Abstract
To obtain a detailed picture of sulfur deprivation-induced H2 production in microalgae, metabolome analyses were performed during key time points of the anaerobic H2 production process of Chlamydomonas reinhardtii. Analyses were performed using gas chromatography coupled to mass spectrometry (GC/MS), two-dimensional gas chromatography combined with time-of-flight mass spectrometry (GCxGC-TOFMS), lipid and starch analysis, and enzymatic determination of fermentative products. The studies were designed to provide a detailed metabolite profile of the solar Bio-H2 production process. This work reports on the differential analysis of metabolic profiles of the high H2-producing strain Stm6Glc4 and the wild-type cc406 (WT) before and during the H2 production phase. Using GCxGC-TOFMS analysis the number of detected peaks increased from 128 peaks, previously detected by GC/MS techniques, to ∼1168. More detailed analysis of the anaerobic H2 production phase revealed remarkable differences between wild-type and mutant cells in a number of metabolic pathways. Under these physiological conditions the WT produced up to 2.6 times more fatty acids, 2.2 times more neutral lipids, and up to 4 times more fermentation products compared with Stm6Glc4. Based on these results, specific metabolic pathways involving the synthesis of fatty acids, neutral lipids, and fermentation products during anaerobiosis in C. reinhardtii have been identified as potential targets for metabolic engineering to further enhance substrate supply for the hydrogenase(s) in the chloroplast.
Keywords: Bioenergetics, Chloroplast, Mass Spectrometry (MS), Metabolism, Photosynthesis, C. reinhardtii, Biofuel, Biohydrogen, Metabolomics, Microalgae
Introduction
Renewable, CO2-free energy is increasingly important due to concerns over fuel security and the increase in atmospheric CO2 concentration. Plants and cyanobacteria use oxygenic photosynthesis to convert sunlight and water into oxygen and chemical energy. A specific group of green microalgae and cyanobacteria, including the microalga Chlamydomonas reinhardtii, have evolved the additional ability to use sunlight for the production of molecular H2 (1–5). The process of H2 production has been described in detail for C. reinhardtii. Under anaerobic conditions two oxygen-sensitive FeFe-hydrogenases (HydA1 and HydA2) are induced (6) catalyzing the reduction of protons to molecular H2. There are two possible sources of electron supply: light energy is needed to generate protons and electrons from the water-splitting reaction (7) and, in parallel, a Photosystem II (PSII)3-independent process uses electrons originating from the breakdown of starch (8, 9). In both cases the reduced ferredoxin serves as electron donor for the FeFe-hydrogenases. Anaerobic culture conditions, required for H2 production, can be achieved by bubbling inert gases through the culture or by sulfur depletion of the culture medium. During sulfur depletion the oxygen in the culture is consumed, and H2 production can be observed (8).
Recently, several molecular approaches have been applied to gain more detailed insights into the H2 production metabolism (10–16), to guide molecular genetics for further improvement of H2 production capacities (9, 17–20). Most recently, more attention was paid to systems biology approaches (21–25) to obtain a more detailed picture of the processes that are interconnected with hydrogenase activity (21, 24, 26, 27). The aim of all of these systematic approaches was to understand the regulatory mechanisms of substrate supply to hydrogenase and to identify bottlenecks of the H2 production process as a precondition for subsequent targeted metabolic engineering approaches.
In particular metabolomics represents an important component of systems biology research. Currently, metabolome analyses are still limited by the number of unequivocally identified compounds, by difficulties in their exact quantification and limited information on the fluxes of intermediates. However, the advanced analysis of complex metabolite mixtures, termed metabolite profiling, has been made possible through recent advances in modern technologies consisting of metabolite separation techniques and high resolution detection systems (28). Advanced metabolite profiling now opens up the possibility to follow the physiological adaptation from the aerobic to the anaerobic H2-producing lifestyle of C. reinhardtii.
This article reports on the differential analysis of metabolite profiles between Stm6Glc4, the newest and most advanced generation of our high H2-producing C. reinhardtii strains (18), and the corresponding WT cc406 before and during H2 production using two-dimensional GC coupled with time of flight MS (GCxGC-TOFMS), which has resolved an order of magnitude more compounds than previously reported (29). Stm6Glc4 is a cell line developed by inserting a hexose symporter into the high H2-producing mutant Stm6. It therefore couples regular photosynthetic H2 production with the ability to convert glucose into H2.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Liquid cultures of C. reinhardtii wild-type (WT) cc406 and mutant strain Stm6Glc4 (18) were grown in Tris acetate phosphate medium (30) under continuous illumination (40 μmol × m−2 × s−1 white light, measured with a QSL 2100 laboratory radiometer (Biospherical Instruments Inc.)). The WT cc406 was selected as reference strain, because Stm6Glc4 originated from a similar genetic background. For H2 production experiments, cultures were grown under continuous illumination in Tris acetate phosphate medium until they reached the late logarithmic growth phase. After changing the medium (triplicate washes with sulfur-depleted Tris acetate phosphate medium followed by centrifugation at 2,500 × g, 4 min, 20 °C), the cultures were adjusted to ∼1.2 × 107 cells per ml (∼27 μg of chlorophyll/ml) and supplemented with 1 mm glucose for optimal H2 production in Stm6Glc4 (18). The gas-tight bioreactors were then illuminated with continuous light (300 μmol × m−2 × s−1) for up to 120 h.
H2 Measurement
H2 production was determined volumetrically, and the gas quality was measured via gas chromatography as described in a previous study (18).
Chlorophyll Fluorescence Measurements
Chlorophyll a fluorescence yields were determined at room temperature (Mini-PAM, Walz, Germany). Photosynthetic quantum yield (ΦPSII) was determined by illuminating cell suspensions with actinic white light (300 μmol × m−2 × s−1, saturation pulse of 15,000 μmol × m−2 × s−1). Fluorescence parameters were recorded, and quantum yield was calculated (ΦPSII = (F′m − Ft)/F′m (31)) and recorded every 2 h.
Metabolite and Fatty Acids Extraction and Preparation of Samples for GC/MS and GCxGC-TOFMS Analysis
Algal culture samples (30 ml) were removed from the bioreactor side-port with a syringe and harvested by centrifugation at 3,000 × g for 1 min and immediately frozen in liquid nitrogen. 200 μl of the supernatant was also frozen in liquid nitrogen and later used for determination of fermentative products (see below).
One milliliter of 80% methanol containing 10 μm ribitol (internal standard) and 500 mg of silica beads (0.1-mm diameter, Roth) were added to 10 mg of freeze-dried cells. Cells were disrupted with a Precellys homogenizer (three times at 6,500 rpm for 45 s, Peqlab) and centrifuged at 14,500 rpm for 20 min. 700 μl of the supernatant was then dried under a nitrogen stream. The samples were incubated with 100 μl of methoxylamine hydrochloride (Sigma-Aldrich) in pyridine (20 mg/ml) for 90 min at 37 °C while stirring. N-Methyl-N-(trimethylsilyl)trifluoroacetamide (Macherey & Nagel) was then added, and the mixture was incubated for 30 min at 37 °C while stirring (32).
GC/MS Instrument Settings
GC/MS analysis was performed with a TraceGC gas chromatograph and a PolarisQ ion trap mass spectrometer equipped with an AS2000 auto sampler (ThermoFinnigan, Dreieich, Germany) according to a previous study (33). Splitless injection of 1-μl sample volume was conducted at 300 °C injector temperature. The gas chromatograph was equipped with a 30-m × 0.25-mm VF-5 ms column coated with 0.25-μm of 5% diphenyl and 95% dimethylsiloxane (Varian Deutschland GmbH, Darmstadt, Germany). The interface temperature was set to 250 °C, and the ion source to 200 °C. Helium was used as carrier gas and was set to a constant flow of 1 ml min−1. The oven temperature was heated for 1 min at 80 °C and was then raised in steps of 6 °C/min to 300 °C. Mass spectra were recorded at 20 scans s−1 using a scanning range of 50–750 m/z.
GC/MS Metabolite Identification and Quantification
Metabolite and fatty acids identification was performed according to a previous study (32). The evaluation of the chromatograms was performed with Xcalibur software (ThermoFinnigan). Metabolites and fatty acids were identified by comparison with the NIST 05 library (National Institute of Standards and Technology, Gaithersburg, MD; ThermoFinnigan), the Golm Metabolome Data base (GMD), a metabolite library obtained from the Max Planck Institute of Molecular Plant Physiology (MPI, Golm, Germany) (34), and verified with purified standards (Sigma-Aldrich). All spectra were manually reviewed. Quantification was based on the internal standard ribitol.
GCxGC-TOFMS Settings and Data Analysis
GCxGC-TOFMS analysis was performed using a LECO (St. Joseph, MI) Pegasus 4D time-of-flight mass spectrometer. The Pegasus 4D system was equipped with an Agilent 6890 gas chromatograph. The following instrument settings were used for analysis.
The inlet temperature was set to 275 °C. A 30-m × 0.25-mm × 0.25-μm film thickness, Rtx-5ms (Restek Corp.), capillary column was used as the primary column. The secondary column was a BPX-50 (SGE) 2-m × 0.1-mm × 0.1-μm capillary column. The temperature program of the primary oven was set to the following conditions: 70 °C for 2 min, 4 °C/min to 180 °C, 2 °C/min to 230 °C, and 4 °C/min to 325 °C, held for 3 min. The secondary oven was programmed with an offset of 15 °C to the primary oven temperature. The thermal modulator was set at 30 °C relative to primary oven and with a modulation time of 5 s and a hot pulse time of 0.4 s.
The mass spectrometer ion source temperature was set at 200 °C, and the ionization was performed at −70 eV. The detector voltage was 1600 V, and the stored mass range was 50–750 m/z with an acquisition rate of 200 spectra/s.
The samples were processed automatically by using the LECO ChromaTOF® software v.4.22 (LECO,) at a signal to noise ratio of 100. The baseline offset was 0.8, and the two peak widths were set at 0.2 s (as measured from baseline to baseline) and 15 s (first dimension). The background peaks originating from the column bleed or solvent tailing were eliminated by the processing method by using the classification feature of the software. Analytes were identified by database searches using the NIST 05 library and the GMD metabolite library. All compounds with a similarity of >800 were selected. For quantification, the peak area was calculated by using specific target ions or the unique mass.
Fisher ratios were calculated by using the “statistical compare” feature of the LECO ChromaTOF® v.4.22. This function calculates differences in the average of specific analytes from different sample groups (e.g. Stm6Glc4 versus WT at Peak H2) and differences in the variance of specific analytes from different sample groups. The Fisher ratio is subsequently calculated by dividing the differences of the average of each analyte by the differences of the variance in each analyte from different sample groups. A high Fisher ratio value indicates great variances between a specific analyte of two different sample groups and vice versa (35). Due to the good reproducibility of two-dimensional retention times, chromatograms were displayed as a contour plot, and an overlay of these plots could be constructed with the open source software OpenOffice Impress.
Nile Red Lipid Analyses
Total neutral lipids amount was evaluated with a Nile Red fluorescence assay, performed as described in a previous study (36), and optimized for Chlamydomonas cells. For lipid analyses, 1 ml of liquid cell culture was harvested by centrifugation (3000 × g for 1 min). The pellet was resuspended in water and diluted to the concentration of 1.2 × 107 cells/ml. 2 ml of the cell suspension was stained with 4 μl of Nile Red (Sigma-Aldrich, 250 μg/ml in acetone), vortexed for 30 s, and transferred into a multiwell plate (200 μl per well). Fluorescence was measured with the Infinite M200 spectrofluorometer (Tecan) using the excitation and emission wavelengths of 530 and 580 nm, respectively. The results are reported as fluorescence units adjusted to cell number: (fluorescence after Nile Red staining − fluorescence without Nile Red staining) × dilution factor × 106 cells per ml.
Determination of Fermentative Products
Determinations of acetate, formate, and ethanol were performed by using the Roche Diagnostics GmbH, Mannheim/R-Biopharm enzymatic assay kits (catalog nos. 10-148-261-035, 10-979-732-035, and 10-176-290-035). 30 μl of thawed and centrifuged supernatant was used in a 1-ml final reaction volume.
Starch Analysis
Algal cultures (1.2 × 107 cells/ml) were centrifuged at 2,000 × g for 5 min, and the pellets were immediately frozen in liquid nitrogen and stored at −80 °C. 1 ml of methanol (100%, −20 °C) was added, and the pellets were thawed on ice. After centrifugation (5 min, 18,000 × g) pellets were washed with 1 ml of sodium acetate buffer (1 m). After centrifugation (1 min, 16,000 × g) the pellets were resuspended in 350 μl of sodium acetate buffer and autoclaved (15 min at 120 °C) for starch solubilization. Cellular debris was removed by centrifugation (18,000 × g, 2 min), and the supernatant was taken for starch analysis with an enzymatic assay kit (catalog no. 10-207-748-035, Roche Diagnostics GmbH, Mannheim/R-Biopharm). 30 μl of supernatant were used in a 1-ml final reaction volume.
Statistical Analysis
A Student's t test was performed using the open source software “R” (37) for statistical analyses of metabolite changes.
Visualization of Cellular Pathways
Metabolic pathways were visualized by using the MapMan platform (38).4 MapMan software version 3.1.0 was used for data processing with the settings adjusted to the MapMan pathway, “Metabolites,” and for MapMan mapping, “C. reinhardtii” (38).
RESULTS
The Interplay between PSII Activity and H2 Production
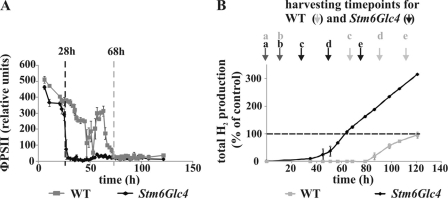
H2 production during anaerobiosis, induced by sulfur depletion, was analyzed in the high H2-producing mutant Stm6Glc4 and compared with the WT. This differential analysis approach was set up to identify in particular those changes in metabolites that are potentially related to the high H2 production phenotype in Stm6Glc4. Photosynthetic efficiency was monitored by measuring the quantum yield of PSII during cultivation in sulfur-depleted medium. Sulfur depletion reduces the rate of PSII repair as it blocks the incorporation of sulfur-containing amino acids into the D1 protein during the repair cycle of the photo-damaged subunit (40, 41). As expected, the quantum yield of PSII in Stm6Glc4 rapidly declined to nearly zero within 28 h after transfer into sulfur-depleted medium. With the drop of photosynthetic activity, anaerobiosis was gradually established in the culture (42). Interestingly, the WT showed a much slower and delayed decline phase lasting over a period of around 68 h (Fig. 1A). This phenotype may be caused by an impaired PSII repair mechanism in the mutant, resulting in an earlier establishment of anaerobiosis. As a consequence, H2 production by the oxygen-sensitive hydrogenase(s) started 51 h earlier in Stm6Glc4 compared with the WT (after 35 h for Stm6Glc4 versus 86 h for WT, Fig. 1B). In addition, the Stm6Glc4 H2 production phase was maintained for a much longer period of time (215 h versus 79 h in WT; data not shown) with higher peak H2 production rates (4.9 ml/h versus 2.8 ml/h in WT; data not shown). The experiment was stopped after 120 h during the peak H2 production phase, before the H2 production cycle was completed. Until then, the WT had produced a total amount of 86 ml H2/liter cell culture (set to 100% in Fig. 1B), whereas Stm6Glc4 had produced 284 ml H2/liter cell culture. Based on these physiological differences distinct harvesting time points for comparative metabolomics had to be selected for the WT and mutant Stm6Glc4 (see Fig. 1B). For both strains, the first samples were taken at a time point directly before the transfer into sulfur-depleted medium (time point a, “Peak O2”) followed by a second sample collection 16 h later. At this time point cell cultures are subjected to sulfur depletion but still contain oxygen produced by the residual PSII activity (time point b). Three additional samples were collected: 28 (Stm6Glc4) or 68 (WT) h after the transfer when anaerobiosis was reached (time point c); 24 h after the establishment of anaerobiosis when H2 production started (time point d); and 48 h after establishment of anaerobiosis during the peak H2 production period (time point e, “Peak H2”).
FIGURE 1.
Correlation between photosynthetic quantum yield and H2 production rates of C. reinhardtii WT and Stm6Glc4 during the shift from aerobiosis to anaerobiosis (induced by S-deprivation). A, quantum yield of PSII (ΦPSII) of sealed, sulfur-depleted C. reinhardtii cultures. B, total H2 production during S-deprivation. Harvesting time points (a–e) are labeled. Each curve represents an average of at least three biological replicates. Error bars indicate standard errors. The amount of H2 produced in the WT was set to 100%.
GC/MS Analysis of the Metabolome of C. reinhardtii WT and Mutant Stm6Glc4
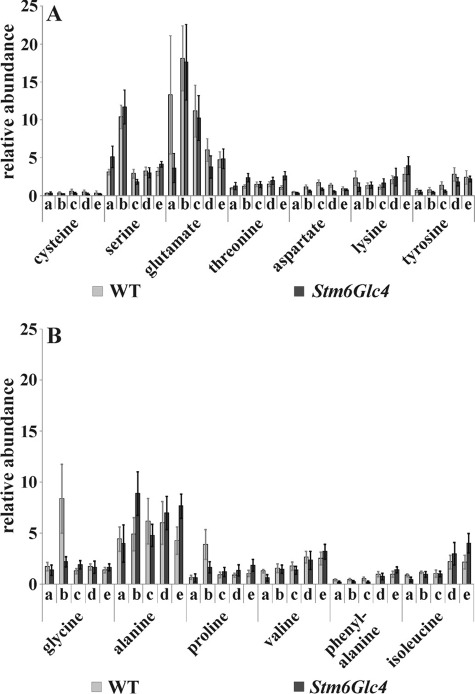
The metabolome of the WT and of Stm6Glc4 was analyzed by GC/MS at the five time points described above. As a result, 128 chromatographic peaks could be detected with 80 compounds being successfully identified. This was achieved through comparison with the GMD and the metabolite library of the National Institute of Standards and Technology (NIST 05). 48 of the identified metabolites were verified by comparison with reference compounds and classified according to their occurrence in distinct metabolic pathways. The relative abundance of each of these metabolites is shown in Figs. 2–4 and supplemental Table S1 and will now be described in more detail.
FIGURE 2.
Relative abundance of amino acids detected by GC/MS at time points before and during H2 production process in the WT (gray) and Stm6Glc4 (black). A, polar amino acids; B, non-polar amino acids. Time points a–e are labeled as described (see Fig. 1B). Each value represents an average of at least nine measurements (three biological replicates and three technical replicates). Error bars indicate standard errors.
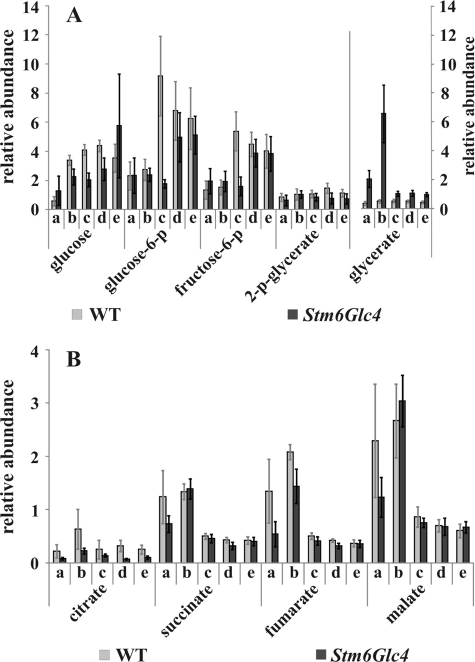
FIGURE 3.
Relative abundance of the intermediates of the glycolysis pathway (A) and the citric acid cycle/glyoxylate cycle (B) detected by GC/MS at time points before and during H2 production process in the WT (gray) and Stm6Glc4 (black). Time points a–e are labeled as described (see Fig. 1B). Each value represents an average of at least nine measurements (three biological replicates and three technical replicates). Error bars indicate standard errors.
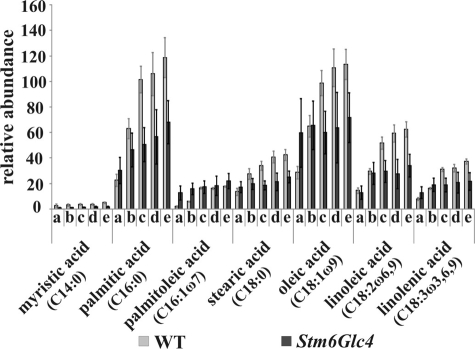
FIGURE 4.
Relative abundance of fatty acids detected by GC/MS at time points before and during H2 production process in the WT (gray) and Stm6Glc4 (black). Time points a–e are labeled as described (see Fig. 1B). Each value represents an average of at least nine measurements (three biological replicates and three technical replicates). Error bars indicate standard errors.
Amino Acid Metabolism and Photorespiration Pathway
Of the 13 amino acids detected, serine, glutamate (Fig. 2A), glycine, and alanine (Fig. 2B) showed the highest abundance, followed by proline, valine, and isoleucine (Fig. 2B). Threonine, aspartate, lysine, tyrosine (Fig. 2A), and phenylalanine (Fig. 2B) were only detected in small quantities. The sulfur-containing amino acid cysteine was only detectable in trace amounts (Fig. 2A) and showed a further decrease during H2 production induced by sulfur depletion.
It could be concluded that the level of most of the amino acids increased significantly (Fig. 2 and supplemental Table S2) during H2 production process (time points a–e) in Stm6Glc4, with the exceptions of glutamate, serine, and glycine. These levels increased significantly at time point b (see supplemental Table S2). The levels of alanine increased significantly during H2 production in Stm6Glc4. In detail, the levels first increased significantly (time point b), then decreased (time point c), and again increased at the last two time points. In contrast, the levels of alanine remained more constant during H2 production in the WT, no significant increase at time points b or e could be observed (Fig. 2, A and B, and supplemental Table S2). For the WT a statistically significant increase could only be determined for the amino acid phenylalanine when time points a–e were compared (Fig. 2, A and B, and supplemental Table S2). However, the level of serine increased significantly at time point b. An increasing level of amino acids during the H2 production phase indicates increased protein-breakdown processes (41) and/or increased de novo synthesis of amino acids (27).
Particularly notable was that Stm6Glc4 cultures showed a higher ratio of increase of the amino acids alanine, isoleucine, lysine, phenylalanine, proline, threonine, and valine, during the overall H2 production process (time points a–e) compared with the WT (supplemental Table S1). In contrast, the WT showed higher amounts of glutamate at the time point preceding the transfer into sulfur-depleted medium (time point a), and of glycine 16 h after transfer into sulfur depletion (time point b), most likely as a consequence of an increase of the photorespiration pathway in sulfur-free, oxygen-containing medium.
Glycine as well as serine and glycerate are synthesized in the course of the photorespiratory glycolate cycle, which is initiated by the oxygenase activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (43). Via this oxygenation reaction Rubisco adds O2 to the ribulose-1,5-bisphosphate (instead of CO2 as occurs during photosynthesis) resulting in the intermediate 2-phosphoglycolate. The latter is converted into glycolate and glyoxylate and subsequently into glycine, serine, pyruvate, glycerate, and 3-p-glycerate. Serine, glycine, and glycerate (Figs. 2A, 2B, and 3A) levels were significantly increased at time point b in Stm6Glc4. The levels of serine and glycine also were found to be highest at time point b in the WT with serine showing a significant increase. From these observations we conclude that the photorespiratory glycolate cycle increases as a result of sulfur depletion, until anaerobiosis is established.
Both strains contained comparably large amounts of glutamate and alanine at time point b (see Fig. 2, A and B). This could be explained as a consequence of a strong increase of the citric acid cycle and/or glyoxylate cycle metabolites (see paragraph below), because the precursor for glutamate synthesis, α-ketoglutarate, is a component of the citric acid cycle, and alanine synthesis is dependent on glutamate as the amino donor during synthesis from pyruvate (43).
Glycolysis
It has been previously reported that starch accumulates in C. reinhardtii cells during the first 24 h after transfer into sulfur-depleted medium and is then used as an energy source under anaerobic conditions (27, 41, 44). Relative amounts of the intermediates of the glycolysis pathway (glucose, glucose 6-phosphate, fructose 6-phosphate, and 2-p-glycerate) are presented in Fig. 3A. A general increase in the levels of all glycolysis intermediates was observed in both strains when H2 production set in. Glucose 6-phosphate and fructose 6-phosphate showed a significant increase in Stm6Glc4, whereas glucose and fructose 6-phosphate levels significantly increased in the WT (Fig. 3A and supplemental Table S2). This indicates that during H2 production glycolysis was enhanced for an increased starch breakdown, most likely to provide an alternative energy source when oxygen-dependent mitochondrial respiration is blocked. Higher levels of glucose, glucose 6-phosphate, and fructose 6-phosphate were detected in the WT compared with mutant Stm6Glc4 during the H2 production period, possibly as a result of a more efficient metabolic flux of glycolysis products toward H2 production in the high H2 production strain Stm6Glc4.
Citric Acid Cycle and Glyoxylate Cycle
In contrast to the metabolites of the glycolysis pathway, the intermediates of the citric acid cycle and/or glyoxylate cycle (Fig. 3B) showed the highest abundance before H2 production set in (at time point b). As soon as anaerobiosis was established (time point c), the amount of these intermediates sharply decreased. This suggests that under anaerobic conditions the citric acid cycle and/or glyoxylate cycle are down-regulated.
The levels of all intermediates of the citric acid cycle and/or glyoxylate cycle increased significantly at time point b and decreased significantly during H2 production in Stm6Glc4 (Fig. 3B, supplemental Table S2). In the WT these levels increased slightly at time point b, but decreased significantly during H2 production (Fig. 3B, supplemental Table S2). Compared with Stm6Glc4, the WT showed higher levels of the two citric acid cycle/glyoxylate cycle intermediates measured, citrate and fumarate, before H2 production set in. These generally lower levels might be caused by the disruption of the nuclear encoded protein MOC1 in Stm6Glc4 (45), resulting in a reduced level of the rotenone-insensitive NADPH dehydrogenase. The high level of the intermediates of the citric acid cycle/glyoxylate cycle prior to H2 production is also reflected by the increase of the amino acids glutamate and alanine (see Fig. 2A & 2B) since glutamate synthesis and citric acid cycle are interconnected via the metabolite α-ketoglutarate, as mentioned earlier. When the cells reached anaerobiosis, the levels of all citric acid/glyoxylate cycle metabolites sharply decreased (Fig. 3B, time point c). This confirmed the results of (27) which also provided evidence for a decrease of the intermediates of the citric acid cycle/glyoxylate cycle 24 h after cultivation in sulfur-depleted medium (physiologically corresponding to our time point c in this study).
Accumulation of Fatty Acids and Neutral Lipids
In both strains, fatty acid levels strongly increased during sulfur depletion (Fig. 4 and supplemental Table S2), except palmitoleic acid, stearic acid, oleic acid, and linolenic acid in Stm6Glc4, which showed only a slight increase. Once anaerobiosis was reached, fatty acids remained at more constant levels (Fig. 4). It is well established that cells start to produce fatty acids and lipids as storage products during environmental stress situations (46). The fatty acids levels increased 2- to 6-fold in both strains. It is of particular note that the increase of the fatty acid levels in the WT was far more pronounced than in the mutant with levels reaching 2.6 times higher values (Fig. 4).
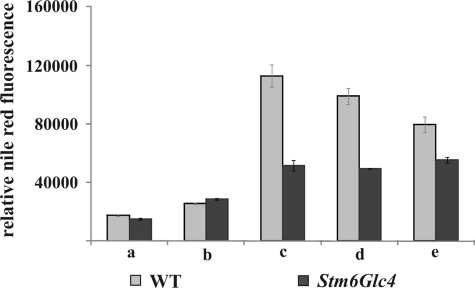
To study this striking phenotypic difference in more detail we determined the amount of neutral lipids in a separate series of experiments (Fig. 5) in addition to the analysis of the fatty acid levels. The highest levels of neutral lipids were detected at time point c when cells reached anaerobiosis. Compared with time point a, the WT reached 6.4-fold higher levels, Stm6Glc4 reached 3.4-fold higher lipid levels (supplemental Table S2). Again, the WT produced significantly more lipids compared with Stm6Glc4 (up to 2.2-fold) (Fig. 5). During the H2 production phase, neutral lipid levels remained constant in Stm6Glc4 and slightly declined in the WT. The fact that fatty acid and lipid levels were generally lower in the high H2 production mutant Stm6Glc4 suggests that fatty acid and lipid accumulation may compete with the substrate supply to the hydrogenases, especially in the WT.
FIGURE 5.
Relative amounts of lipids measured by relative fluorescence after Nile Red staining at time points before and during H2 production process in the WT (gray) and Stm6Glc4 (black). 1.2 × 107 cells per ml were used. Time points a–e are labeled as described (see Fig. 1B). Each value represents an average of at least eight measurements (two biological replicates, four technical replicates). Error bars indicate standard errors.
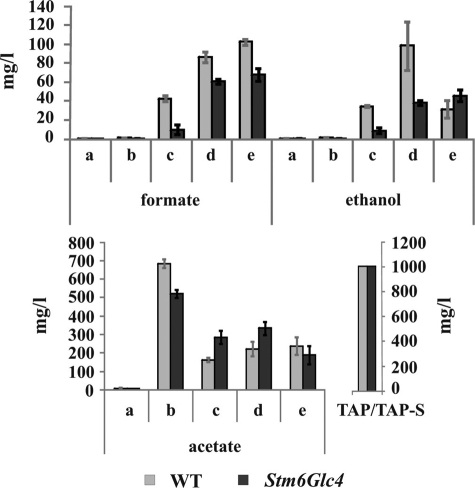
Enzymatic Analysis of Fermentative Products
The amounts of the fermentation products formate, ethanol, and acetate in the cell-free culture medium were analyzed enzymatically (Fig. 6). Formate and ethanol production and secretion into the medium could be detected in both strains immediately after the cultures reached anaerobiosis (Fig. 6, time point c) and the levels increased significantly over time (supplemental Table S2). Higher amounts of formate and ethanol were produced by the WT during anaerobiosis compared with Stm6Glc4.
FIGURE 6.
Enzymatic analysis of fermentative products during H2 production process in WT (gray) and Stm6Glc4 (black). Time points are labeled with a–e (see Fig. 1B). Each value represents an average of at least three measurements (three biological replicates). Error bars indicate standard errors.
Acetate levels were very low in the culture medium before the cells were transferred into sulfur-free medium (containing 1 g/liter acetate) (Fig. 6, time point a), because Chlamydomonas consumes acetate during heterotrophic/mixotrophic growth, presumably by incorporation into acetyl-CoA. The replenished acetate of the sulfur-free medium was consumed in both strains but only until the culture reached anaerobiosis. This suggests that acetate was consumed rapidly by the respiratory processes that lead to anaerobiosis. Once anaerobiosis was reached, the concentration of acetate remained relatively constant, indicating no net take-up or secretion of acetate during anaerobiosis (Fig. 6). Consequently, these data suggest that the acetate from the medium is only used as long as mitochondrial respiration takes place and is not used as a substrate for H2 production (8).
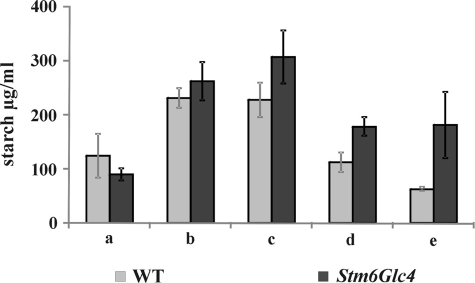
Starch Analysis of WT and Stm6Glc4
The intracellular starch content of WT and Stm6Glc4 was determined enzymatically. For both strains, samples were taken at time points a–e (Fig. 7). Both strains accumulated starch during the first hours of sulfur depletion (time points b and c). Interestingly, mutant Stm6Glc4 showed a significantly higher increase (3–4 times) of the starch levels during this pre-H2 production phase compared with WT (1.8 times) (Fig. 7 and supplemental Table S2). Stm6Glc4 accumulated a maximum of 308 μg/ml starch, whereas the WT only reached maximal starch levels of 232 μg/ml (both at time point c). A significantly stronger decrease of the accumulated starch during the H2 production process was observed in the WT (Fig. 7, time points c–e, and supplemental Table S2). Here, 70% of the starch amount was metabolized, whereas only 42% of the starch was metabolized in Stm6Glc4.
FIGURE 7.
Starch content in WT (gray) and Stm6Glc4 (black) monitored during the H2 production process. 1.2 × 107 cells per ml were used. Time points are labeled with a–e (see Fig. 1B). Each value represents an average of at least four biological replicates. Error bars indicate standard errors.
GCxGC-TOFMS Analysis of the Metabolome of C. reinhardtii WT and Stm6Glc4
To obtain a more detailed picture of metabolic changes during the H2 production process we applied GCxGC-TOFMS, a novel and powerful metabolic profiling method to analyze the metabolome of C. reinhardtii WT and Stm6Glc4. In contrast to GC/MS analysis (27), which only allows the separation of a limited number of metabolites, two-dimensional GCxGC-TOFMS (29) enhanced the analytical resolution dramatically. GCxGC-TOFMS analysis was performed with samples from both strains collected at time points a (“Peak O2”) and e (“Peak H2”). Because no fully automated processing of the massive amount of data generated by the new GCxGC-TOFMS method has been established so far, we restricted the experimental setup to these two time points.
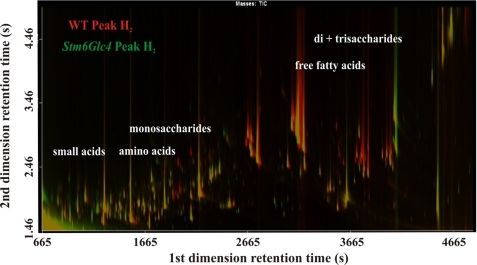
Metabolites were identified by their specific mass and subsequently compared (see supplemental Table S1). For visualization of the differences between WT and mutant Stm6Glc4 during peak H2 production, total ion current chromatograms were superimposed (Fig. 8), allowing easy detection of strongly up- and down-regulated metabolites. As a result, a variety of amino acids, fatty acids, and other compounds were detected, which showed great differences in abundance during H2 production. These compounds of interest were annotated using two independent libraries (GMD and NIST 05 libraries) (supplemental Table S1).
FIGURE 8.
Overlay of total ion current chromatograms displayed as a contour plot of two-dimensional GCxGC-TOFMS analysis. Chromatograms of WT and Stm6Glc4 during peak H2 production (time point e) were selected. Red or green peaks represent metabolites with significant differences in abundance with “green” representing higher amounts in Stm6Glc4 and “red” representing higher amounts in the WT. Yellow peaks represent metabolites that were equally abundant in both strains. Regions corresponding to specific compounds (e.g. small organic acids, amino acids, free fatty acids, and monosaccharides) are indicated. Differences between two samples regarding the amount of specific compounds can be easily detected and subsequently analyzed by this method.
As shown in Fig. 8, the amounts of free fatty acids at peak H2 production were higher in the WT compared with the mutant (Fig. 8, region for free fatty acids is indicated), confirming the GC/MS data described previously (Fig. 4, time points e). The higher levels of some amino acids in Stm6Glc4 compared with WT at peak H2 production could also be detected (Fig. 2, time point e and Fig. 8, the region for amino acids is indicated).
By using GCxGC-TOFMS we were able to detect up to 1168 peaks with a low signal to noise ratio (>100) and 553 chromatographic signals with a high signal to noise ratio (>800) (Table 1). Up to 775 of the peaks were co-eluting in the first GC dimension and could only be assigned after separation in the second GC dimension. Therefore, a great number of previously undetected compounds could be identified, indicating the great potential of the GCxGC-TOFMS technique. Interestingly, a number of unique peaks were detected for each strain at a given time point (Table 1).
TABLE 1.
Summary of the peak detection by GCxGC-TOFMS of WT and Stm6Glc4 samples before and during peak H2 production
Unknown compounds are shown in parentheses.
| WT peak O2 | WT peak H2 | Stm6Glc4 peak O2 | Stm6Glc4 peak H2 | |
|---|---|---|---|---|
| Total amount of significant peaks | 1168 | 1089 | 1078 | 1145 |
| Co-eluting peaks | 812 | 712 | 726 | 775 |
| Peaks detected with S/N ratio 800 | 553 | 521 | 525 | 541 |
| Peaks only detected in this sample | 161 (27) | 96 (5) | 157 (11) | 166 (52) |
Peak tables of the four sample groups “WT Peak O2,” “WT Peak H2,” “Stm6Glc4 Peak O2,” and “Stm6Glc4 Peak H2” were generated by comparison with the GMD and NIST 05 libraries and subsequently aligned with the software ChromaTOF (35). This software matches peaks by comparing their relative masses, peak height, peak area, retention time, and other individual parameters. Fisher ratios were calculated and used as a marker to highlight the chemical differences between different sample groups. The Fisher ratio is a statistical calculation, which is a feature of the “statistical compare” function of the LECO ChromaTOF. When two chromatographic peaks were compared, high Fisher ratios indicated a high probability of statistically significant differences of the corresponding analyte level (see “Experimental Procedures”). The Fisher ratios are shown in plots (see Fig. 9) and in tables (supplemental Tables S3–S6).
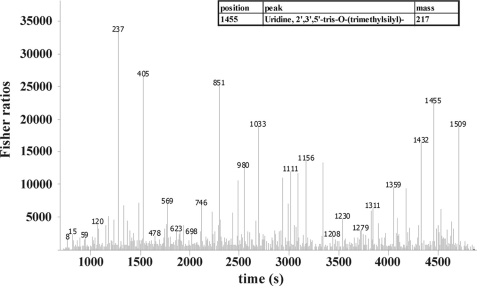
FIGURE 9.
Statistical comparison of GCxGC-TOFMS chromatograms using Fisher ratios. This plot shows the analysis of two processed chromatograms from both, WT at peak H2 and Stm6Glc4 at peak H2. The y axis represents the values for the Fisher ratio, the x axis the retention time scale. Peak heights in the Fisher ratio plot represent compounds with the highest chemical differences between the analytes. This figure represents an example of the analysis of the fisher ratio calculation. The high peak labeled with “1455” represents uridine as an example of a compound with a high Fisher ratio. Numeric values at each peak represent the position of the analyte in the processed compound table used for statistical comparison (compound table not shown).
When WT at Peak O2 was compared with Stm6Glc4 at Peak O2, 246 peak differences were detected. Comparison of both strains at Peak H2 resulted in the identification of 349 peak differences. 172 peak differences were detected when time points Peak O2 versus Peak H2 were compared in the WT, whereas 209 peaks could be identified to be different in mutant Stm6Glc4 at Peak O2 versus Peak H2. The supplemental Tables S3–S6 show an extract of important metabolites that were identified based on Fisher ratios higher than 1000.
Table 2 provides a comparison of all compounds, which were detected by GCxGC-TOFMS and GC/MS (see also supplemental Table S1). In this table the increase or decrease (“enrichment factors” (e.f.)) of individual metabolite levels at peak H2 production were compared with levels before H2 production. Metabolites identified based on the Fisher ratios higher than 1000 are indicated. The ratio of the relative changes (Stm6Glc4 e.f./WT e.f.) was also calculated (Table 2) with the idea that strong enrichment of a certain compound in the mutant as opposed to minor enrichment in the WT could indicate a special importance of the substance for the H2 production process.
TABLE 2.
Increase/decrease of metabolite levels at peak H2 production compared to levels before H2 production in WT and mutant Stm6Glc4
The first two columns show the enrichment factors (e.f.) of each metabolite at peak H2 production compared to the respective metabolite levels before H2 production. The third column shows the ratio of the enrichment factors (Stm6Glc4/WT) to illustrate the relative differences between the WT and the high H2 producing strain Stm6Glc4. The numbers indicate: 1, compound detected by GC/MS; 2, compound detected by GCxGC-TOFMS; 3, verified by comparison with standard references; 4, identified by database searches; and 5, Fisher ratio higher than 1000. Asterisks indicate whether the normalization was performed with GC/MS data (*) or with GCxGC-TOFMS data (**).
| Compound | WT | Stm6Glc4 | Ratio Stm6Glc4/WT |
|---|---|---|---|
| e.f. | e.f. | ||
| Glycolysis intermediates, sugars, and sugar alcohols | |||
| Galactose1,2,3,* | 1.24 | 2.27 | 1.83 |
| Glucose1,2,3,5,* | 6.13 | 4.43 | 0.72 |
| Glucose-6-p1,3,* | 2.69 | 2.18 | 0.81 |
| Fructose-6-p1,3,* | 2.98 | 1.95 | 0.65 |
| Dihydroxyacetone-p1,3,* | 2.80 | 0.79 | 0.28 |
| Glycerate-2-p1,3,* | 1.33 | 1.15 | 0.86 |
| p-Enolpyruvate1,3,* | 1.08 | 1.78 | 1.64 |
| Inositol1,3,* | 0.90 | 1.25 | 1.38 |
| Inositol-2-p1,3,* | 1.27 | 1.31 | 1.03 |
| Citric acid cycle intermediates | |||
| Citrate1,3,* | 1.14 | 1.19 | 1.05 |
| Succinate1,2,3,5,* | 0.34 | 0.55 | 1.63 |
| Fumarate1,3,* | 0.28 | 0.67 | 2.43 |
| Malate1,2,3,5,* | 0.26 | 0.54 | 2.06 |
| Pentose phosphate pathway | |||
| Ribose2,4,** | 0.23 | 0.24 | 1.02 |
| Amino acids | |||
| Alanine1,2,3,5,* | 0.97 | 1.92 | 1.98 |
| Asparagine2,4,** | 0.11 | 0.41 | 3.85 |
| Aspartate1,2,3,* | 1.86 | 2.36 | 1.27 |
| β-Alanine1,2,3,* | 1.66 | 3.40 | 2.05 |
| Citrulline2,4,** | 0.71 | 1.33 | 1.89 |
| Cysteine1,2,3,* | 1.17 | 0.67 | 0.57 |
| Glutamate1,2,3,* | 0.36 | 1.34 | 3.74 |
| Glycine1,2,3,* | 0.80 | 1.20 | 1.50 |
| Homocysteine1,3,* | 1.50 | 0.73 | 0.48 |
| Homoserine1,3,* | 0.31 | 0.65 | 2.14 |
| Isoleucine1,2,3,5,* | 2.44 | 8.48 | 3.47 |
| Leucine2,4,** | 2.22 | 3.06 | 1.38 |
| Lysine1,3,* | 1.21 | 3.52 | 2.91 |
| Methionine2,4,** | 0.13 | 0.18 | 1.36 |
| Ornithine2,4,** | NDa | 1.00 | 1.00 |
| Phenylalanine1,2,3,5,* | 2.15 | 6.83 | 3.18 |
| Proline1,3,* | 1.58 | 2.93 | 1.85 |
| Serine1,3,* | 1.02 | 0.81 | 0.79 |
| Threonine1,2,3,* | 1.10 | 2.05 | 1.86 |
| Tyrosine1,3,* | 3.43 | 4.57 | 1.33 |
| Organic acids | |||
| Benzoate2,4,** | 0.84 | 0.89 | 1.05 |
| Glycerate1,3,* | 1.19 | 0.48 | 0.40 |
| Oxalate2,4,** | ND | ND | ND |
| Ribonate2,4,** | 0.98 | 0.19 | 0.19 |
| Fatty acids | |||
| Linoleic acid1,3,5,* | 4.28 | 2.63 | 0.62 |
| Linolenic acid1,3,* | 4.76 | 1.68 | 0.35 |
| Oleic acid1,3,* | 3.95 | 1.20 | 0.30 |
| Palmitelaidic acid1,3,5,* | 8.83 | 1.73 | 0.20 |
| Palmitic acid1,2,3,5,* | 5.25 | 2.26 | 0.43 |
| Stearic acid1,2,3,5,* | 3.13 | 1.46 | 0.47 |
| Terpenoids, steroids, and vitamins | |||
| α-Tocopherol1,4,* | 1.91 | 0.90 | 0.47 |
| Cholesterol1,4,* | 0.92 | 0.53 | 0.58 |
| Dehydroergosterol2,4,** | 0.73 | 0.63 | 0.86 |
| Ergosterol1,4,* | 0.89 | 0.50 | 0.56 |
| Lycopene2,4,5,** | 0.25 | 0.33 | 1.31 |
| Pantothenate2,4,5,** | ND | 0.57 | 1.00 |
| Nucleosides and nucleobases | |||
| Adenine1,2,4,* | 0.55 | 0.63 | 1.16 |
| Adenosine1,2,4,5,* | 0.90 | 1.06 | 1.18 |
| Guanine1,2,4,5,* | 4.76 | 1.62 | 0.34 |
| Guanosine1,4,* | 1.58 | 1.30 | 0.82 |
| Purin-2-amine2,4,** | 0.72 | 2.62 | 3.62 |
| e.f. | e.f. | ||
| Pyrimidine2,4,** | 0.58 | 0.46 | 0.79 |
| Uracil2,4,5,** | 0.35 | 0.90 | 2.59 |
| Uridine2,4,5,** | 0.10 | 0.30 | 3.06 |
| Others | |||
| Calystegine1,4,* | 1.91 | 1.22 | 0.64 |
| Disulfide2,4,** | ND | ND | ND |
| Ethanolamine1,2,4,* | 1.01 | 0.52 | 0.51 |
| Ethanolaminephosphate2,4,5,** | 0.08 | 0.15 | 1.89 |
| Galactosylglycerol1,2,4,* | 1.16 | 0.64 | 0.55 |
| Glucuronolactone2,4,** | 0.29 | ND | ND |
| Glycerol-2-p1,3,* | 1.52 | 2.32 | 1.53 |
| Glycerol-3-p1,3,* | 1.34 | 1.17 | 0.88 |
| Phenylethanolamine2,4,** | ND | ND | ND |
a ND, not determined.
As a general conclusion, the metabolites detected by GCxGC-TOFMS confirmed the results obtained previously by our GC/MS study, e.g. the increase of glycolysis intermediates, sugars and sugar alcohols, the decrease of citric acid cycle/glyoxylate cycle intermediates, and the increase of the amount of many amino acids and fatty acids during the H2 production process (Figs. 2–4 and Table 2).
In addition, we were able to detect further compounds within the groups of amino acids, sugars, terpenoids, sugar alcohols, organic acids, and nucleosides by GCxGC-TOFMS, which could not be detected by one-dimensional GC/MS analyses. Amino acids like ornithine, asparagine, citrulline, leucine, and the sulfur-containing amino acid methionine, which was present during aerobic growth but not detectable during the H2 production phase, could only be identified via the two-dimensional GCxGC-TOFMS approach. Most of the newly identified amino acids showed an increased abundance during the peak H2 production phase (Table 2). When compared with the WT, the overall abundance of amino acids in Stm6Glc4 was clearly higher (Table 2). Of particular note was that ribose, a compound derived from the pentose phosphate pathway, was only detected by using GCxGC-TOFMS (Table 2). The observed decrease of ribose during H2 production suggests that phosphorylated sugars are only metabolized via glycolysis under these conditions. In addition some organic acids, nucleosides or nucleobases, terpenoids, carotenoids, and steroids such as α-tocopherol and ergosterol as well as ethanolamine, ethanolaminephosphate, phenylethanolamine, and lycopene were exclusively detected by using the advanced technique of GCxGC-TOFMS (Table 2).
Some of these components are highly interesting for H2 production in C. reinhardtii. α-Tocopherol is involved as an antioxidant in photoprotection reactions against high light (47). The level of this isoprenoid increased during H2 production in the WT, whereas it slightly decreased in Stm6Glc4, implying differences in stress related signaling between the two strains during anaerobiosis. It can be noted that in both strains the amount of steroids, which are synthesized by acetyl-CoA and the mevalonate pathway, generally decreased during H2 production. The amount of lycopene, as a precursor for β-carotene, also decreased during anaerobiosis. The first intermediate of carotenoid synthesis, isoprene, is synthesized exclusively by pyruvate and glyceraldehyde 3-phosphate in the non-mevalonate pathway (also referred to as the 2-C-methyl-d-erythritol-4-phosphate/1-deoxy-d-xylulose-5-phosphate pathway or MEP/DOXP pathway) (48). Ergosterol could only be detected by GCxGC-TOFMS, and its level showed a decrease during H2 production. This is interesting, because ergosterol is the predominant sterol in the membrane of C. reinhardtii (49) and indicates that important factors for membrane assembly do not seem to be required during H2 production as much as in steady-state cultures. This assumption was confirmed by the fact that ethanolamine, ethanolamine phosphate, and phenylethanolamine could only be detected during Peak O2 and not during H2 production.
Assignment of the Strain-dependent Metabolic Differences at Peak H2 Production to Major Cellular Pathways
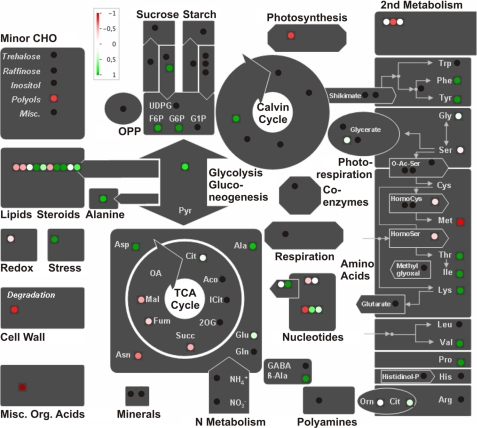
The data obtained by GC/MS and GCxGC-TOFMS were processed using the MapMan software platform (38)4 to summarize and visualize the metabolic changes in Stm6Glc4 during the H2 production phase according to their cellular pathways (Fig. 10). The data sets shown in Table 2 and supplemental Table S1 are summarized in Fig. 10 and therefore allow a rapid evaluation of the importance of the major cellular pathways for H2 production. All metabolite levels of the peak H2 state (time point e) were compared with the metabolite levels of peak O2 state (time point a). As a result, this illustration points to key metabolic pathways that are heavily affected during H2 production and are consequently putative targets for subsequent manipulation approaches to further enhance H2 production capacities of the cell. In particular, the higher lipid amounts at peak H2 production suggest a strong increase in fatty acid and lipid synthesis activity during the H2 production process (see Fig. 10, green spots in the “lipids/steroids” box; see also Figs. 4 and 5). Similar conclusions could be drawn for the increasing levels of amino acids (see Fig. 2). In contrast, the pink spots in the “lipids/steroid” box represent steroids that are slightly down-regulated (see Table 2). The red and pink spots for intermediates of the citric acid cycle also point to the fact that these compounds decreased during the H2 production process, suggesting a controlled down-regulation during anaerobiosis (see Fig. 3B, time points a and e).
FIGURE 10.
Schematic representation of the relative changes of metabolite levels during H2 production in the mutant strain Stm6Glc4. The figure was prepared with the MapMan visualization platform (38).4 Sixty metabolites were assigned to the major cellular pathways. Metabolites that increased during H2 production are marked in green; metabolites that decreased are marked in red. Metabolites with no changes are marked in white. Black points represent metabolites present in the MapMan platform, which were not detected in our metabolomic study.
DISCUSSION
The aim of this work was to intensively characterize the metabolic profiles of C. reinhardtii WT (cc406) and the high H2-producing mutant strain (Stm6Glc4) during defined stages of the H2 production process to identify potential key components that are crucial for efficient hydrogenase activity. A better understanding of the metabolic adaptation and especially of the differences between WT cells and high H2-producing strains could help to improve H2 production in C. reinhardtii. The direct parental strain of mutant Stm6 (and derivates), cc1618, shows an arginine auxotrophy, and for this reason could not be used for the comparative physiological studies. We therefore used strain cc406 as the WT control strain, which has a similar genetic background to that of Stm6 (i.e. strain cc124 with a cell wall deficiency “cw15” phenotype) for our experiments. It should be noted that the H2 production kinetics of the WT control and strain Stm6Glc4 differ significantly in that the WT reaches anaerobiosis and subsequent H2 production 40 h later than the mutant (Fig. 1, time point c). These kinetic differences made it necessary to determine time points c, d, and e individually for each strain. Although these time points should reflect comparable physiological states in terms of H2 production, we cannot exclude the possibility that the differences of the incubation period could have had some additional, unknown effects on the cells.
Metabolic compound levels were monitored with standard GC/MS techniques and with the newly developed high resolution GCxGC-TOFMS (29) technique. Recently the metabolome of Stm6, the first generation of high H2 production mutants (9), was characterized at lower resolution via GC/MS and by NMR studies (27). By applying GCxGC-TOFMS in our improved experimental setup, the quality of the analysis is taken to the next level because chromatographic peak detection is increased by ∼10-fold.
Two-dimensional GCxGC-TOFMS Analysis during H2 Production Revealed a Variety of Metabolic Changes
The improved approach used to analyze the cellular metabolome is based on two-dimensional GC coupled with time-of-flight MS (GCxGC-TOFMS) and increases the amount of detectable peaks dramatically (29). The combined GCxGC-TOFMS and GC/MS data of our study generally were in good agreement with results obtained previously (27). However, as a consequence of the improved separation of substances, which co-elute in conventional GC, the two-dimensional GCxGC-TOFMS allowed us to detect nearly ten times as many metabolic peaks. Therefore a number of new compounds were found, which had not been described before, generating a much more detailed metabolic map of the H2 production process. By applying GCxGC-TOFMS we were for the first time able to identify ribose, a compound derived exclusively from the pentose phosphate pathway, and obtained more detailed information on the terpenoids and steroids metabolism and of many amino acids, organic acids, and compounds of membrane synthesis. It has to be mentioned that, up to now, no fully automated annotation program has been available for GCxGC-TOFMS, making the processing of massive amounts of data generated by this new method rather time-consuming. Nevertheless, the “statistical compare” feature of the LECO ChromaTOF® software affords a good possibility of analyzing these data.
The combination of GC/MS, GCxGC-TOFMS, enzymatic determination of fermentation products, starch, and the analyses of neutral lipids revealed a variety of metabolic changes in the green algal cells during sulfur-induced H2 production. A general trend observed was that, during the H2 production process, intermediates of the citric acid cycle/glyoxylate cycle decreased, whereas the levels of intermediates of the glycolysis, amino acids, fermentative products, fatty acids, and lipids increased. Metabolic processes like steroid, terpenoid, and isopropenoid synthesis were reduced during H2 production, as were most of the pathways for nucleoside and nucleobase synthesis. These results confirm that, under anaerobic H2-producing conditions, only a selected number of metabolic processes are maintained to ensure survival of the organism.
Several switches within the intracellular metabolism are observed when cells are transferred into sulfur-depleted conditions, including a sharp decline of Rubisco protein levels during the first 24 h, which results in an inhibition of CO2 fixation (7). Under these conditions, acetate from the medium is consumed (Fig. 6) and starch accumulates (27, 41). Starch subsequently appears to serve as a substrate source for H2 production (41, 44). Starch levels decreased to 70% of the maximum levels in the WT and to 42% of the maximum level in Stm6Glc4 (Fig. 7). As a consequence the intermediates of the glycolysis pathway increased (Fig. 3A). It has been reported previously that metabolite levels of fermentative products and transcript levels of genes of fermentative pathways increase during anaerobic cultivation in the dark (21). Recent metabolome studies have demonstrated that these fermentative compounds are also produced during photobiological H2 production in the light (27). Our data confirm these results, because formate and ethanol levels were increased under these conditions (Fig. 6). Detailed microarray studies during sulfur depletion also demonstrated the increase of transcript levels of genes associated with the fermentative and the starch metabolism (26, 50) as well as the increase of hydrogenase-encoding transcripts HydA1, HydA2, and HydEF (26). The increase of neutral lipids (triacylglycerides), sterol esters, and fatty acids reported previously (27) could also be verified by our data (Figs. 4 and 5 and Table 2).
The Competition for Pyruvate during Anaerobiosis Is a Key Factor for Efficient H2 Production
The degradation of starch as a feedstock for glycolysis has been reported to play a major role in H2 production in C. reinhardtii (27, 41, 44). Our metabolic studies revealed that compared with Stm6Glc4, the WT surprisingly accumulated less starch in the early sulfur depletion phase but metabolized more accumulated starch during the peak H2 production phase. The fact that apparently more starch was consumed in the WT, but at the same time less H2 was produced compared with Stm6Glc4, clearly indicates that starch degradation can be replaced by external glucose feeding. Stm6Glc4 has the ability of limited glucose uptake from the medium and glucose is needed for producing pyruvate as a key intermediate product via glycolysis.
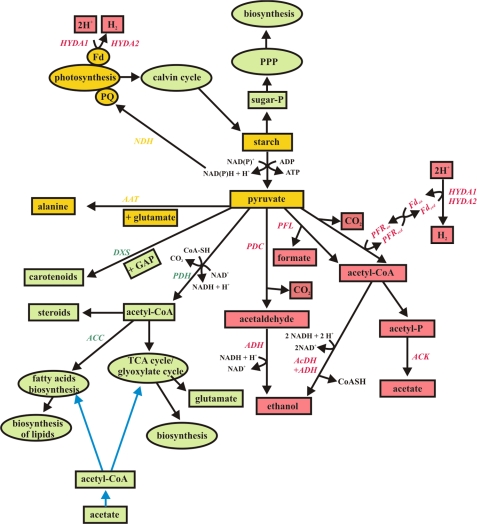
During aerobic growth, pyruvate is used for many biosynthesis pathways, as illustrated in Fig. 11. When the culture is subjected to sulfur limitation but still aerobic, starch, fatty acid, and lipid synthesis are strongly induced. In contrast when anaerobic conditions are established, it seems that pyruvate is mainly used for amino acid synthesis and fermentative pathways to control the cellular redox balance (21, 24, 25, 46) (Figs. 2 and 4–6). We conclude from our data that, during anaerobic conditions, accumulated starch and phosphorylated sugars are preferentially catabolized via the glycolysis pathway and not via the pentose phosphate pathway, because the detected compound of the latter decreased significantly. This conclusion is confirmed by the fact that the intermediates of the glycolysis pathway showed a strong increase during the H2 production process, during which accumulated starch reserves were consumed. During glycolytic starch breakdown, NAD(P)+ is reduced to NAD(P)H + H+, which can then serve as the electron source for H2 production, because it can be used to reduce the PQ-pool, catalyzed by the NAD(P)H dehydrogenase (NDH, Fig. 11). Pyruvate, generated by glycolysis, is then used for fermentation (Fig. 6), and/or for amino acid biosynthesis, especially alanine (Fig. 2B).
FIGURE 11.
Visualization of the pyruvate metabolism before and during H2 production. Green color represents metabolites/pathways present during the aerobic state; red color represents metabolites/pathways present during the anaerobic state, yellow color represents pathways occurring in both states. Blue arrows symbolize acetate uptake and feeding. AAT, alanine aminotransferase; ACC, acetyl-CoA carboxylase; AcDH, acetaldehyde dehydrogenase; ACK, acetate kinase; ADH, alcohol dehydrogenase; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; GAP, glyceraldehyde-3-phosphate; Fd, ferredoxin; HYDA1 and HYDA2, hydrogenases; NDH, NAD(P)H dehydrogenase; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase complex; PFL, pyruvate formate lyase; PFR, pyruvate ferredoxin oxidoreductase; PPP, pentose phosphate pathway; PQ, plastochinon pool; and TCA cycle, citric acid cycle.
Alanine is synthesized by transamination of pyruvate with glutamate being the amino donor, a process catalyzed by the alanine aminotransferase (43). In both strains, increased levels of alanine were monitored already during the first hours of sulfur depletion. However, a clear difference between WT and mutant could be detected during the H2 production phase with only Stm6Glc4 exhibiting continuously increasing levels of alanine (see Fig. 2B). Glutamate is used as the amino donor for amino acid synthesis and is in turn converted into α-ketoglutarate, releasing ammonium. To prevent loss of the ammonium and for regeneration of glutamate, two ferredoxins are thought to be oxidized, controlling the redox balance of the glycolysis under anaerobic conditions. This process can also be interpreted as a direct competition for hydrogenases activity (25).
Pyruvate can also be fermented to formate (catalyzed by the pyruvate formate lyase), converted to acetyl-CoA (catalyzed by the pyruvate ferredoxin oxidoreductase), or metabolized to acetate or ethanol (catalyzed by the acetate kinase), or the acetaldehyde dehydrogenase/alcohol dehydrogenase (ADH) (21, 24) (Fig. 11). Pyruvate ferredoxin oxidoreductase reduces ferredoxin, which can potentially serve as the electron donor for the hydrogenases (24). Our results confirmed that formate and ethanol started to be produced as soon as anaerobiosis was established in both strains, and the levels kept increasing when H2 production set in (see Fig. 6). The amount of fermentative products was clearly higher in WT than in Stm6Glc4 (1.6–4.2 times more). In contrast, no net acetate production could be observed during anaerobiosis and H2 production. A sharp decrease of the total amount of acetate in the sulfur-depleted medium was detected after cells were transferred, but the levels remained constant during anaerobiosis. These data suggest that, during the first hours of sulfur depletion, the acetate from the medium is taken up by the cells and converted into acetyl-CoA (51), which is subsequently metabolized via the citric acid cycle and/or the glyoxylate cycle. However, when oxygen is consumed and anaerobiosis sets in, acetate does not seem to be incorporated anymore (see Fig. 6). In the past a major point of discussion has been whether acetate acts as a direct substrate for H2 production (8, 25, 52). Our results show that acetate is consumed only during the initial, aerobic phase and that it appears necessary for an efficient transition into the anaerobic, H2 production phase. After this transition, acetate levels remained unchanged suggesting that acetate does not serve as a direct substrate for H2 production. Acetate consumption from the medium and starch accumulation within the cells coincides. A possible explanation is that acetate is first converted into acetyl-CoA after cellular uptake, which has been shown to preferentially be metabolized via the glyoxylate cycle (51), generating succinate as a product. Subsequently, succinate can be converted into malate and oxaloacetate. Oxaloacetate is converted into phosphoenolpyruvate and then into sugar phosphates, which are the basis for starch synthesis (51).
Some of the acetyl-CoA is also likely to be fed into the citric acid cycle, where it can be used for energy production and to generate carbon skeletons. Finally, the observed accumulation of fatty acids and lipid (Figs. 4 and 5) in the plastid, which is a well reported cellular reaction during environmental stress conditions (46), could also be mediated by acetate uptake with acetyl-CoA being the initial building unit.
The accumulation of starch and the production of fatty acids was observed during the first hours of sulfur depletion and strongly increased until anaerobiosis was reached. Of particular note was that the WT showed a much higher tendency to accumulate fatty acids and neutral lipids during H2 production process compared with Stm6Glc4. In contrast, mutant Stm6Glc4 has the tendency to build up relatively more starch then the WT.
This study and an earlier study of Wang and co-workers (46) indicate that biosynthesis of fatty acids/lipids and starch are competing pathways. Wang et al. showed that blocking of the starch biosynthesis in the strain sta6 resulted in an increased lipid body formation (up to 30-fold, compared with a 15-fold increase in the WT) during nitrogen deprivation. 3-Phosphoglycerate is the precursor for starch synthesis as well as for triacylglyceride lipid biosynthesis. If starch production is blocked more carbon skeletons are used for triacylglyceride biosynthesis (46). This is of particular importance, because it is unlikely that neutral lipids are used as a substrate for H2 production. The reason is that during anaerobic conditions no energy can be generated via the oxidation of fatty acids. For example, 7NAD+, 7FAD, and one molecule ATP are needed to oxidize palmitic acid, generating a lot of reduction potential, which cannot be used efficiently, because ATP generation from NADH and FADH2 via mitochondrial respiration is blocked during anaerobiosis.
In summary, the consumption of acetate under aerobic conditions (Fig. 6), the increase of the intermediates of the citric acid cycle/glyoxylate cycle (Fig. 3B), the increase of fatty acids and lipids up until anaerobiosis is reached (Figs. 4 and 5), and the starch accumulation during the first 24 h of sulfur-induced H2 production (27, 41, 44) all indicate that the competition for pyruvate as a product of glycolysis is a key factor for H2 production activity. The results also indicate that acetate is most likely used for respiratory processes, which induce anaerobiosis (through O2 consumption), starch accumulation, and fatty acid and neutral lipids synthesis during the aerobic phase of the process. Acetate appears not to be used as a direct substrate supply for the hydrogenase(s).
CONCLUSIONS
Combination of GC/MS and GCxGC-TOFMS within this work made it possible to resolve almost an order of magnitude more metabolic compounds compared with previous studies, therefore providing a much more detailed picture of important metabolic pathways during H2 production of both WT and the high H2-producing Stm6Glc4 strain (Figs. 10 and 11). From the differential analysis performed in this work important information about bottlenecks of the H2 production process could be obtained and potential key targets for subsequent bioengineering can now be determined. In particular, remarkable differences between WT and mutant Stm6Glc4 obtained in the anaerobic metabolic pathways of starch degradation, fatty acids and lipid synthesis, and formate/ethanol production are of interest for future improvement strategies. Because the production of fatty acids, lipids, and fermentative products are possible competitors for H2 production, it is a goal for the future to diminish these competitors by molecular engineering. Fatty acids and lipids are produced under a number of stress conditions (46). The regulatory key enzyme of the fatty acids synthesis is the acetyl-CoA carboxylase. This enzyme is a biotin-containing multienzyme complex with two catalytic activities, a biotin carboxylase and an acetyl-CoA carboxyltransferase activity. It catalyzes two reactions, in which bicarbonate is combined with acetyl-CoA to yield malonyl-CoA. Malonyl-CoA is then used for the following steps of fatty acids and lipids biosynthesis. To avoid fatty acid and lipid biosynthesis and potentially increase H2 production in C. reinhardtii, a future goal will be to down-regulate the activity of acetyl-CoA carboxylase, e.g. by artificial microRNA (39).
Pathways leading to synthesis of amino acids (especially alanine) could also be a potential target for improvement strategies, because NADH is consumed, therefore potentially competing with electron supply for the hydrogenase (25).
Another interesting candidate in this respect could be the enzyme pyruvate ferredoxin oxidoreductase, which is involved in the conversion of pyruvate into acetyl-CoA. During this reaction, reduced ferredoxin is generated, which could serve as a direct electron donor for the hydrogenase (24). Therefore an up-regulation of this enzyme and the down-regulation of the pyruvate formate lyase, which also uses the reducing equivalents, could be a promising approach to positively affect the H2 production process. Finally, enzymes of pathways leading to the production of ethanol could be potential targets, because (a) NADH is consumed (24, 25) and (b) ethanol could become toxic at increasing concentrations thus preventing an extended H2 production phase. The down-regulation of ADH would therefore be a further promising approach to increase photobiological H2 production.
Supplementary Material
This work was supported in part by the Deutsche Bundesstiftung Umwelt (Grant DBU 20006/828 to A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6.
MapMan Site of Analysis, Genomanalyse im Biologischen System Pflanze, available on-line.
- PSII
- Photosystem II
- ADH
- alcohol dehydrogenase
- GCxGC-TOFMS
- two-dimensional gas chromatography combined with time-of-flight mass spectrometry
- GMD
- Golm Metabolom Database
- NIST
- National Institute of Standards and Technology
- Stm6
- state transition mutant 6
- e.f.
- enrichment factor(s).
REFERENCES
- 1.Melis A., Happe T. (2001) Plant Physiol. 127, 740–748 [PMC free article] [PubMed] [Google Scholar]
- 2.Rupprecht J., Hankamer B., Mussgnug J. H., Ananyev G., Dismukes C., Kruse O. (2006) Appl. Microbiol. Biotechnol. 72, 442–449 [DOI] [PubMed] [Google Scholar]
- 3.Hankamer B., Lehr F., Rupprecht J., Mussgnug J. H., Posten C., Kruse O. (2007) Physiol. Plant 131, 10–21 [DOI] [PubMed] [Google Scholar]
- 4.Gaffron H., Rubin J. (1942) J. Gen. Physiol. 26, 219–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruse O., Rupprecht J., Mussgnug J. H., Dismukes G. C., Hankamer B. (2005) Photochem. Photobiol. Sci. 4, 957–970 [DOI] [PubMed] [Google Scholar]
- 6.Forestier M., King P., Zhang L., Posewitz M., Schwarzer S., Happe T., Ghirardi M. L., Seibert M. (2003) Eur. J. Biochem. 270, 2750–2758 [DOI] [PubMed] [Google Scholar]
- 7.Melis A. (2007) Planta. 226, 1075–1086 [DOI] [PubMed] [Google Scholar]
- 8.Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. (2000) Plant Physiol. 122, 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruse O., Rupprecht J., Bader K. P., Thomas-Hall S., Schenk P. M., Finazzi G., Hankamer B. (2005a) J. Biol. Chem. 280, 34170–34177 [DOI] [PubMed] [Google Scholar]
- 10.Makarova V. V., Kosourov S., Krendeleva T. E., Semin B. K., Kukarskikh G. P., Rubin A. B., Sayre R. T., Ghirardi M. L., Seibert M. (2007) Photosynth. Res. 94, 79–89 [DOI] [PubMed] [Google Scholar]
- 11.Surzycki R., Cournac L., Peltier G., Rochaix J. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17548–17553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desplats C., Mus F., Cuiné S., Billon E., Cournac L., Peltier G. (2009) J. Biol. Chem. 284, 4148–4157 [DOI] [PubMed] [Google Scholar]
- 13.Hemschemeier A., Jacobs J., Happe T. (2008) Eukaryot. Cell 7, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009) FEBS Lett. 583, 325–329 [DOI] [PubMed] [Google Scholar]
- 15.Rühle T., Hemschemeier A., Melis A., Happe T. (2008) BMC Plant Biol. 8, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chochois V., Dauvillée D., Beyly A., Tolleter D., Cuiné S., Timpano H., Ball S., Cournac L., Peltier G. (2009) Plant Physiol. 151, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posewitz M. C., Smolinski S. L., Kanakagiri S., Melis A., Seibert M., Ghirardi M. L. (2004) Plant Cell 16, 2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doebbe A., Rupprecht J., Beckmann J., Mussgnug J. H., Hallmann A., Hankamer B., Kruse O. (2007) J. Biotechnol. 131, 27–33 [DOI] [PubMed] [Google Scholar]
- 19.Beckmann J., Lehr F., Finazzi G., Hankamer B., Posten C., Wobbe L., Kruse O. (2009) J. Biotechnol. 142, 70–77 [DOI] [PubMed] [Google Scholar]
- 20.Torzillo G., Scoma A., Faraloni C., Ena A., Johanningmeier U. (2009) Int. J. Hyd. Energy 34, 4529–4536 [Google Scholar]
- 21.Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. (2007) J. Biol. Chem. 282, 25475–25486 [DOI] [PubMed] [Google Scholar]
- 22.Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 23.Rupprecht J. (2009) J. Biotechnol. 142, 10–20 [DOI] [PubMed] [Google Scholar]
- 24.Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C. (2009) J. Biol. Chem. 284, 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posewitz M. C., Dubini A., Meuser J. E., Seibert M., Ghirardi M. L. (2009) in The Chlamydomonas Sourcebook. Organellar and Metabolic Processes (Stern D. B., Harris E. H. eds) Vol. 2, 2nd Ed., pp. 217–256, Academic Press, San Diego, CA [Google Scholar]
- 26.Nguyen A. V., Thomas-Hall S. R., Malnoë A., Timmins M., Mussgnug J. H., Rupprecht J., Kruse O., Hankamer B., Schenk P. M. (2008) Eukaryot. Cell 7, 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmins M., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., Schenk P. M. (2009) J. Biol. Chem. 284, 23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiehn O., Kopka J., Dörmann P., Altmann T., Trethewey R. N., Willmitzer L. (2000) Nat. Biotechnol. 18, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 29.Welthagen W., Shellie R. A., Spranger J., Ristow M., Zimmermann R., Fiehn O. (2005) Metabolomics 1, DOI: 10.1007/s11306-005-1108-2 [DOI] [PubMed] [Google Scholar]
- 30.Harris E. (1989) in The Chlamydomonas Sourcebook, pp. 25–52, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 31.Maxwell K., Johnson G. N. (2000) J Exp. Bot. 51, 659–668 [DOI] [PubMed] [Google Scholar]
- 32.Barsch A., Patschkowski T., Niehaus K. (2004) Funct. Integr. Genomics. 4, 219–230 [DOI] [PubMed] [Google Scholar]
- 33.Watt T. F., Vucur M., Baumgarth B., Watt S. A., Niehaus K. (2008) J. Biotechnol. 140, 59–67 [DOI] [PubMed] [Google Scholar]
- 34.Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmüller E., Dörmann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A. R., Steinhauser D. (2005) Bioinformatics 21, 1635–1638 [DOI] [PubMed] [Google Scholar]
- 35.Heim J. (2009) Urine: Diabetic vs. Non-diabetic Using Statistical Compare, LECO Corp., 3000 Lakeview Avenue, St. Joseph, MI 49085: (application note) [Google Scholar]
- 36.Chen W., Zhang C., Song L., Sommerfeld M., Hu Q. (2009) J. Microbiol. Methods 77, 41–47 [DOI] [PubMed] [Google Scholar]
- 37.R Development Core Team (2009) R Foundation for Statistical Computing, Vienna, Austria, ISBN 3-900051-07-0 [Google Scholar]
- 38.Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., Selbig J., Hannemann J., Piques M. C., Steinhauser D., Scheible W. R., Gibon Y., Morcuende R., Weicht D., Meyer S., Stitt M. (2005) Plant Physiol. 138, 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molnár A., Schwach F., Studholme D. J., Thuenemann E. C., Baulcombe D. C. (2007) Nature 447, 1126–1129 [DOI] [PubMed] [Google Scholar]
- 40.Wykoff D. D., Davies J. P., Melis A., Grossman A. R. (1998) Plant Physiol. 117, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L., Melis A. (2002) Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1499–1507; discussion 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosourov S., Patrusheva E., Ghirardi M. L., Seibert M., Tsygankov A. (2007) J. Biotechnol. 128, 776–787 [DOI] [PubMed] [Google Scholar]
- 43.Vallon H., Spalding M. H. (2009) in The Chlamydomonas Sourcebook, Organellar and Metabolic Processes (Stern D. B., Harris E. H. eds) Vol. 2, 2nd Ed., pp. 115–158, Academic Press, San Diego, CA [Google Scholar]
- 44.Tsygankov A., Kosourov S., Seibert M., Ghirardi M. L. (2002) Int. J. Hyd. Energy 27, 1239–1244 [Google Scholar]
- 45.Schönfeld C., Wobbe L., Borgstädt R., Kienast A., Nixon P. J., Kruse O. (2004) J. Biol. Chem. 297, 50366–50374 [DOI] [PubMed] [Google Scholar]
- 46.Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. (2009) Eukaryot. Cell 8, 1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niyogi K. K. (2009) in The Chlamydomonas Sourcebook, Organellar and Metabolic Processes (Stern D. B., Harris E. H. eds) Vol. 2, 2nd Ed., pp. 847–870, Academic Press, San Diego, CA [Google Scholar]
- 48.Lohr M. (2009) in The Chlamydomonas Sourcebook, Organellar and Metabolic Processes (Stern D. B., Harris E. H. eds) Vol. 2, 2nd Ed., pp. 799–817, Academic Press, San Diego, CA [Google Scholar]
- 49.Brumfield K. M., Moroney J. V., Moore T. S., Simms T. A., Donze D. (2010) PLoS ONE 5, e8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z., Shrager J., Jain M., Chang C. W., Vallon O., Grossman A. R. (2004) Eukaryot. Cell 3, 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spalding M. H. (2009) in The Chlamydomonas Sourcebook, Organellar and Metabolic Processes (Stern D. B., Harris E. H. eds) Vol. 2, 2nd Ed., pp. 257–301, Academic Press, San Diego, CA [Google Scholar]
- 52.Happe T., Mosler B., Naber J. D. (1994) Eur. J. Biochem. 222, 769–774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.