Abstract
Missense mutations in the human C10orf2 gene, encoding the mitochondrial DNA (mtDNA) helicase, co-segregate with mitochondrial diseases such as adult-onset progressive external ophthalmoplegia, hepatocerebral syndrome with mtDNA depletion syndrome, and infantile-onset spinocerebellar ataxia. To understand the biochemical consequences of C10orf2 mutations, we overproduced wild type and 20 mutant forms of human mtDNA helicase in Escherichia coli and developed novel schemes to purify the recombinant enzymes to near homogeneity. A combination of molecular crowding, non-ionic detergents, Mg2+ ions, and elevated ionic strength was required to combat insolubility and intrinsic instability of certain mutant variants. A systematic biochemical assessment of the enzymes included analysis of DNA binding affinity, DNA helicase activity, the kinetics of nucleotide hydrolysis, and estimates of thermal stability. In contrast to other studies, we found that all 20 mutant variants retain helicase function under optimized in vitro conditions despite partial reductions in DNA binding affinity, nucleotide hydrolysis, or thermal stability for some mutants. Such partial defects are consistent with the delayed presentation of mitochondrial diseases associated with mutation of C10orf2.
Keywords: DNA Helicase, DNA Replication, Mitochondria, Mitochondrial DNA, Neurodegeneration, Mitochondrial DNA Replication, Progressive External Ophthalmoplegia
Introduction
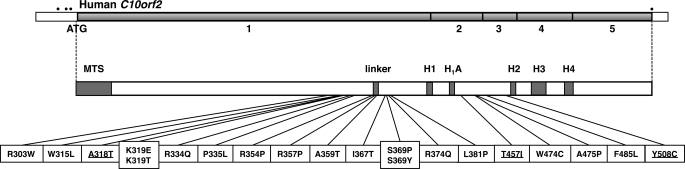
Chronic disruption of mitochondrial function can result in a broad array of neuromuscular degenerative disorders known as mitochondrial diseases, including progressive external ophthalmoplegia (PEO),2 Alpers syndrome, parkinsonism, and several complex ataxia neuropathy syndromes (1–3). A heritable form of PEO was first mapped to a chromosomal interval near position 10q24 in a Finnish pedigree (4). One molecular hallmark of PEO was the age-dependent accumulation of mtDNA deletions in somatic tissues of affected individuals, which suggested that this locus was needed for replication or maintenance of mtDNA (4). In 2001, autosomal dominant PEO with mtDNA deletions was shown to co-segregate with 11 different missense mutations in the C10orf2 gene (5), which encoded a protein containing predicted amino acid sequences homologous to portions of the bacteriophage T7 gene product 4 and other superfamily 4 DNA helicases (5, 6). Since that time, numerous reports have identified 23 additional missense mutations in C10orf2 associated with heritable mitochondrial diseases such as adPEO, hepatocerebral mtDNA depletion syndrome (MDS), and infantile-onset spinocerebellar ataxia (IOSCA) (7, 8) (see Fig. 1).
FIGURE 1.
Schematic of the human C10orf2 gene shows the positions of missense mutations associated with disease. The five exons of C10orf2 are preceded by three in-frame stop codons (dots) (5). The positions of the amino-terminal mitochondrial targeting sequence (amino acids 1–42) and the conserved motifs of superfamily 4 DNA helicases are shown. Missense mutations associated with PEO (plain text) and MDS (underlined) are indicated (5, 14, 44, 45, 47, 51–62). The Y508C substitution is also associated with IOSCA (43, 44, 46, 47).
Several lines of evidence more directly link C10orf2 function to maintenance of mtDNA. The C10orf2 gene product dynamically colocalizes with mtDNA in nucleoprotein structures known as mitochondrial nucleoids (5, 9), and knocking down expression of C10orf2 by RNAi results in the rapid decrease in mtDNA copy number in cultured human osteosarcoma (143B) cells (10). Overexpression of catalytic mutants and dominant disease variants of the mtDNA helicase in cultured human or Schneider cells results in stalled mtDNA replication or depletion of mtDNA (11–13), which emulates the disease state. However, only two of five adPEO mutants exhibited a dominant negative phenotype with mtDNA depletion in Schneider cells (12). Overexpression of two adPEO variants in a mouse transgenic model recapitulated many of the symptoms observed in human PEO, including accumulation of multiple mtDNA deletions, progressive respiratory dysfunction, and cytochrome c oxidase deficiency (14, 15).
Like the replicative DNA helicases Escherichia coli DnaB, bacteriophage T7 gene product 4, bacteriophage T4 gene product 41, and the prokaryotic RepA protein, the mitochondrial enzyme is an hexameric helicase that catalyzes hydrolysis of nucleoside triphosphates to unwind DNA substrates with a 5′ to 3′ polarity (16). Also known as Twinkle, the mitochondrial DNA helicase can function in concert with polymerase γ and the mitochondrial single-stranded DNA binding protein to synthesize multikilobase DNA products in vitro (17). The majority of pathogenic C10orf2 mutations cluster in a region of the protein linking the N-terminal primase-like domain and the C-terminal helicase domain (see Fig. 1). Comparison of sequence homology among superfamily 4 helicases suggests that this linker region participates in subunit interaction and facilitates the formation of functional hexamers. The existing literature contains conflicting descriptions of the specific enzymatic activities and biochemical properties of the wild type (WT) and certain mutant variants of human mtDNA helicase. Variability in protocols for protein purification and differences in DNA substrates and assay conditions make it difficult to reconcile these discrepancies.
To begin to determine how biochemical defects in the mitochondrial DNA helicase may be contributing to diseases of mtDNA maintenance, we developed a scheme to overproduce wild type (WT) and mutant variants of the mitochondrial DNA helicase in E. coli and to purify the recombinant enzymes to near homogeneity. We found that the solubility and long term stability of the enzyme were significantly enhanced by a combination of molecular crowding, non-ionic detergents, Mg2+ ions, and elevated ionic strength, which has allowed the enzymatic properties of some of the less stable helicase variants to be determined. Biochemical characterization of the enzyme collection included analysis of DNA binding affinity, DNA helicase activity, the kinetics of nucleotide hydrolysis, and estimates of thermal stability.
EXPERIMENTAL PROCEDURES
Subcloning and Expression of C10orf2 in E. coli
A cDNA for the human C10orf2 gene (5) was amplified from a HeLa cDNA library (Stratagene) by PCR with 5′-TATAGAGCTCCCTCCTCGCAGACGTTACAGGAAGGAG-3′ and 5′-TATAAAGCTTTCACTTTGAACGCTTGGAGGTGTCTGGCTGG-3′ as the forward and reverse primers, respectively, and the DNA sequence was found to match that of GenBankTM accession number AF292004. To remove the predicted 42-amino acid mitochondrial targeting sequence from the N terminus (18), the coding sequence was amplified with 5′-TATACATATGGAGACTCTCCAAGCCTTGGATATGCCA-3′ (forward) and 5′-TATAAAGCTTTCACTTTGAACGCTTGGAGGTGTCTGGCTGG-3′ (reverse) primers, digested with NdeI and HindIII, and transferred to the E. coli expression vector pET21a (Novagen). The sequence of the resulting plasmid, pT684D42, was confirmed and shown to encode the mature form of the mitochondrial helicase (amino acids 43–684, predicted molecular weight 72,202). A second plasmid, pT684D42C-His, was constructed to introduce an affinity tag by appending the amino acids Ala-Ala-Ala-Leu-Glu-His6 to the C terminus (predicted molecular weight 73,479). Missense mutations in C10orf2 associated with mitochondrial diseases were generated in pT684D42C-His using the QuikChange site-directed mutagenesis kit (Stratagene). Primers used for generation of the mutant forms are listed in the supplemental material.
Overnight cultures of E. coli strain BL21(DE3) CodonPlus-RIPL (Stratagene) that had been freshly transformed with each plasmid were grown at 37 °C in Luria-Bertani medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Prewarmed Luria-Bertani medium (1 liter) supplemented only with 100 μg/ml ampicillin was inoculated with 11 ml of each overnight culture and incubated with rotary shaking at 37 °C. Once the optical density (600 nm) of cultures reached 0.6, flasks were chilled on ice to 20 °C, and IPTG was added to a final concentration of 1 mm. Incubation was continued at 20 °C for 18 h, after which cells were harvested by centrifugation, washed once in 10 mm Tris-HCl (pH 7.5) with 0.1 mm EDTA, frozen on liquid nitrogen, and stored at −80 °C.
Purification of His-tagged Recombinant Mitochondrial DNA Helicase
All steps were performed at 4 °C. Frozen pellets of induced cells (5–6 g) were thawed on wet ice and resuspended in 25 ml of a lysis buffer containing 50 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 10% glycerol, 0.5% Nonidet P-40, 20 mm imidazole, 5 mm MgCl2, 0.1 mm PMSF, and 1 mm ATP. The cell suspension was twice passed through a 40-ml French pressure cell (Amicon) at 18,000 p.s.i., and the resulting whole cell lysate was clarified by centrifugation at 12,000 × g for 15 min. This soluble lysate was immediately applied by peristaltic pump at 30 ml/h to a HisTrap HP column (1 ml, GE Healthcare) previously equilibrated in lysis buffer. The column was washed with 12 ml of a buffer consisting of 30 mm Tris-HCl (pH 7.5), 20% glycerol, 0.5% Nonidet P-40, 1 mm 2-mercaptoethanol, 5 mm MgCl2, 0.1 mm PMSF, 0.2 mm ATP (buffer T) also containing 0.50 m NaCl and 20 mm imidazole followed by a 12-ml wash with buffer T containing 0.31 m NaCl and 20 mm imidazole. Then the column was eluted with 7 ml of buffer T containing 0.31 m NaCl and 250 mm imidazole. Protein-containing fractions were applied to a HiTrap Q HP column (1 ml, GE Healthcare) equilibrated in buffer T also containing 0.31 m NaCl. Under these conditions, the enzyme remained in the unbound fraction, and was applied directly to a HiTrap Heparin HP column (1 ml, GE Healthcare) previously equilibrated in buffer T containing 0.31 m NaCl. The heparin column was washed with 10 ml of equilibration buffer and eluted with an 11-ml linear gradient of NaCl (0.31–1.0 m) in buffer T. The helicase eluted at ∼0.55 m NaCl and was frozen in small aliquots with liquid nitrogen and stored at −80 °C. Materials bound to the HiTrap Q HP column were eluted with buffer T containing 1.0 m NaCl and saved for analysis.
Purification of Recombinant Mitochondrial DNA Helicase Lacking an Affinity Tag
Soluble lysates of cells induced to overexpress the untagged mitochondrial helicase were prepared as before, except that the lysis buffer consisted of 50 mm Tris-HCl (pH 7.5), 0.2 m NaCl, 10% glycerol, 0.5% Nonidet P-40, 1 mm EDTA, 5 mm 2-mercaptoethanol, and 0.1 mm PMSF. Soluble lysates were supplemented with 1 m MgCl2 to a final concentration of 5 mm, and solid ammonium sulfate was added to 34% saturation (0.202 g/ml solution). After 30 min of slow stirring, the precipitate was collected by centrifugation at 27,000 × g for 15 min. The well drained precipitate was resuspended in a sufficient volume (4–5 ml) of buffer T to make the conductivity of the sample equivalent to buffer T also containing 0.35 m NaCl. The sample was applied to a 10-ml column of heparin-Sepharose HP (GE Healthcare) equilibrated in buffer T also containing 0.35 m NaCl. The column was washed with 50 ml of equilibration buffer and developed with a 110-ml gradient of NaCl (0.35–1.0 m) in buffer T. Fractions containing the recombinant protein eluted at ∼0.55 m NaCl, as identified by SDS-PAGE. Pooled fractions were diluted with buffer T until their conductivity matched that of buffer T containing 0.35 m NaCl. This protein fraction was passed first through a 1-ml HiTrap Q HP column equilibrated in buffer T containing 0.35 m NaCl before being applied to a 2.2-ml column of hydroxylapatite (BioGel HT, Bio-Rad) equilibrated in the same buffer. After washing with 10 ml of equilibration buffer, the HiTrap Q HP column was removed, and protein was step-eluted from the hydroxylapatite column with 4 ml of buffer T also containing 0.35 m NaCl and 0.5 m potassium phosphate (pH 7.5). The enzyme was further purified by size exclusion chromatography by passage at 1 ml/min through a column of Sephacryl S-200 HR (26 × 5.3 cm2) equilibrated in a buffer containing 25 mm HEPES-OH (pH 7.5), 20% glycerol, 1 mm 2-mercaptoethanol, 5 mm MgCl2, 0.01% Nonidet P-40, and 0.5 m NaCl. The single, symmetrical peak of highly purified protein eluted 55 ml after sample injection (0.41 column volumes). As needed, the enzyme could be concentrated by cation exchange chromatography. Protein samples were diluted or dialyzed against buffer T containing 0.2 m NaCl, and then the samples were applied to a 1 ml HiTrap SP HP column equilibrated in this buffer. After washing the column to obtain a stable baseline, the column was developed with a 10-ml linear gradient of NaCl (0.2–0.7 m) in buffer T. The enzyme eluted at ∼0.32 m NaCl and was frozen in small aliquots with liquid nitrogen and stored at −80 °C. As needed, the helicase can be concentrated successfully by ultrafiltration.
Exonuclease and Phosphatase Assays
Exonuclease activity was determined in reaction mixtures (15 μl) containing 25 mm HEPES-OH (pH 7.5), 5 mm 2-mercaptoethanol, 5 mm MgCl2, 50 μg/ml BSA, 1 mm ATP, 0.1 pmol of the 5′-32P-labeled oligonucleotide 5′-GTATGTTCGCCTGTAATATTGAACGTAGGTGCGA-3′, and 0.43 pmol of purified protein. Activity on a double-stranded DNA substrate was also determined with the labeled oligonucleotide hybridized to the second oligonucleotide 5′-TTATCGCACCTACGTTCAATATTACAGGCGAACATACT-3′. Reactions were incubated at 37 °C for 30 min prior to analysis on denaturing 15% polyacrylamide gels, as described previously (19).
Oligonucleotide Substrates
DNA substrates utilized to assay DNA binding and DNA helicase activity were constructed from synthetic oligonucleotides. Three 40-nucleotide oligomers TD1 (5′-ATGCTAGCTTGGCTGTGACTTTAAACCTGTCGTGCCAGCT-3′), TD2 (5′-AGCTGGCACGACAGGTTTAAAGTCACAGCCAAGCTAGCAT-3′), and TD3 (5′-AGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTG-3′) were obtained from Oligos Etc. (Wilsonville, OR) and were quantified by absorbance at 260 nm. An internally modified version of TD1 with fluorescein covalently attached to the thymine at position 21 (TD1-F) was purchased from Sigma-Aldrich. The concentration of TD1-F was determined spectrophotometrically at 260 nm and adjusted slightly downward to compensate for intrinsic absorbance of fluorescein at 260 nm (20). TD1 was utilized directly as a single-stranded DNA substrate, whereas TD1 hybridized to the fully complementary oligonucleotide TD2 served as the double-stranded DNA substrate. Annealing TD1 and TD3 generated a forked DNA substrate with 18 nt at the 3′-end of TD1 being fully complementary to 18 nt at the 5′-end of TD3, leaving 22-nt single-stranded tails at the 5′-end of TD1 and the 3′-end of TD3.
DNA Binding
Fluorescent substrates were hybridized in a solution containing 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 0.1 mm EDTA, and 10 μm each of the indicated oligonucleotides by heating to 85 °C followed by slow cooling to room temperature. Immediately prior to use, the substrates were warmed to 42 °C for 5 min and held at room temperature. Steady state fluorescence anisotropy was measured with an Olis RSM1000 spectrofluorometer (Bogart, GA) equipped with a 1.24-mm slit and a temperature-controlled cell set to 22 °C. Incident light at a 480 nm excitation wavelength was vertically plane-polarized and passed through a T-format quartz fluorometer cell, and fluorescence was measured at two identical detectors fitted with 530-nm high pass filters and vertical or horizontal polarizing filters. Binding mixtures (200 μl) contained 30 mm Tris-HCl (pH 7.5), 1 mm 2-mercaptoethanol, 5 mm MgCl2, 20% (v/v) glycerol, 0.1% Nonidet P-40, and a 15 nm concentration of the specified fluorescein-conjugated oligonucleotide substrate. Changes in fluorescence polarization were measured in response to the stepwise addition of purified helicase in buffer T that also contained 0.35 m NaCl. Following a 1-min equilibration period after each addition, anisotropy data were collected in triplicate with a 10-s integration time. Changes in anisotropy were plotted against the total concentration of protein, expressed as hexamers. To correct for the ligand depletion effect caused by non-trivial concentrations of protein-DNA complex relative to the total protein concentration, binding isotherms were fit to a quadratic equation by non-linear regression analysis to calculate apparent Kd(DNA) values (21). Intrinsic fluorescence of buffer components was undetectable at wavelengths relevant to fluorescein. Volume-corrected total fluorescence intensity varied less than 4% during all titrations and additions, indicating that changes in anisotropy were minimally affected by light scattering due to protein aggregation or by fluorescence quenching due to protein binding directly to the fluorescein instead of to the DNA substrate.
DNA Helicase Assay
The substrate chosen for DNA helicase assays was patterned after the oligonucleotide-based, forked DNA substrate used to characterize the T7 gp4 DNA helicase (22). T4 polynucleotide kinase was utilized to 5′-end-label oligonucleotide TD1, and the forked DNA substrate was constructed by annealing radiolabeled TD1 with the partially complementary oligonucleotide TD3. Helicase reaction mixtures (12 μl) contained 20 mm HEPES-OH (pH 7.5), 5 mm dithiothreitol, 50 μg/ml BSA, 7% glycerol, 5 mm MgCl2, 4.5 mm ATP, 40 nm [32P]TD1-TD3 forked DNA substrate, and 12 nm of the indicated protein (as hexamers). Reactions were incubated at 37 °C for the indicated times and were terminated by the addition (3 μl) of a stop solution composed of 0.1 m EDTA, 0.5% SDS, 30% glycerol, and a trace quantity of bromphenol blue. Double-stranded DNA substrates and single-stranded DNA products were resolved by native PAGE. Gels (17 × 14 × 0.075 cm) contained 62 mm Tris, 62 mm boric acid, 1.7 mm EDTA, and 12% polyacrylamide (19:1 acrylamide:bis-acrylamide ratio), and electrophoresis was performed at room temperature for 2 h at 150 V. Gels were dried onto DE81 paper (Whatman), and the relative amounts of substrate and product in each lane were quantified with a Typhoon 9400 PhosphorImager (GE Healthcare) and NIH ImageJ software. The percentage of displaced oligonucleotide was calculated as described previously (23).
ATPase Assay
Enzymatic hydrolysis of ATP was determined in reaction mixtures (20 μl) containing 20 mm Tris-Cl (pH 7.8), 4.5 mm MgCl2, 15% glycerol, 0.3 mg/ml BSA, 0.8 μCi of [α-32P]ATP (3000 Ci/mmol), 0.7 mm ATP, and the indicated quantities of helicase protein (0.2–0.4 μm monomers). As needed, enzymes were diluted in buffer T also containing 0.5 m NaCl and 0.1 mg/ml BSA. Unless otherwise indicated, 0.2 μg of activated calf thymus DNA (Invitrogen) was included in each reaction. Unlabeled ATP was varied from 0 to 5 mm for kinetic analyses. Reactions were incubated at 37 °C for 25 min, and reactions were stopped by chilling on ice followed by the addition of 0.5 μl of 0.5 m EDTA. Samples (1.6 μl) of each reaction were spotted onto polyethyleneimine-cellulose thin layer chromatography plates (Merck) and air-dried. Plates were rinsed in absolute methanol to remove buffer components and developed for 30 min by ascending TLC with a mobile phase consisting of 0.5 m LiCl, 1.0 m acetic acid. Dried plates were exposed to phosphor storage screens, and radiolabeled ATP substrate and ADP product were quantified with a Typhoon 9400 PhosphorImager (GE Healthcare) and NIH ImageJ software.
Other Methods
Protein concentrations were determined by SDS-PAGE of multiple sample dilutions, staining with Coomassie Brilliant Blue, and quantitative digital imaging of protein bands relative to BSA standards in neighboring lanes. Polyclonal antibodies were raised in a New Zealand White rabbit against denatured, SDS-PAGE purified, affinity-tagged mitochondrial DNA helicase. Freund's complete adjuvant was included in the first immunization. For immunoblot analysis, proteins were resolved by SDS-PAGE, electrotransferred to Immobilon P membranes (Millipore), and sequentially incubated with rabbit anti-helicase antiserum (1:100 dilution) and alkaline phosphatase-conjugated second antibody. Cross-reactive protein bands were visualized with the Western Blue reagent (Promega).
RESULTS
Mitochondrial disease phenotypes co-segregate with mutations in C10orf2 (Fig. 1). Heritable clinical and molecular phenotypes include adult-onset PEO with mtDNA deletions and dominant inheritance, infantile-onset spinocerebellar ataxia with a recessive mode of inheritance, and hepatocerebral mtDNA depletion. The deletion and depletion of mtDNA that are hallmarks of these disease pedigrees may arise from defects in mtDNA replication and repair. A biochemical approach to determine the molecular mechanisms underlying dysfunctional mtDNA replication requires purified mitochondrial DNA helicases for in vitro studies.
Overexpression and Purification of Human C10orf2
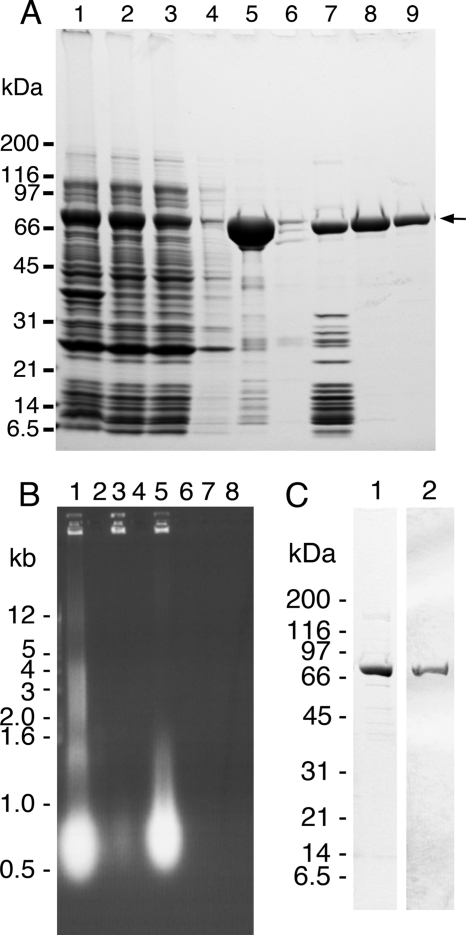
An E. coli expression system was chosen to maximize the yield of recombinant proteins and to simplify the production of mutant forms of the mitochondrial helicase. A cDNA encoding the human C10orf2 gene was amplified and placed under the control of an inducible promoter on the E. coli expression vector pET21a, and overproduction of recombinant helicase lacking the mitochondrial targeting sequence was induced with IPTG. Conditions for expression were routinely screened by SDS-PAGE analysis of whole cell lysates, and the highest production of WT enzyme was observed for the BL21(DE3) CodonPlus-RIPL host strain with a reduced temperature of induction. Analysis of whole cell lysates by SDS-PAGE showed that more than 20 mg of the 72-kDa protein (p72) could be produced from 1 liter of culture (Fig. 2A, lane 1). The His-tagged form of the enzyme was purified to apparent homogeneity by immobilized metal ion affinity chromatography (IMAC) followed by anion exchange and heparin affinity chromatography, as described under “Experimental Procedures” (Fig. 2A). Initial attempts to purify the enzyme from whole cell lysates resulted in precipitation of the recombinant protein. Trial experiments demonstrated that precipitation could be minimized by including 10% glycerol, 0.5% Nonidet P-40, 5 mm MgCl2, and 0.5 m NaCl in the lysis buffer. The high concentration of NaCl also helped to release p72 from the nucleic acid debris upon clearing the lysate by centrifugation (Fig. 2A, lane 2). Aggregation of the protein was completely eliminated by raising the glycerol concentration to 20% for the chromatographic steps. Although immobilized nickel ion chromatography was remarkably efficient at capturing p72 from the soluble lysate (Fig. 2A, lane 5), contamination of this fraction with nucleic acid was revealed by agarose gel electrophoresis and staining with ethidium bromide (Fig. 2B, lane 3). Accordingly, the NaCl concentration of the IMAC elution buffer was adjusted to 0.31 m NaCl because nucleic acids are retained quantitatively by the subsequent HiTrap Q HP column under these buffer conditions (Fig. 2B, lane 5). Several low molecular weight protein contaminants and roughly 10% of the available p72 are also retained on this column, possibly through interaction with the bound nucleic acids (Fig. 2A, lane 7). The final purification step produced p72 that was >95% pure (Fig. 2A, lanes 8 and 9) and free of nucleic acids (Fig. 2B, lanes 6–8). Plasmids bearing missense variants of p72 (Fig. 1) were prepared by site directed mutagenesis, as described under “Experimental Procedures.” The 20 mutant forms of p72 exhibited chromatographic behavior that was identical to WT p72 when purified by this rapid protocol. The purity of the mutant variants was uniformly ≥95%, although the yield of each preparation varied somewhat from 0.7 to 3.1 mg of purified protein.
FIGURE 2.
Purification of recombinant human mitochondrial DNA helicase from E. coli. A, samples of affinity-tagged p72 were taken at each stage of purification and analyzed by SDS-PAGE on a 4–20% gradient gel. Proteins were stained with Coomassie Brilliant Blue. Lane 1, crude cell lysate after induction; lane 2, soluble lysate; lane 3, IMAC unbound fraction; lane 4, IMAC wash step; lane 5, IMAC-bound fraction; lane 6, heparin unbound fraction; lane 7, HiTrap Q-bound fraction; lanes 8 and 9, heparin peak fractions. The positions of molecular mass markers (kDa) are indicated. The arrow shows the position of the recombinant 72-kDa protein. B, samples of the indicated fractions were analyzed by electrophoresis through 1% agarose gels in buffer containing 40 mm Tris base, 20 mm acetic acid, and 2 mm EDTA. Nucleic acids were stained with ethidium bromide. Lane 1, IMAC unbound fraction; lane 2, IMAC wash step; lane 3, IMAC-bound fraction; lane 4, heparin unbound fraction; lane 5, HiTrap Q-bound fraction; lanes 6–8, heparin peak fractions. The positions of size markers (kb) are indicated. C, the purity of untagged p72 protein (∼3.3 μg) after size exclusion chromatography was assessed by 4–20% SDS-PAGE and by staining with Coomassie Brilliant Blue (lane 1) and by immunoblot analysis with polyclonal rabbit antiserum (lane 2), as described under “Experimental Procedures.”
To assess any effects of the affinity tag on the activities of the recombinant enzyme, the recombinant helicase lacking an affinity tag was also prepared, as described under “Experimental Procedures.” As observed for the affinity-tagged protein, the untagged enzyme also had a propensity to precipitate in the absence of a relatively high concentration of NaCl, and this tendency increased after removal of residual nucleic acid contaminants by passage through the HiTrap Q anion exchange column (data not shown). Nevertheless, the untagged enzyme could be purified through a series of steps that did not rely on lowering ionic strength, including ammonium sulfate precipitation, heparin affinity and hydroxylapatite chromatography, and size exclusion chromatography. The purity of untagged WT p72 was high after the size exclusion step (Fig. 2C, lane 1), and the yield of protein was generally 1.1–1.3 mg from 1 liter of induced culture. Cation exchange chromatography could be used to achieve higher protein concentrations, as described under “Experimental Procedures,” although losses associated with falling below ∼0.3 m NaCl during sample preparation significantly reduce the overall yield of purified protein. Interestingly, diluting the enzyme to lower salt concentrations is not harmful when DNA is present, such as during the assembly of reactions with DNA substrates (see below). Immunoblot analysis confirmed the identify and purity of non-affinity-tagged protein (Fig. 2C, lane 2). Enzyme preparations were screened for contaminating exonuclease and phosphatase activities, as described under “Experimental Procedures.” Reactions were designed with long incubation times and a stoichiometric excess of enzyme, and neither phosphatase nor exonuclease activity could be detected.
DNA Binding by the Mitochondrial Helicase
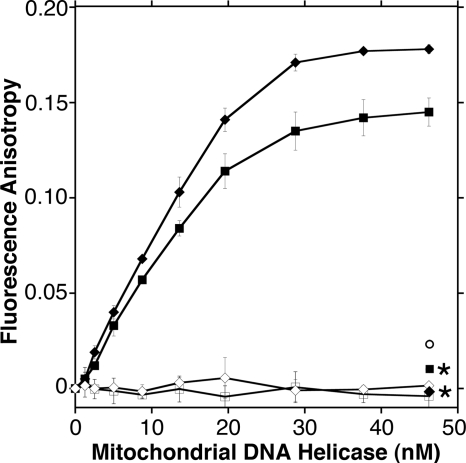
The affinity of p72 for oligomeric DNA substrates was assessed to aid in the development of biochemical assays. Fluorescence polarization methods were chosen to permit the rapid determination of binding affinity in solution with free and bound species at equilibrium. The rotational diffusion of the DNA substrates is reduced dramatically upon binding to protein, which allows the formation of protein-DNA complexes to be measured as an increase in fluorescence anisotropy. Fluorescein-labeled single-stranded or double-stranded DNA oligonucleotides were mixed with increasing concentrations of wild type p72 protein, and changes in anisotropy were plotted as a function of total protein concentration (Fig. 3). Binding isotherms were fit with a quadratic equation to estimate Kd(DNA) values, as described under “Experimental Procedures.” Wild type p72 exhibited a Kd of 5.7 ± 2.5 nm on our single-stranded DNA substrate and 4.9 ± 1.9 nm on our double-stranded DNA substrate. Both of these values are in good agreement with the ∼5 nm affinity of p72 hexamers for 30-nt single-stranded or double-stranded oligonucleotide substrates or for an oligo(dT)30 homopolymer, as estimated by electrophoretic mobility shift assay (24, 25). The replicative DNA helicase of bacteriophage T7, gp4, also exhibits a similar Kd(DNA) of 6.4 nm for a single-stranded 15-mer oligonucleotide in a filter binding assay (26).
FIGURE 3.
DNA binding affinity of mitochondrial DNA helicase proteins. Changes in the anisotropy of fluorescein-labeled oligonucleotide substrates were measured in response to the stepwise addition of p72 protein, as described under “Experimental Procedures.” Protein concentrations are expressed as hexamers. Baseline anisotropies of single-stranded (TD1-F) and double-stranded (TD1-F·TD2) substrates were 0.072 and 0.039, respectively, reflecting conformational differences in each unbound substrate. Changes in anisotropy of TD1-F (squares) or TD1-F·TD2 (diamonds) due to the addition of wild type p72 protein (closed symbols) or buffer only (open symbols) are shown. Error bars are standard deviations of triplicate determinations. Residual anisotropy following the addition of NaCl to 0.42 m (asterisks) or EDTA to 40 mm (circle) is shown.
The effects of small molecules on binding equilibria can be tested easily because optical methods do not require physical separation of bound and unbound species. For example, the addition of NaCl to a final concentration of 0.42 m restored anisotropy to baseline values (±0.01 units) with no change in total fluorescence intensity (Fig. 3), indicating disassociation of the protein from the single-stranded and double-stranded DNA substrates. Inclusion of 1 mm ATP in the binding buffer resulted in no change in the apparent Kd(DNA) for either substrate (data not shown), although the presence of ∼20 μm residual ATP carried forward from our protein storage buffer prevented testing of a strict nucleotide requirement for DNA binding. The addition of excess EDTA restored anisotropy to near the baseline value (Fig. 3), which supports the prior observation that p72 requires Mg2+ and ATP to bind to single-stranded DNA (25). The T7 gp4 helicase requires both Mg2+ ions and nucleotide triphosphates to bind single-stranded DNA (27), and the bound protein species are primarily hexamers, suggesting that hexamerization significantly enhances ssDNA binding (28). Interestingly, titration of free Mg2+ in our binding buffer by the stepwise addition of EDTA resulted in temporary but significant light scattering, and titration beyond this transient stage revealed a steady decrease in anisotropy, whereas total fluorescence intensity remained stable (supplemental Fig. S1). One interpretation of this pattern is that Mg2+ ions are required not only to facilitate DNA binding but also to stabilize the unbound p72 protein, which is consistent with the need to include Mg2+ in all buffers to prevent aggregation during protein purification.
A DNA binding analysis was also performed on the WT p72 protein bearing a His6 affinity tag, and calculated affinities for both single-stranded and double-stranded DNA substrates were almost identical to the Kd(DNA) values for the untagged protein (Table 1). As an initial screen for enzyme dysfunction, DNA binding affinity was also determined for each of the mutant forms of p72. Among the 20 mutant forms, binding affinities varied from 1.0 to 10.8 nm for single-stranded DNA and from 0.7 to 21.4 nm for double-stranded DNA (Table 1). Although slightly greater variability was observed with double-stranded DNA, all of the mutant proteins exhibited uniformly tight binding to both DNA substrates, like the WT enzyme. To the extent that the binding of a protein to heparin-Sepharose mimics binding to DNA, the relatively high affinities of the mutant forms to DNA were not unexpected because the mutant proteins exhibited very consistent elution profiles during heparin-Sepharose chromatography. Despite the 3–4-fold increases in Kd values for the A359T, S369Y, R374Q, F485L, and Y508C variants, disassociation constants in the nanomolar range predict high affinity interactions with DNA substrates.
TABLE 1.
Binding affinities of C10orf2 variants to single-stranded and double-stranded DNA
Fluorescence anisotropy was monitored during the stepwise addition of the indicated p72 proteins (calculated as hexamers), as described under “Experimental Procedures.” Binding isotherms for single-stranded (TD1-F) and double-stranded (TD1-F·TD2) oligonucleotide substrates were fit with a quadratic equation to calculate apparent Kd(DNA) values. Uncertainties are standard errors of least square residual values.
| Enzyme | Kd(ssDNA) | Kd(dsDNA) |
|---|---|---|
| nm | nm | |
| WT without His | 5.7 ± 2.5 | 4.9 ± 1.9 |
| WT | 6.0 ± 1.1 | 6.5 ± 1.9 |
| R303W | 1.4 ± 1.2 | 1.0 ± 0.5 |
| W315L | 1.6 ± 0.2 | 1.1 ± 0.2 |
| A318T | 1.4 ± 0.4 | 1.4 ± 0.3 |
| K319E | 2.7 ± 0.9 | 2.9 ± 0.5 |
| K319T | 4.1 ± 0.8 | 3.4 ± 0.7 |
| R334Q | 2.8 ± 0.8 | 2.2 ± 0.2 |
| P335L | 3.5 ± 0.7 | 5.6 ± 1.7 |
| R354P | 3.0 ± 0.8 | 1.7 ± 0.4 |
| R357P | 1.1 ± 0.5 | 3.2 ± 0.6 |
| A359T | 5.5 ± 2.3 | 17.5 ± 3.0 |
| I367T | 1.0 ± 0.9 | 0.7 ± 0.5 |
| S369P | 1.4 ± 0.7 | 0.8 ± 0.3 |
| S369Y | 6.2 ± 1.1 | 11.4 ± 4.1 |
| R374Q | 10.8 ± 1.6 | 6.8 ± 1.9 |
| L381P | 1.5 ± 0.5 | 1.8 ± 0.2 |
| T457I | 3.6 ± 0.4 | 6.7 ± 0.9 |
| W474C | 3.2 ± 0.5 | 3.7 ± 0.9 |
| A475P | 5.6 ± 1.6 | 4.2 ± 1.1 |
| F485L | 8.4 ± 2.7 | 21.4 ± 4.8 |
| Y508C | 7.3 ± 2.1 | 9.3 ± 2.4 |
DNA Helicase Activity of p72 in Vitro
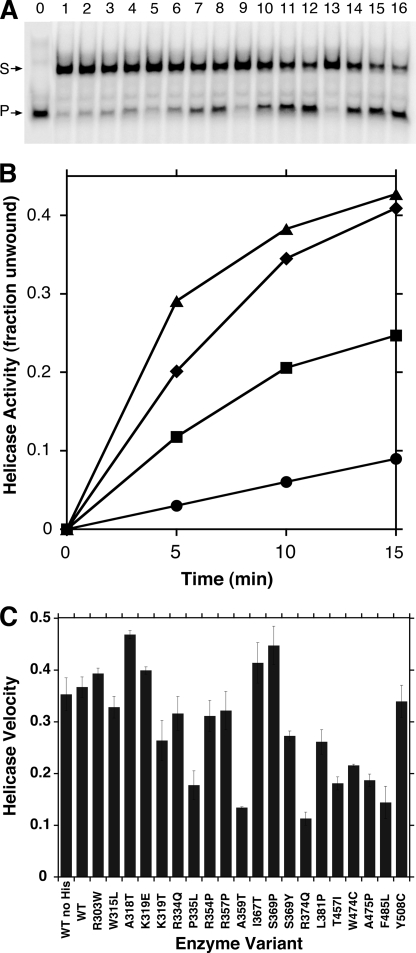
The substrate chosen to assess helicase function consisted of radiolabeled TD1 oligonucleotide hybridized to unlabeled TD3 oligonucleotide, forming a partially complementary, forked DNA with a 22-nt single-stranded tail at the 5′-end of the labeled strand. The DNA binding affinity of WT p72 for a fluorescein-tagged version of this substrate (TD1-F·TD3) was determined by changes in steady state fluorescence anisotropy, as described under “Experimental Procedures.” ATP was not included in the binding buffer to prevent translocation during the measurement. Baseline anisotropy was 0.10, reflecting slow rotational diffusion of this branched oligonucleotide substrate in comparison with the single-stranded or double-stranded oligonucleotides. Binding isotherms were determined in triplicate by titrating WT p72, and the calculated disassociation constant was Kd = 2.3 ± 1.4 nm. Knowledge of such high affinity binding informed the design of a quantitative DNA helicase assay, and a fixed substrate concentration of 40 nm, well above the Kd, was chosen to maximize the fraction of protein bound to the forked DNA helicase substrate at equilibrium. The dynamic range of the helicase assay was examined in standard helicase reactions at 37 °C with 3, 6, 12, or 20 nm concentrations of WT p72 hexamers. Samples were removed at 5-min intervals and analyzed by native PAGE (Fig. 4A). A heat-denatured sample revealed rapid migration of the single-stranded DNA product (lane 0), and time-dependent unwinding activity was apparent at each concentration of enzyme (lanes 1–4, 5–8, 9–12, and 13–16, respectively). Similar reactions assembled without ATP showed no activity, and spontaneous disassociation of the DNA strands was undetectable in 30-min reactions lacking enzyme (data not shown). When reaction products were quantified and plotted against time, the unwinding response was virtually linear for at least 10 min at the 3, 6, and 12 nm enzyme concentrations (Fig. 4B). The relative slopes of the lines showed that helicase activity was also proportional to enzyme concentration, although the substrate began to saturate at 20 nm p72 hexamers. Helicase activity increased incrementally with more enzyme, and maximal activity was observed at a 7-fold stoichiometric excess of enzyme over DNA substrate (supplemental Fig. S2). Additional enzyme was inhibitory, perhaps due to non-productive competition of enzyme for the substrate or by excess enzyme facilitating strand reannealing, although these ideas were not tested experimentally. Subsequent helicase reactions included 40 nm substrate and 12 nm p72 hexamers to ensure a stoichiometric excess of DNA and a catalytic quantity of protein. Considering the measured disassociation constant, theory predicts that 28% of the substrate and 93% of the enzyme should be bound at equilibrium in vitro under these standardized conditions. With ATP fixed at 4.5 mm, the helicase exhibited peak activity at 5 mm MgCl2 with a broad optimum between 3 and 8 mm. Product inhibition did not substantially interfere with the assay because inclusion of up to 1 mm ADP did not diminish helicase function (data not shown).
FIGURE 4.
DNA helicase activity of mitochondrial DNA helicase proteins. A, the ability of WT p72 to unwind a forked oligonucleotide substrate was determined as described under “Experimental Procedures,” except that reactions contained 3 (lanes 1–4), 6 (lanes 5–8), 12 (lanes 9–12), or 20 nm (lanes 13–16) WT p72 hexamers. Reactions were incubated at 37 °C for 0 (lanes 1, 5, 9, and 13), 5 (lanes 2, 6, 10, and 14), 10 (lanes 3, 7, 11, and 15), or 15 min (lanes 4, 8, 12, and 16) before single-stranded [32P]TD1 products (P, arrow) and [32P]TD1·TD3 forked DNA substrates (S, arrow) were resolved by native gel electrophoresis. Substrate denatured by boiling is shown (lane 0). B, products of helicase reactions were quantified as described under “Experimental Procedures” and plotted as a function of time. Reactions contained 3 (circles), 6 (squares), 12 (diamonds), or 20 nm (triangles) WT p72 hexamers. Reactions without enzyme did not exhibit a detectable increase in products after a 15-min incubation. C, DNA helicase activity was determined in standard helicase reactions containing 40 nm forked oligonucleotide substrate and 12 nm of the indicated p72 variant, as described under “Experimental Procedures.” The slopes of independent time course reactions were calculated in duplicate, and helicase velocities were expressed as the fraction of total substrate unwound in 10 min. Error bars are standard deviations of multiple determinations.
We wanted to compare the intrinsic catalytic efficiencies of the helicases independent of DNA binding affinity, and the high substrate concentration utilized in the standard assay served to minimize variability due to differential DNA binding. Helicase reactions were incubated for 0, 5, or 10 min to remain in the linear portion of the reaction profile, and reaction velocities were calculated in duplicate from the slopes of independent time course reactions (Fig. 4C). WT p72 unwound 35 ± 3% of the substrate in 10 min, which represents 170 ± 15 fmol of oligonucleotide displaced in the standard reaction. WT p72 bearing a His6 affinity tag consumed 37 ± 2% (176 ± 10 fmol) of the substrate, which was essentially indistinguishable from the activity of the untagged enzyme. Notably, all 20 mutant forms of p72 exhibited helicase activity under standard reaction conditions. Specific activities varied from 54 ± 6 fmol displaced/10 min for R374Q variant (31% of WT activity) to 225 ± 4 fmol displaced/10 min for the A318T variant (128% of WT activity). In all cases, helicase activity was proportional to time of incubation. The demonstrated DNA helicase activities of the W315L, K319E, I367T, S369P, R374Q, and L381P variants (Fig. 4C) disagree with published reports that these amino acid substitutions significantly impair or even inactivate helicase function (13, 24, 29).
ATPase Activity of p72 in Vitro
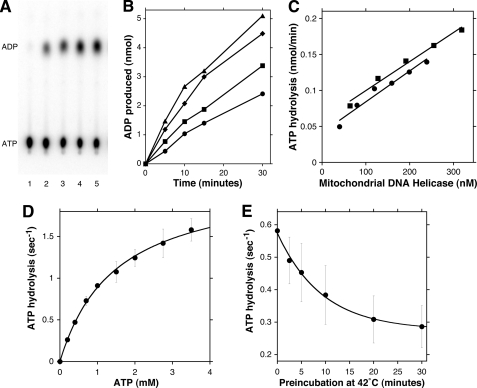
As DNA helicase activity is driven by hydrolysis of nucleoside 5′-triphosphates (30), biochemical characterization of nucleotide hydrolysis is another means to identify potential dysfunction within the collection of mutant proteins. The ability to vary ATP concentration over a very broad range, which can be impractical for a DNA substrate, also makes evaluation of ATPase activity more flexible than the DNA helicase assay. ATP hydrolysis by WT p72 was measured as described under “Experimental Procedures.” Reactions were terminated at 5-min intervals, and radiolabeled ATP substrate and ADP product were resolved by thin layer chromatography (Fig. 5A). Parallel reactions lacking enzyme did not cause a detectable increase in the ADP signal within 30 min (data not shown). Although plots of ADP produced as a function of time were linear for at least 10 min, reaction rates began to diminish at longer times of incubation (Fig. 5B). In the absence of added DNA, WT p72 hydrolyzed ATP with an initial rate (0–10 min) of 0.42 ± 0.05 s−1 for each p72 monomer. Rates of ATP hydrolysis were higher when otherwise identical reactions also contained DNA, and the degree of stimulation depended on the specific type of DNA included in each reaction. For example, including double-stranded plasmid DNA enhanced ATPase activity by 157% (0.65 ± 0.03 s−1), whereas activated calf thymus DNA (with a mixed single-stranded and double-stranded DNA character) stimulated ATPase activity by 233% (0.97 ± 0.09 s−1). The addition of purely single-stranded M13mp18 DNA stimulated ATPase activity by 294% (1.22 ± 0.09 s−1). Maximal stimulation of ATPase activity by single-stranded, circular DNA is noteworthy because it suggests that a free 5′-end is not needed for the helicase to bind to DNA. These observations highlight a similarity between the mitochondrial DNA helicase and T7 gene 4 protein, insofar as DNA stimulates NTP hydrolysis although it is not required for the reaction (22, 26). Our observations conflict with a previous report that the ATPase activity of p72 is stimulated 2-fold by ssDNA but specifically is not stimulated by dsDNA (25). We cannot easily explain this discrepancy, although we note fundamental differences in the two assay methods. Although we directly monitor conversion of [α-32P]ATP to [α-32P]ADP by TLC, the earlier assay detects cleavage of the γ-32P-O4 from [γ-32P]ATP by capturing unhydrolyzed ATP with acidic activated charcoal (Norit A).
FIGURE 5.
ATPase activity of the mitochondrial DNA helicase. A, ATPase activity was assessed as described under “Experimental Procedures.” Reactions (20 μl) contained 191 nm WT p72 monomers (3.82 pmol) and were incubated at 37 °C for 0, 5, 10, 15, or 30 min (lanes 1–5, respectively) prior to thin layer chromatography. Relative mobilities of the ATP substrate and ADP product are indicated. B, ATPase reactions were assembled as in A and included no supplement (circles), 0.2 μg of double-stranded replicative form M13mp18 DNA (squares), 0.2 μg of calf thymus DNA (diamonds), or 0.2 μg of single-stranded viral form M13mp18 DNA (triangles). The time course of each ATPase reaction was quantified as described under “Experimental Procedures.” C, ATPase reactions were supplemented with calf thymus DNA and incubated 30 min with the indicated concentrations of p72 monomers. ATP hydrolysis by WT p72 with (square) and without (circle) a C-terminal His6 affinity tag was quantified as described under “Experimental Procedures.” D, standard ATPase reactions containing the indicated concentrations of ATP and 245 nm WT p72 monomers (lacking an affinity tag) were incubated for 25 min. Reaction rates are expressed as turnover number for p72 monomers, and data were fit to the Michaelis-Menten steady state rate equation. E, duplicate samples of WT p72 protein were preincubated at 42 °C in the absence of DNA for the indicated times, and ATPase activity was determined in standard reactions incubated for 25 min. Reaction rates are expressed as turnover number for p72 monomers, and data were fit to an equation for exponential decay. Error bars in D and E are standard deviations of at least three determinations.
Under standardized reaction conditions, p72 ATPase activity was proportional to enzyme concentration over at least a 5-fold range (Fig. 5C). This relationship was true for both the His6 affinity-tagged form of p72 and the untagged form, and the virtually superposable plots of enzymatic specific activity for the two forms indicate that the affinity tag is not detrimental to ATPase activity. Although salt was not included in standard ATPase reactions, enzyme preparations contributed ∼25 mm NaCl to the mixtures. ATPase activity decreased steadily with additional NaCl, with 0.1, 0.2, 0.3, and 0.4 m NaCl inhibiting activity to 87, 66, 48, and 26% full activity, respectively. Excess salt likely interferes with ATP binding as well as causing a loss of stimulation due to reduced DNA binding.
The ATPase activity of the mitochondrial helicase was quantified more rigorously by steady state kinetic analysis. Enzyme was incubated in standard ATPase reactions with variable concentrations of ATP, and steady state kinetic parameters were calculated from curves fit to the Michaelis-Menten equation by a non-linear least squares algorithm (Fig. 5D). WT p72 catalyzed ATP hydrolysis with a maximal turnover rate of 2.18 ± 0.08 s−1, as calculated for p72 monomers, and the Km(ATP) was 1.43 ± 0.15 mm. Kinetic parameters were also determined for each of the mutant forms of p72 to probe for subtle defects in ATPase function (Table 2). Km(ATP) values among the 20 mutant forms ranged from 0.84 to 4.55 mm, and the I367T, R374Q, and L381P variants exhibited notably higher Km values (>1 S.D. above median Km). Greater variability was observed for kcat values in the mutant collection, and Vmax values ranged from 0.66 to 8.31 s−1. The A475P and W474C variants exhibited the lowest ATPase rates, and the I367T and L381P variants had turnover numbers even higher than WT p72. The overall catalytic efficiency as estimated by the specificity constant (kcat/Km) is also a useful parameter for comparing the enzymes. The mutant collection displayed a median kcat/Km ratio (1.07 ± 0.32 s−1 · mm−1) that was 69% of the WT value. Although a majority of the variants displayed kcat/Km ratios that clustered near the median value, the A475P and W474C variants were only 35 and 42% as efficient as the WT enzyme. The L381P variant exhibited interesting behavior; despite several assays with multiple protein preparations, L381P p72 always generated high kcat and Km values. These values may actually underestimate the kcat and Km for the L381P variant due to depletion of Mg2+ cofactor at higher ATP concentrations. The I367T and R374Q variants behaved in a similar fashion but to a lesser extent (Table 2). This effect is discussed in more detail below.
TABLE 2.
Steady state kinetic parameters for ATPase activity of mitochondrial helicase enzymes
ATPase activity of p72 proteins was measured as described under “Experimental Procedures.” Reactions contained 0.2 μg of activated calf thymus DNA, and ATP concentrations were varied from 0 to 5 mm. Steady state kinetic parameters were derived from non-linear curve fitting to the Michaelis-Menten equation. Uncertainties represent standard errors. Reaction velocity at 0.7 mm ATP was calculated from the Michaelis-Menten equation and the kinetic parameters of each enzyme. Turnover numbers are calculated for p72 monomers.
| Enzyme | Km(ATP) | kcat at Vmax | kcat/Km | Velocity at 0.7 mm ATP |
|---|---|---|---|---|
| mm | s−1 | s−1 · mm−1 | s−1 | |
| WT without His | 1.43 ± 0.15 | 2.18 ± 0.08 | 1.53 | 0.72 |
| WT | 1.22 ± 0.14 | 1.90 ± 0.08 | 1.55 | 0.69 |
| R303W | 1.81 ± 0.15 | 2.29 ± 0.08 | 1.27 | 0.64 |
| W315L | 1.00 ± 0.12 | 1.13 ± 0.05 | 1.13 | 0.47 |
| A318T | 0.84 ± 0.12 | 1.19 ± 0.05 | 1.42 | 0.54 |
| K319E | 1.82 ± 0.21 | 2.24 ± 0.11 | 1.23 | 0.62 |
| K319T | 1.02 ± 0.13 | 0.98 ± 0.04 | 0.96 | 0.40 |
| R334Q | 1.04 ± 0.18 | 1.01 ± 0.06 | 0.97 | 0.40 |
| P335L | 1.32 ± 0.12 | 1.08 ± 0.04 | 0.82 | 0.38 |
| R354P | 1.03 ± 0.19 | 1.18 ± 0.08 | 1.15 | 0.48 |
| R357P | 1.24 ± 0.09 | 1.52 ± 0.04 | 1.23 | 0.55 |
| A359T | 1.30 ± 0.24 | 1.06 ± 0.08 | 0.81 | 0.37 |
| I367T | 2.23 ± 0.15 | 2.89 ± 0.09 | 1.30 | 0.69 |
| S369P | 1.89 ± 0.14 | 2.09 ± 0.06 | 1.11 | 0.57 |
| S369Y | 1.17 ± 0.15 | 1.94 ± 0.10 | 1.67 | 0.73 |
| R374Q | 2.48 ± 0.32 | 2.17 ± 0.13 | 0.88 | 0.48 |
| L381P | 4.55 ± 0.49 | 8.31 ± 0.50 | 1.83 | 1.11 |
| T457I | 1.08 ± 0.08 | 0.88 ± 0.03 | 0.81 | 0.35 |
| W474C | 1.10 ± 0.16 | 0.70 ± 0.04 | 0.64 | 0.27 |
| A475P | 1.22 ± 0.24 | 0.66 ± 0.04 | 0.54 | 0.24 |
| F485L | 1.44 ± 0.06 | 1.15 ± 0.03 | 0.80 | 0.38 |
| Y508C | 1.96 ± 0.16 | 2.04 ± 0.08 | 1.04 | 0.54 |
Protein Stability Assessed by Heat Inactivation of ATPase Activity
As determined above, none of the amino acid substitutions associated with mitochondrial disease caused profound defects in DNA binding, DNA helicase function, or ATPase activity. The relatively minor changes we observed suggest that the mutations may cause only subtle structural changes to the helicase. Because catalysis is sensitive to protein conformation, one simple measure of enzyme stability is the ability to resist inactivation from repeated freeze/thaw cycles. However, the ATPase activity of WT p72 remained completely undiminished following four consecutive cycles of freezing on liquid nitrogen and thawing on wet ice (data not shown). Therefore, resistance to thermal inactivation was chosen as a more stringent test of enzyme stability. Helicase preparations were incubated at 42 °C for increasing intervals of time, and residual ATPase activity was assessed by standard assay (Fig. 5E). Remaining activity was plotted against time of heat treatment, and the data were fit to an equation for exponential decay (Nt = Noe−λt). WT p72 had an average decay constant (λ) of 0.087 min−1, which gives a calculated half-life of 7.9 min at 42 °C (t½ = ln 2/λ). Next, the stabilities of the substituted p72 proteins were estimated by thermal inactivation (Table 3). Decay constants varied from 0.081 to 0.273 min−1, which translate to half-lives ranging from 8.6 min down to a minimum of 2.5 min. The R303W, W315L, A359T, S369Y, R374Q, and F485L variants were much more sensitive to heat inactivation than WT p72. Curves for all of the disease variants exhibited excellent agreement with the first order decay equation, indicating that inactivation occurs by single step, which we assume is an irreversible, heat-induced conformational change.
TABLE 3.
Heat inactivation of ATPase activity
Thermostability of p72 variants was determined by pretreating the proteins in a 42 °C water bath for 0, 2.5, 5, 10, 20, or 30 min in the absence of DNA followed by standard ATPase assay with 0.7 mm ATP, as described under “Experimental Procedures.” Exponential decay constants were calculated from plots of remaining ATPase activity as a function of time. Uncertainties represent standard errors. Half-lives were calculated as ln 2 divided by average decay constants.
| Enzyme | Decay constant | τ½ at 42 °C |
|---|---|---|
| min−1 | min | |
| WT without His | 0.087 ± 0.021 | 7.9 |
| WT | 0.097 ± 0.021 | 7.2 |
| R303W | 0.203 ± 0.016 | 3.4 |
| W315L | 0.162 ± 0.021 | 4.3 |
| A318T | 0.122 ± 0.011 | 5.7 |
| K319E | 0.134 ± 0.010 | 5.2 |
| K319T | 0.150 ± 0.012 | 4.6 |
| R334Q | 0.081 ± 0.038 | 8.6 |
| P335L | 0.111 ± 0.013 | 6.2 |
| R354P | 0.081 ± 0.013 | 8.6 |
| R357P | 0.151 ± 0.030 | 4.6 |
| A359T | 0.251 ± 0.039 | 2.8 |
| I367T | 0.096 ± 0.005 | 7.2 |
| S369P | 0.127 ± 0.014 | 5.5 |
| S369Y | 0.217 ± 0.025 | 3.2 |
| R374Q | 0.158 ± 0.010 | 4.4 |
| L381P | 0.131 ± 0.018 | 5.3 |
| T457I | 0.116 ± 0.008 | 6.0 |
| W474C | 0.137 ± 0.008 | 5.1 |
| A475P | 0.152 ± 0.010 | 4.6 |
| F485L | 0.273 ± 0.030 | 2.5 |
| Y508C | 0.115 ± 0.014 | 6.0 |
DISCUSSION
Biochemical characterization of the mitochondrial helicase variants in vitro completely relies on the ability to maintain the stability and solubility of the enzymes. Previous strategies for recombinant p72 have included incorporation of single or dual affinity tags to facilitate rapid purification or supplementing buffers with glycerol, NaCl or 0.1 m arginine to stabilize the protein (11, 13, 24, 31). Nevertheless, preparation of certain adPEO variants has proven difficult, such as the inability to make R354P p72 in soluble form with baculovirus-infected insect cells (24). To establish an E. coli expression system for recombinant p72, we surveyed a variety of conditions by trial and error to maximize expression, solubility, and activity of the protein. The most favorable conditions included induction at a reduced temperature and inclusion of NaCl, glycerol, the non-ionic detergent Nonidet P-40, Mg2+ ions, and the ATP cofactor in all buffers throughout purification. Optimization of conditions for protein purification was instructive to the subsequent design of in vitro assays. For example, the enzyme cannot tolerate dialysis or dilution into buffers containing <0.3 m NaCl without suffering irreversible precipitation. Sensitivity to aggregation is increased greatly once contaminating nucleic acid fragments have been removed by passage through a HiTrap Q column, and aggregation is mitigated by Mg2+, glycerol, and Nonidet P-40. Consequently, stabilizing additives were included in enzyme dilution buffers, and enzymatic reactions with low ionic strength were assembled so that DNA substrates were always added prior to enzymes.
The existing scientific literature contains significant discrepancies regarding various physical and biochemical properties of the wild type and mutant variants of the p72 protein, and examination of specific methods and conditions utilized in these studies helps to resolve much of the conflicting information. For example, published reports of the DNA binding strength of certain mutant forms of p72 are inconsistent. As determined by electrophoretic mobility shift assay (EMSA), the S369P protein was reported to have low affinity for single-stranded DNA relative to the wild type enzyme (24). However, another group reported higher than wild type DNA binding affinity for this protein using comparable experimental conditions (13). In the present work, we also found the affinity of the S369P variant to be higher than that of the WT enzyme for both single-stranded DNA and double-stranded DNA (Table 2). Similarly, single-stranded DNA binding by the R374Q and W315L forms is reported to be both deficient (24, 29) and proficient (13) by EMSA. Although the current work concurs with the near WT binding affinity previously observed for the A359T (13, 24) and R334Q (29) variants, the weak DNA binding reported for the K319E (13), L381P and I367T (24), and P335L (29) variants is difficult to reconcile with the high affinity binding that we observed (Table 1). Possible explanations include fundamental differences in the methodologies used to estimate binding and differential stability of some of the mutant variants. Methods to determine DNA binding affinity generally utilize a stoichiometric excess of protein over DNA. However, published methods for EMSA of p72 utilized absolute DNA concentrations 4–12-fold lower than the accepted equilibrium binding constant for p72 and ssDNA (13, 24). Less than 10% of available protein can be bound to DNA at equilibrium under these conditions. During protein purification, p72 has a tendency to aggregate after removal of nucleic acids. Also, DNA stimulates the ATPase activity of p72. These observations suggest that the free form of the mtDNA helicase is less stable than the form bound to DNA. Taken together, the low DNA concentrations utilized for EMSA are expected to underestimate DNA affinity for the least stable p72 variants. In contrast, fluorescence anisotropy measurements utilized 15 nm DNA, which is ∼3-fold higher than the Kd(DNA), for WT p72. Also, fluorescence anisotropy measures steady state DNA binding in solution with the free and bound forms of protein in rapid equilibrium, whereas EMSA necessarily partitions free and bound species during electrophoresis. This partitioning, combined with the long time interval needed for electrophoresis when compared with optical methods, may exacerbate the instability effect and underestimate binding affinity for some mutant variants.
Published reports that the W315L, K319E, R334Q, P335L, I367T, S369P, R374Q, and L381P p72 variants are deficient for DNA helicase function (13, 24, 29) also disagree with our results (Fig. 4C). In consideration of the instability of the enzyme at low ionic strength when not bound to DNA, we designed our in vitro helicase assay with a DNA substrate concentration >4-fold higher than the Kd(DNA) for WT p72. This served both to maximize the fraction of enzyme bound to DNA and to render the assay insensitive to the small variations in DNA binding affinity among the mutants. Under optimized conditions, DNA helicase activity was easily detected with catalytic quantities of enzyme. Although ∼4-fold variability in helicase function was observed within the mutant collection, none of the p72 variants displayed a profound defect in helicase activity. In contrast, published DNA helicase assays utilized only 0.075–0.33 nm oligonucleotide substrate (13, 16, 24, 25, 29), conditions under which unwinding activity is directly proportional to single-stranded DNA binding (25). At such low substrate concentrations, greater than 90% of the enzyme population is not bound to the DNA at equilibrium. Accordingly, comparing enzymatic specific activities reveals that the measured DNA helicase activity of WT p72 is at least 200-fold lower with 0.33 nm substrate than with 40 nm substrate (compare Refs. 16, 24, 25, and 29 with Fig. 4). Comparisons of specific activity are further complicated because the previous studies employed assays with a 30–90-fold stoichiometric excess of p72 hexamers over the DNA substrate (16, 24, 25, 29). Although excess enzyme would facilitate the formation of sufficient enzyme-DNA complex to produce a detectable helicase signal, we found the addition of surplus enzyme to be counterproductive, if not inhibitory to helicase function (see supplemental Fig. S2). Certain amino acid substitutions, including W315L, K319T, R334Q, P335L, I367T, R374Q, and L381P, are reported to result in concomitant, severe reductions in DNA binding, ATP hydrolysis, and DNA helicase function (24, 29). Molecular modeling of these residues based on the three-dimensional structure of bacteriophage T7 gene 4 helicase has invoked alteration of specific side chain interactions as possible explanations for the reported dysfunctions. However, insufficient sequence identity (19%) makes the reliability of this structural homology model difficult to assess, and the absence of separation of function mutations among the seven widely distributed substitutions is inadequately explained. In any event, our observation that recombinant enzymes with the same substitutions retain these activities when the proteins are purified and stored under our conditions strongly suggests that substitution at these residues promotes a more global structural instability rather than deficiencies specifically targeted to DNA binding or ATP hydrolysis.
The mitochondrial helicase belongs to the superfamily 4 of hexameric helicases and may share conformational dynamics similar to AAA+ ATPases (6, 32, 33). Recombinant human p72 was first shown to form higher order multimers by minimally denaturing SDS-PAGE analysis of whole cell extracts (5). Velocity sedimentation and gel filtration analyses proved the purified, recombinant enzyme to be a stable hexamer with a native molecular mass of 420,000 Da (31). In our E. coli expression system, the recombinant helicase elutes as a single peak at the void volume of a preparative Sephacryl S200 HR size exclusion column, indicating a native molecular weight in the presence of 5 mm Mg2+ and 0.5 m NaCl that exceeds the exclusion limit of the resin (molecular weight >250,000). A very recent report from Kaguni and co-workers (33) also documents the effects of salt and Mg2+ on the stability, oligomeric state, and conformation of the WT enzyme. Through electron microscopy, glutaraldehyde cross-linking, and gel filtration analyses, they demonstrate that low salt conditions disrupt hexameric structure and cause p72 to aggregate. Interestingly, they also show that p72 monomers can form heptamers in 0.1 m NaCl when Mg2+ and ATPγS are present (33). This situation is reminiscent of bacteriophage T7gp4 DNA helicase, the prototypical superfamily 4 helicase, in which hexamerization is significantly stimulated by binding to nucleotide cofactors and single-stranded DNA (34) and the ratio of heptamer and hexamer conformations of T7gp4 subunits is modulated by dTDP and single-stranded DNA (35, 36). DnaB, the replicative helicase from E. coli, also requires Mg2+ or a high concentration of NaCl to maintain a stable hexameric structure (37). A common feature of superfamily 4 helicases is a linker region separating the N-terminal primase/primase-interacting domain and the C-terminal helicase domain (38, 39). This linker region is important for subunit association in T7gp4 (22, 40). Of the 20 C10orf2 mutations in the current study, 15 of the amino acid substitutions reside in the linker region of the p72 protein (Fig. 1). Six substitutions in a narrow zone (codons 359–381) within the linker region consistently resulted in lower yields of purified recombinant p72 despite ample overexpression and the presence of 5 mm Mg2+ and >0.3 m NaCl throughout purification. Lower yields for the A359T, I367T, S369P, S369Y, R374Q, and L381P variants suggest that this 22-amino acid zone contributes to stability of the hexamer. In support of this idea, a previous study of p72 variants bearing linker region mutations revealed that the I367T and R374Q variants behaved as monomers when analyzed by gel filtration in the absence of Mg2+, and the L381P variant was an unstable hexamer under these conditions (24). However, the L381P and I367T variants were not more sensitive to thermal inactivation of ATPase activity than WT p72 (Table 3), implying that the thermally disrupted conformation needed for ATPase function is distinct from the quaternary structure that is destabilized by the L381P and I367T substitutions. In any event, all 20 mutant variants bind DNA with high affinity (Table 1) and exhibit both ATPase and DNA helicase activities approaching that of the wild type enzyme (Table 2 and Fig. 4), which supports the argument that these p72 variants are capable of hexamerization.
Hexameric helicases have a number of common attributes, including the formation of multiple nucleotide binding sites at the interface of neighboring subunits, passage of single-stranded DNA through the central channel of the toroid, and contact with the encircled DNA by hairpin loops extending inward from each subunit (38). Several models for unwinding nucleic acids exist that couple nucleotide hydrolysis to strand separation and unidirectional translocation along single-stranded DNA (38, 41). Although mechanistic details for the mitochondrial helicase are not yet known, the comparable affinities of p72 for both single-stranded and double-stranded DNA suggest that the enzyme may interact with both the displaced strand and the duplex portion of the helicase substrate. Also, the I367T, R374Q, and L381P variants displayed potentially informative behavior in our kinetic analysis of ATPase function. Elevated Km values reflect partial nucleotide binding defects for all three enzymes, and the I367T and L381P variants exhibited maximal rates of ATP hydrolysis equal to or greater than WT p72. However, this elevated ATP hydrolysis was not accompanied by proportionately higher DNA helicase activity in a separate assay, implying that nucleotide hydrolysis and DNA translocation may be partially uncoupled in these variants. Although I367T and L381P p72 had near WT thermostability and helicase function in vitro (Table 3 and Fig. 4C), a previous study indicated that the I367T, R374Q, and L381P variants were unstable hexamers (24). We propose that weakened interaction between neighboring subunits bearing these substitutions is linked to the altered nucleotide binding and hydrolysis. Future experiments may determine whether interaction with polymerase γ relieves the apparent uncoupling observed for these variants.
Utilizing clinically identified p72 variants to decipher enzyme mechanisms through subunit-poisoning experiments is complicated because our observation that homohexamer mutant proteins retain ATPase and DNA helicase activities predicts that heterohexamers composed from mixed WT and mutant monomers will also retain activity in vitro. In fact, evaluation of mtDNA copy number in cultured Schneider cells following overexpression of Drosophila mtDNA helicase genes bearing missense mutations suggested that heterohexamers with only one or two mutant subunits retained some helicase activity in vivo (12, 42). However, targeted mutations that substantially inactivate catalytic function may prove enlightening in subunit mixing experiments. The three extreme C-terminal substitutions that largely inactivate ATPase activity of human p72 in vitro and greatly reduce mtDNA copy number in cultured Schneider cells are excellent candidates for such a study (42).
The widely variable clinical presentation of disease associated with mutation of C10orf2 is very intriguing. Other investigators identified 17 separate heterozygous mutations of C10orf2 in individuals with adult-onset PEO with multiple mtDNA deletions (Fig. 1). In most cases, co-segregation of disease with the mutant allele revealed autosomal dominant inheritance, indicating that one mutant allele is sufficient to develop PEO in adulthood. Homozygous individuals generally have a more severe phenotype with an earlier onset PEO than their heterozygous relatives (5). In contrast, mutations in C10orf2 causing the T457I, Y508C, and A318T substitutions are associated with hepatocerebral MDS and IOSCA. These severe neurodegenerative disorders, which present in the first weeks or month of life, are characterized by tissue specific depletion of mtDNA without the accumulation of deletions in mtDNA (43, 44). Three related individuals who were homozygous for a mutation causing dual T457I substitutions developed severe hepatocerebral MDS and died within the first three years of life. The heterozygous parents and grandparents in this consanguineous pedigree were unaffected, which established a recessive mode of inheritance (45). Similarly, homozygous Y508C substitution results in IOSCA, which is a special form of MDS with a specific pattern of neuronal degeneration, and heterozygotes are unaffected (46). These investigators further demonstrated that recombinant Y508C p72 hexamers have near WT DNA helicase activity with a slight reduction in single-stranded DNA binding affinity (43), results that corroborate the current work. The A318T substitution is only found as a compound heterozygous mutation with Y508C in a severe early onset encephalopathy with liver involvement and depletion of mtDNA in the liver (44, 47). These observations that one A318T-, T457I-, or Y508C-substituted copy of C10orf2 is insufficient to induce disease strongly suggests that the remaining WT allele is sufficient for health or is able to rescue any biochemical deficiencies of the protein derived from the mutant allele. The description of a heterozygous patient (WT/Y508C) affected with IOSCA due to reduced expression of the WT allele supports this view (46). Attempts to correlate biochemical dysfunction with the clinical phenotypes associated with specific C10orf2 mutations are problematic because the degree of helicase dysfunction needed to induce a phenotype is unknown. Overall DNA helicase function is affected by the affinity of a given enzyme to DNA, efficiency of ATP hydrolysis, and sensitivity to inactivation. Although the helicase activity of each p72 variant (Fig. 4C) is generally proportional to the ATPase specificity constant (Table 2), this single parameter is insufficient to account for the linked contributions of differential DNA binding and protein stability. For example, R303W p72 has ∼80% of WT ATPase function (Table 2) and a low half-life of heat inactivation in the absence of DNA (Table 3), yet it also displays tight DNA binding (Table 1). Elevated binding likely compensates for the reduced stability, resulting in net helicase function in vitro similar to the WT enzyme (Fig. 4C). Similarly, R334Q p72 has only ∼60% WT ATPase function, yet it exhibits WT resistance to heat inactivation and a high affinity to DNA relative to WT, again resulting in near WT helicase function. In contrast, the near WT DNA binding affinity of A359T p72 was insufficient to compensate for only ∼50% ATPase function combined with a pronounced sensitivity to thermal inactivation, and this enzyme suffered a 3-fold reduction in helicase activity. Similar logic applies to the other enzymes, but it must be stressed that the spectrum of dysfunction within this mutant collection was <5-fold for any individual biochemical parameter, and none of the p72 variants exhibited a profound loss of function in vitro. In the simplest sense, partial biochemical dysfunction is consistent with a delayed presentation of disease, such as adult-onset PEO, but it is inadequate to explain the extreme variability in clinical severity, age of onset, or tissue specificity observed in patients.
In vivo ATP concentrations have been estimated in studies that targeted recombinant luciferase to various intracellular compartments. The concentration of ATP in the mitochondrial matrix of cultured osteosarcoma is estimated to be as low as 0.22 mm (48), whereas MIN6 pancreatic beta cells have a mitochondrial ATP concentration of 1.2 mm (49). The average of these two values (0.7 mm ATP) is ∼2-fold lower than the Km for ATP hydrolysis determined for the WT mitochondrial helicase, which predicts that helicase activity in vivo may be reduced relative to ideal conditions in vitro. The relative rate of ATP hydrolysis at 0.7 mm ATP for each p72 variant is estimated in Table 2. The adenine nucleotide translocator, encoded by ANT1 in humans, is the mitochondrial enzyme responsible for exchanging ADP and ATP between the mitochondrial matrix and the cytoplasm and is also a disease locus for PEO (50). Variability in the concentration of available ATP in different tissues and subcellular compartments, possibly linked to the function of the ANT, may affect helicase function. Unequal expression of heterozygous C10orf2 alleles in different individuals or tissues would affect the composition of p72 heterohexamers and could affect mtDNA replication in complex ways, such as through altered functional and physical interaction with other proteins at the mtDNA replication fork. Similarly, the underlying mechanisms influencing the progressive formation of multiple mtDNA deletions in PEO and the tissue-specific depletion of mtDNA in MDS and IOSCA are not currently known. Our biochemical analysis of C10orf2 proteins emphasized the importance of experimental design and stabilizing the enzymes, which permitted the quantitative biochemical characterization of some of the less stable mutant variants. Nevertheless, we recognize that the dynamic effects of mitochondrial fission and fusion, mitotic segregation of mitochondrial nucleoids, environmental stressors, and purifying selection of altered mitochondrial genomes contribute to the complexity of mitochondrial disease.
Acknowledgments
We thank Drs. Rajesh Kasiviswanathan and Scott Lujan for critical evaluation of the manuscript.
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health through the NIEHS (Grant ES 065078) (to W. C. C.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and primers.
- PEO
- progressive external ophthalmoplegia
- adPEO
- autosomal dominant PEO
- MDS
- mtDNA depletion syndrome
- IOSCA
- infantile-onset spinocerebellar ataxia
- IMAC
- immobilized metal ion affinity chromatography
- nt
- nucleotides
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Wallace D. C. (2005) Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace D. C., Fan W., Procaccio V. (2010) Annu. Rev. Pathol. 5, 297–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland W. C. (2008) Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suomalainen A., Kaukonen J., Amati P., Timonen R., Haltia M., Weissenbach J., Zeviani M., Somer H., Peltonen L. (1995) Nat. Genet. 9, 146–151 [DOI] [PubMed] [Google Scholar]
- 5.Spelbrink J. N., Li F. Y., Tiranti V., Nikali K., Yuan Q. P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., Santoro L., Toscano A., Fabrizi G. M., Somer H., Croxen R., Beeson D., Poulton J., Suomalainen A., Jacobs H. T., Zeviani M., Larsson C. (2001) Nat. Genet. 28, 223–231 [DOI] [PubMed] [Google Scholar]
- 6.Leipe D. D., Aravind L., Grishin N. V., Koonin E. V. (2000) Genome Res 10, 5–16 [PubMed] [Google Scholar]
- 7.Van Hove J. L., Cunningham V., Rice C., Ringel S. P., Zhang Q., Chou P. C., Truong C. K., Wong L. J. (2009) Am. J. Med. Genet. A 149A, 861–867 [DOI] [PubMed] [Google Scholar]
- 8.Fratter C., Gorman G. S., Stewart J. D., Buddles M., Smith C., Evans J., Seller A., Poulton J., Roberts M., Hanna M. G., Rahman S., Omer S. E., Klopstock T., Schoser B., Kornblum C., Czermin B., Lecky B., Blakely E. L., Craig K., Chinnery P. F., Turnbull D. M., Horvath R., Taylor R. W. (2010) Neurology 74, 1619–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A. M., Spelbrink J. N. (2003) Mol. Biol. Cell 14, 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J. N., Holt I. J., Suomalainen A. (2004) Hum. Mol. Genet. 13, 3219–3227 [DOI] [PubMed] [Google Scholar]
- 11.Wanrooij S., Goffart S., Pohjoismäki J. L., Yasukawa T., Spelbrink J. N. (2007) Nucleic Acids Res. 35, 3238–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima Y., Kaguni L. S. (2007) J. Biol. Chem. 282, 9436–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffart S., Cooper H. M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J. N. (2009) Hum. Mol. Genet. 18, 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyynismaa H., Mjosund K. P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J. N., Paetau A., Suomalainen A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17687–17692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyynismaa H., Suomalainen A. (2009) EMBO Rep. 10, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korhonen J. A., Gaspari M., Falkenberg M. (2003) J. Biol. Chem. 278, 48627–48632 [DOI] [PubMed] [Google Scholar]
- 17.Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 19.Longley M. J., Ropp P. A., Lim S. E., Copeland W. C. (1998) Biochemistry 37, 10529–10539 [DOI] [PubMed] [Google Scholar]
- 20.Heyduk T., Ma Y., Tang H., Ebright R. H. (1996) Methods Enzymol. 274, 492–503 [DOI] [PubMed] [Google Scholar]
- 21.Heyduk T., Lee J. C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1744–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notarnicola S. M., Park K., Griffith J. D., Richardson C. C. (1995) J. Biol. Chem. 270, 20215–20224 [DOI] [PubMed] [Google Scholar]
- 23.Debyser Z., Tabor S., Richardson C. C. (1994) Cell 77, 157–166 [DOI] [PubMed] [Google Scholar]
- 24.Korhonen J. A., Pande V., Holmlund T., Farge G., Pham X. H., Nilsson L., Falkenberg M. (2008) J. Mol. Biol. 377, 691–705 [DOI] [PubMed] [Google Scholar]
- 25.Farge G., Holmlund T., Khvorostova J., Rofougaran R., Hofer A., Falkenberg M. (2008) Nucleic Acids Res. 36, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S. J., Qimron U., Richardson C. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8908–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matson S. W., Richardson C. C. (1985) J. Biol. Chem. 260, 2281–2287 [PubMed] [Google Scholar]
- 28.Hingorani M. M., Patel S. S. (1993) Biochemistry 32, 12478–12487 [DOI] [PubMed] [Google Scholar]
- 29.Holmlund T., Farge G., Pande V., Korhonen J., Nilsson L., Falkenberg M. (2009) Biochim. Biophys. Acta 1792, 132–139 [DOI] [PubMed] [Google Scholar]
- 30.Matson S. W., Kaiser-Rogers K. A. (1990) Annu. Rev. Biochem. 59, 289–329 [DOI] [PubMed] [Google Scholar]
- 31.Ziebarth T. D., Farr C. L., Kaguni L. S. (2007) J. Mol. Biol. 367, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilyina T. V., Gorbalenya A. E., Koonin E. V. (1992) J. Mol. Evol. 34, 351–357 [DOI] [PubMed] [Google Scholar]
- 33.Ziebarth T. D., Gonzalez-Soltero R., Makowska-Grzyska M. M., Núñez-Ramírez R., Carazo J. M., Kaguni L. S. (2010) J. Biol. Chem. 285, 14639–14647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel S. S., Picha K. M. (2000) Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 35.Crampton D. J., Ohi M., Qimron U., Walz T., Richardson C. C. (2006) J. Mol. Biol. 360, 667–677 [DOI] [PubMed] [Google Scholar]
- 36.Satapathy A. K., Kochaniak A. B., Mukherjee S., Crampton D. J., van Oijen A., Richardson C. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6782–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bujalowski W., Klonowska M. M., Jezewska M. J. (1994) J. Biol. Chem. 269, 31350–31358 [PubMed] [Google Scholar]
- 38.Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 39.Toth E. A., Li Y., Sawaya M. R., Cheng Y., Ellenberger T. (2003) Mol. Cell 12, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 40.Guo S., Tabor S., Richardson C. C. (1999) J. Biol. Chem. 274, 30303–30309 [DOI] [PubMed] [Google Scholar]
- 41.Donmez I., Patel S. S. (2006) Nucleic Acids Res. 34, 4216–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsushima Y., Farr C. L., Fan L., Kaguni L. S. (2008) J. Biol. Chem. 283, 23964–23971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hakonen A. H., Goffart S., Marjavaara S., Paetau A., Cooper H., Mattila K., Lampinen M., Sajantila A., Lönnqvist T., Spelbrink J. N., Suomalainen A. (2008) Hum. Mol. Genet. 17, 3822–3835 [DOI] [PubMed] [Google Scholar]
- 44.Lönnqvist T., Paetau A., Valanne L., Pihko H. (2009) Brain 132, 1553–1562 [DOI] [PubMed] [Google Scholar]
- 45.Sarzi E., Goffart S., Serre V., Chrétien D., Slama A., Munnich A., Spelbrink J. N., Rötig A. (2007) Ann. Neurol. 62, 579–587 [DOI] [PubMed] [Google Scholar]
- 46.Nikali K., Suomalainen A., Saharinen J., Kuokkanen M., Spelbrink J. N., Lönnqvist T., Peltonen L. (2005) Hum. Mol. Genet. 14, 2981–2990 [DOI] [PubMed] [Google Scholar]
- 47.Hakonen A. H., Isohanni P., Paetau A., Herva R., Suomalainen A., Lönnqvist T. (2007) Brain 130, 3032–3040 [DOI] [PubMed] [Google Scholar]
- 48.Gajewski C. D., Yang L., Schon E. A., Manfredi G. (2003) Mol. Biol. Cell 14, 3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy H. J., Pouli A. E., Ainscow E. K., Jouaville L. S., Rizzuto R., Rutter G. A. (1999) J. Biol. Chem. 274, 13281–13291 [DOI] [PubMed] [Google Scholar]
- 50.Kaukonen J., Juselius J. K., Tiranti V., Kyttälä A., Zeviani M., Comi G. P., Keränen S., Peltonen L., Suomalainen A. (2000) Science 289, 782–785 [DOI] [PubMed] [Google Scholar]
- 51.Agostino A., Valletta L., Chinnery P. F., Ferrari G., Carrara F., Taylor R. W., Schaefer A. M., Turnbull D. M., Tiranti V., Zeviani M. (2003) Neurology 60, 1354–1356 [DOI] [PubMed] [Google Scholar]
- 52.Virgilio R., Ronchi D., Hadjigeorgiou G. M., Bordoni A., Saladino F., Moggio M., Adobbati L., Kafetsouli D., Tsironi E., Previtali S., Papadimitriou A., Bresolin N., Comi G. P. (2008) J. Neurol. 255, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 53.Hudson G., Deschauer M., Busse K., Zierz S., Chinnery P. F. (2005) Neurology 64, 371–373 [DOI] [PubMed] [Google Scholar]
- 54.Deschauer M., Kiefer R., Blakely E. L., He L., Zierz S., Turnbull D. M., Taylor R. W. (2003) Neuromuscul. Disord. 13, 568–572 [DOI] [PubMed] [Google Scholar]
- 55.Van Goethem G., Löfgren A., Dermaut B., Ceuterick C., Martin J. J., Van Broeckhoven C. (2003) Hum. Mutat. 22, 175–176 [DOI] [PubMed] [Google Scholar]
- 56.Vandenberghe W., Van Laere K., Debruyne F., Van Broeckhoven C., Van Goethem G. (2009) Mov. Disord. 24, 308–309 [DOI] [PubMed] [Google Scholar]
- 57.Lewis S., Hutchison W., Thyagarajan D., Dahl H. H. (2002) J. Neurol. Sci. 201, 39–44 [DOI] [PubMed] [Google Scholar]
- 58.Rivera H., Blázquez A., Carretero J., Alvarez-Cermeño J. C., Campos Y., Cabello A., Gonzalez-Vioque E., Borstein B., Garesse R., Arenas J., Martín M. A. (2007) Neuromuscul. Disord. 17, 677–680 [DOI] [PubMed] [Google Scholar]
- 59.Naïmi M., Bannwarth S., Procaccio V., Pouget J., Desnuelle C., Pellissier J. F., Rötig A., Munnich A., Calvas P., Richelme C., Jonveaux P., Castelnovo G., Simon M., Simon M., Clanet M., Wallace D., Paquis-Flucklinger V. (2006) Eur. J. Hum. Genet. 14, 917–922 [DOI] [PubMed] [Google Scholar]
- 60.Baloh R. H., Salavaggione E., Milbrandt J., Pestronk A. (2007) Arch. Neurol. 64, 998–1000 [DOI] [PubMed] [Google Scholar]
- 61.Houshmand M., Panahi M. S., Hosseini B. N., Dorraj G. H., Tabassi A. R. (2006) Neurol. India 54, 182–185 [PubMed] [Google Scholar]
- 62.Kiechl S., Horváth R., Luoma P., Kiechl-Kohlendorfer U., Wallacher-Scholz B., Stucka R., Thaler C., Wanschitz J., Suomalainen A., Jaksch M., Willeit J. (2004) J. Neurol. Neurosurg. Psychiatry 75, 1125–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]