the odds of FIH-1 encountering Mint3 localized in the vicinity of MT1-MMP. Indeed, the overexpression of MT1-MMP CPT induces the suppression of FIH-1 activity by Mint3. The suppression of FIH-1 enhances HIF-1 activity and augments ATP generation by glycolysis in macrophages.
DISCUSSION
A Key Role for MT1-MMP in Maintaining Aerobic Glycolysis and ATP Production in Macrophages
MT1-MMP can release FIH-1-mediated suppression of HIF-1α even during normoxia. Activation of HIF-1 by MT1-MMP increases the rate of aerobic glycolysis of macrophages and boosts ATP production. A lack of MT1-MMP in macrophages leads to a 60% reduction in ATP levels, which is reminiscent of the reported 80% reduction in ATP levels in HIF-1α-deficient macrophages (6). Therefore, MT1-MMP plays a major role in regulation of HIF-1 activity and the production of ATP in macrophages. It was unexpected that MT1-MMP could activate HIF-1α in MEFs and thereby increase glycolytic activity, because MT1-MMP deficiency had no effect upon the level of ATP in MEFs. The negligible impact of MT1-MMP deficiency on ATP production in MEFs is probably due to the reliance of these cells on oxidative phosphorylation for ATP production during normoxia. As such, the physiological importance of MT1-MMP-dependent HIF-1 activation in MEFs and other cell types is unclear. However, the HIF-1-activating activity of MT1-MMP is presumably relevant to the previous observation that expression of MT1-MMP enhances the expression of VEGF, which is also a target gene product of HIF-1 (30, 31).
In contrast to the findings in MEFs, the oxidative phosphorylation pathway was constitutively inactive in macrophages because it was clear that MT1−/− macrophages were unable to use oxidative phosphorylation for ATP production rather than glycolysis. Pyruvate dehydrogenase is a key enzyme regulating the consumption of pyruvate, a product of glycolysis, by the TCA cycle. The expression of PDK-1 (pyruvate dehydrogenase kinase-1) is regulated by HIF-1, and PDK-1 inhibits the activity of pyruvate dehydrogenase so as to shut down the TCA cycle (32, 33). Therefore, activation of HIF-1 can suppress oxidative phosphorylation concomitant with activation of glycolysis. However, it is clear that up-regulation of HIF-1 activity is not the reason for the constitutive suppression of oxidative phosphorylation in macrophages because oxidative phosphorylation is not activated in MT1−/− macrophages when HIF-1 activity is reduced. Thus, the oxidative phosphorylation pathway appears to become suppressed during myeloid cell differentiation by an unknown mechanism.
The Unique Mechanism by Which MT1-MMP Inhibits FIH-1 via Mint3
We reported previously that Mint3 binds and inactivates FIH-1 because Mint3 competes for HIF-1α binding to FIH-1 (19). Mint3 localizes to the perinuclear region most likely in the Golgi (19). Co-localization of FIH-1 and Mint3 within the perinuclear region of WT macrophages (Fig. 6) coincided well with the observations that a complex between FIH-1 and Mint3 could be detected by immunoprecipitation (Fig. 5D) and that HIF-1 was activated in the cells (Figs. 1 and 2). Therefore, it is reasonable to postulate that FIH-1 is inactivated by Mint3 when FIH-1 localizes within the perinuclear region. Although we believe this is the major mechanism by which Mint3 inactivates FIH-1 activity, translocation of FIH-1 from the cytoplasm to the Golgi may contribute to the reduction of the level of active FIH-1 in the cytoplasm so as to modify HIF-1α to some extent.
In the absence of MT1-MMP, FIH-1 and Mint3 localize to different subcellular compartments, and Mint3 thereby cannot inhibit FIH-1. FIH-1 localizes diffusely throughout the cytoplasm, whereas Mint3 localizes focally within the perinuclear region (Fig. 6A, MT1−/−), which is stained by a Golgi marker, as reported previously (19). In contrast, FIH-1 co-localizes with Mint3 within the perinuclear region in WT macrophages (Fig. 6A, WT), and expression of MT1-MMP in MT1−/− macrophages induces co-localization of FIH-1 with Mint3. Because the CPT of MT1-MMP can bind FIH-1 in a sequence-specific manner, it is plausible that the CPT of MT1-MMP mediates the initial localization of FIH-1 to the surface of the perinuclear membrane. Indeed, the mutant MT1-MMP CPT (R/A) that cannot bind FIH-1 fails to mediate co-localization of FIH-1 with Mint3 (Fig. 6, B–D, and supplemental Fig. S6). Whereas the CPT of MT1-MMP plays an important role in mediating co-localization of FIH-1 with Mint3, Mint3 itself is also indispensable to retain FIH-1 at the Golgi in WT macrophages, as demonstrated by our previous study showing that Mint3 knockdown in WT macrophages releases FIH-1 from the Golgi to the cytoplasm (19).
Interestingly, FIH-1 does not co-immunoprecipitate MT1-MMP in the presence or absence of Mint3 (data not shown), whereas it does with Mint3. Although the MT1-MMP CPT can bind FIH-1 in vitro, the interaction may not be sufficiently stable to allow co-immunoprecipitation of the complex in the cell lysate. FIH-1 may associate with MT1-MMP only transiently until it is transferred to Mint3, which can form a stable complex with FIH-1. It is of note that both MT1-MMP and Mint3 interact with furin, a resident protein in the Golgi (28, 29). Therefore, FIH-1 bound to the CPT is expected to be localized in close proximity to Mint3. The MT1-MMP CPT appears to act as a siren drawing cytoplasmic FIH-1 into closer contact with Mint3.
Consequently, MT1-MMP activates HIF-1 by suppressing FIH-1 activity using Mint3, but this activity of MT1-MMP can be observed only during normoxia. During hypoxia, both FIH-1 and PHDs are inactive because of low oxygen levels (5), and therefore the effect of MT1-MMP and Mint3 to suppress FIH-1 activity is overwhelmed by the effect of hypoxia (Fig. 1, H–K).
The activity of MT1-MMP to regulate HIF-1α via modulation of FIH-1 was observed not only in macrophages but also in MEFs and HEK293 cells. Although MT1-MMP can regulate HIF-1α in different types of cells, its impact on ATP production was specifically observed with macrophages, as discussed above.
Emerging Roles of the Cytoplasmic Tail of MT1-MMP
The CPT of MT1-MMP comprises only 20 amino acids, but it interacts with multiple proteins and is also a target of post-translational modifications. For example, the CPT is reported to interact with caveolin-1 (34), the μ2 subunit of the AP-2 complex (35), MTCBP-1/hADIi (36), p130cas (37), and p27RF-Rho (38). Modifications include palmitoylation at Cys574 (39), and phosphorylation at Tyr573 (40) and Thr567 (41). Internalization of MT1-MMP is mediated by the CPT through interaction with the AP-2 complex and caveolin-1. MTCB-1 affects MT1-MMP-dependent cell motility by an unknown mechanism. p27RF-Rho regulates formation of invadopodia by regulating RhoA activation and consequently modulates the invasion-promoting activity of MT1-MMP. However, recent studies have uncovered new roles for the CPT of MT1-MMP in regulating various cell signaling pathways independently from its protease activity (9, 37, 42, 43). We observed that the CPT of MT1-MMP can stimulate macrophage migration without requiring its protease activity (9). The CPT is also reported to activate the ERK pathway via an unknown mechanism, and this activity is enhanced by TIMP-2 independently of the protease inhibition (42). Very recently, Gonzalo et al. (37) reported that binding of the CPT to p130cas induced activation of Rac1 in myeloid cells and regulated membrane ruffling. Because of this activity, lack of MT1-MMP in macrophages abrogated migration and fusion of the cells and prevented osteoclast formation. Activation of Rac1 by the CPT is also independent of the protease activity. Taken together with the present findings, it is conceivable that a reduction in ATP and the activation of Rac-1 due to MT1-MMP deficiency may collectively contribute to macrophage functions as well as the abrogation of osteoclast formation. However, the exact relevance of the activation of Rac1 and HIF-1 by MT1-MMP is not yet clear, although both events occur in macrophages. Rac1 activation is expected to occur at the inner surface of the peripheral plasma membrane, whereas inhibition of FIH-1 occurs within the perinuclear region. Thus, these two functions of the CPT appear to be site-specific due to interaction with distinct cytoplasmic proteins in different membrane compartments. Analysis of membrane trafficking that conveys MT1-MMP from the Golgi to the cell surface may provide further insight that may allow us to interconnect these apparently independent functions of the CPT.
MT1-MMP Activates HIF-1 but Other Related MMPs Do Not
MT2-, MT3-, and MT5-MMP also contain a CPT that comprises 20 amino acids and the Arg residue crucial for the binding of the MT1-MMP CPT to FIH-1 is conserved among them (supplemental Fig. S8A). Interestingly, however, only MT1-MMP can stimulate HIF-1α CAD in HEK293 cells (supplemental Fig. S9). The other amino acids surrounding the Arg residue are not necessarily conserved among the related MT-MMPs, and presumably these substitutions affect the ability of the CPT to bind FIH-1. It is also noteworthy that all of the MT1-MMP-related MMPs contain a PDZ-binding motif at their C termini, and one of them, MT5-MMP, is reported to bind the PDZ domains of Mint3 (44). Although we confirmed binding of Mint3 to MT5-MMP, it did not bind MT1-MMP in the same pull-down assay (supplemental Fig. S8B). The Mint3-binding activity of MT5-MMP had no effect on activation of the HIF-1α CAD (supplemental Fig. S9).
In conclusion, the MT1-MMP CPT triggers a cascade of protein-protein interactions that results in activation of HIF-1α and thereby regulates ATP production in macrophages. These results have addressed the question of how the HIF-1 activity of macrophages is maintained even during normoxia. The extracellular protease domain is also important to degrade the extracellular matrix during macrophage invasion. Taken together, MT1-MMP is an intriguing membrane protease that regulates important macrophage functions in an integrated manner through domain-specific functions.
Supplementary Material
Acknowledgments
We thank Ikuo Yana and Yohei Ohtake for helpful discussions and Akiko Rikimaru, Makoto Nagano, Daisuke Hoshino, and Nagayasu Egawa for technical assistance. We also thank Dr. Mitsuaki Yoshida for support to T. S., Dr. Roy Zent for helpful discussion, and Drs. Robert Whittier and Erik Thompson for careful checking of the manuscript.
This work was supported by the Specific Coordination Fund for Promoting Science (to T. S.), by a grant-in-aid for scientific research on priority areas (i.e. the “Integrative Research toward the Conquest of Cancer”) (to M. S.), and by the Global Centers of Excellence Program “Center of Education and Research for the Advanced Genome-Based Medicine: For personalized medicine and the control of worldwide infectious diseases” (to T. S. and M. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
- HRE
- hypoxia response element
- CPT
- cytoplasmic tail
- MMP
- matrix metalloproteinase
- MEF
- mouse embryo fibroblast
- CBP
- CREB-binding protein
- PHD
- prolyl hydroxylase
- CAD
- C-terminal activation domain.
REFERENCES
- 1.Coussens L. M., Werb Z. (2002) Nature 420, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard J. W. (2009) Nat. Rev. Immunol. 9, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condeelis J., Pollard J. W. (2006) Cell 124, 263–266 [DOI] [PubMed] [Google Scholar]
- 4.Denko N. C. (2008) Nat. Rev. Cancer 8, 705–713 [DOI] [PubMed] [Google Scholar]
- 5.Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 6.Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S. (2003) Cell 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinckerhoff C. E., Matrisian L. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 207–214 [DOI] [PubMed] [Google Scholar]
- 8.Joyce J. A., Pollard J. W. (2009) Nat. Rev. Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamoto T., Seiki M. (2009) Genes Cells 14, 617–626 [DOI] [PubMed] [Google Scholar]
- 10.Itoh Y., Seiki M. (2006) J. Cell. Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 11.Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 12.Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 14.Chun T. H., Hotary K. B., Sabeh F., Saltiel A. R., Allen E. D., Weiss S. J. (2006) Cell 125, 577–591 [DOI] [PubMed] [Google Scholar]
- 15.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) Cell 99, 81–92 [DOI] [PubMed] [Google Scholar]
- 16.Ohtake Y., Tojo H., Seiki M. (2006) J. Cell Sci. 119, 3822–3832 [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z., Apte S. S., Soininen R., Cao R., Baaklini G. Y., Rauser R. W., Wang J., Cao Y., Tryggvason K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider F., Sukhova G. K., Aikawa M., Canner J., Gerdes N., Tang S. M., Shi G. P., Apte S. S., Libby P. (2008) Circulation 117, 931–939 [DOI] [PubMed] [Google Scholar]
- 19.Sakamoto T., Seiki M. (2009) J. Biol. Chem. 284, 30350–30359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniwaki K., Fukamachi H., Komori K., Ohtake Y., Nonaka T., Sakamoto T., Shiomi T., Okada Y., Itoh T., Itohara S., Seiki M., Yana I. (2007) Cancer Res. 67, 4311–4319 [DOI] [PubMed] [Google Scholar]
- 21.Semenza G. L. (2010) Curr. Opin. Genet. Dev. 20, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan D. A., Sutphin P. D., Yen S. E., Giaccia A. J. (2005) Mol. Cell. Biol. 25, 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahon P. C., Hirota K., Semenza G. L. (2001) Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasper L. H., Boussouar F., Boyd K., Xu W., Biesen M., Rehg J., Baudino T. A., Cleveland J. L., Brindle P. K. (2005) EMBO J. 24, 3846–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zucker S., Hymowitz M., Conner C. E., DiYanni E. A., Cao J. (2002) Lab. Invest. 82, 1673–1684 [DOI] [PubMed] [Google Scholar]
- 28.Han J., Wang Y., Wang S., Chi C. (2008) J. Cell Sci. 121, 2217–2223 [DOI] [PubMed] [Google Scholar]
- 29.Mayer G., Boileau G., Bendayan M. (2003) J. Cell Sci. 116, 1763–1773 [DOI] [PubMed] [Google Scholar]
- 30.Deryugina E. I., Soroceanu L., Strongin A. Y. (2002) Cancer Res. 62, 580–588 [PubMed] [Google Scholar]
- 31.Sounni N. E., Devy L., Hajitou A., Frankenne F., Munaut C., Gilles C., Deroanne C., Thompson E. W., Foidart J. M., Noel A. (2002) FASEB J. 16, 555–564 [DOI] [PubMed] [Google Scholar]
- 32.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 33.Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 34.Labrecque L., Nyalendo C., Langlois S., Durocher Y., Roghi C., Murphy G., Gingras D., Béliveau R. (2004) J. Biol. Chem. 279, 52132–52140 [DOI] [PubMed] [Google Scholar]
- 35.Uekita T., Itoh Y., Yana I., Ohno H., Seiki M. (2001) J. Cell Biol. 155, 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uekita T., Gotoh I., Kinoshita T., Itoh Y., Sato H., Shiomi T., Okada Y., Seiki M. (2004) J. Biol. Chem. 279, 12734–12743 [DOI] [PubMed] [Google Scholar]
- 37.Gonzalo P., Guadamillas M. C., Hernández-Riquer M. V., Pollán A., Grande-García A., Bartolomé R. A., Vasanji A., Ambrogio C., Chiarle R., Teixidó J., Risteli J., Apte S. S., del Pozo M. A., Arroyo A. G. (2010) Dev. Cell 18, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshino D., Tomari T., Nagano M., Koshikawa N., Seiki M. (2009) J. Biol. Chem. 284, 27315–27326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anilkumar N., Uekita T., Couchman J. R., Nagase H., Seiki M., Itoh Y. (2005) FASEB J. 19, 1326–1328 [DOI] [PubMed] [Google Scholar]
- 40.Nyalendo C., Michaud M., Beaulieu E., Roghi C., Murphy G., Gingras D., Béliveau R. (2007) J. Biol. Chem. 282, 15690–15699 [DOI] [PubMed] [Google Scholar]
- 41.Moss N. M., Wu Y. I., Liu Y., Munshi H. G., Stack M. S. (2009) J. Biol. Chem. 284, 19791–19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Alessio S., Ferrari G., Cinnante K., Scheerer W., Galloway A. C., Roses D. F., Rozanov D. V., Remacle A. G., Oh E. S., Shiryaev S. A., Strongin A. Y., Pintucci G., Mignatti P. (2008) J. Biol. Chem. 283, 87–99 [DOI] [PubMed] [Google Scholar]
- 43.Rozanov D. V., Ghebrehiwet B., Ratnikov B., Monosov E. Z., Deryugina E. I., Strongin A. Y. (2002) FEBS Lett. 527, 51–57 [DOI] [PubMed] [Google Scholar]
- 44.Wang P., Wang X., Pei D. (2004) J. Biol. Chem. 279, 20461–20470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.