Abstract
The c-type cytochromes are metalloproteins with a heme molecule covalently linked to the sulfhydryls of a CXXCH heme-binding site. In plastids, at least six assembly factors are required for heme attachment to the apo-forms of cytochrome f and cytochrome c6 in the thylakoid lumen. CCS5, controlling plastid cytochrome c assembly, was identified through insertional mutagenesis in the unicellular green alga Chlamydomonas reinhardtii. The complementing gene encodes a protein with similarity to Arabidopsis thaliana HCF164, which is a thylakoid membrane-anchored protein with a lumen-facing thioredoxin-like domain. HCF164 is required for cytochrome b6f biogenesis, but its activity and site of action in the assembly process has so far remained undeciphered. We show that CCS5 is a component of a trans-thylakoid redox pathway and operates by reducing the CXXCH heme-binding site of apocytochrome c prior to the heme ligation reaction. The proposal is based on the following findings: 1) the ccs5 mutant is rescued by exogenous thiols; 2) CCS5 interacts with apocytochrome f and c6 in a yeast two-hybrid assay; and 3) recombinant CCS5 is able to reduce a disulfide in the CXXCH heme-binding site of apocytochrome f.
Keywords: Chloroplast, Cytochromes, Disulfide, Heme, Photosynthesis, Reductase, Thiol
Introduction
The c-type cytochromes are functionally versatile hemoproteins housed in energy-transducing membranes of bacteria, mitochondria, and plastids where they participate in electron transfer reactions (1). Conversion of apo- to holocytochrome c requires the covalent and stereospecific attachment of the heme cofactor to a CXXCH motif on the apoprotein, a process that was discovered to take place via at least three distinct assembly mechanisms, the so-called systems I–III (2–7). System IV is a maturation pathway required for the covalent attachment of heme Ci to cytochrome b6 in the b6f complex of cyanobacteria and photosynthetic eukaryotes (8–10). This pathway is not a cytochrome c maturation system because cytochrome b6 is very distinct from a typical cytochrome c. The definition of systems I–III is based on the occurrence of prototypical assembly components that are the hallmark of each maturation pathway (2, 3, 5).
In all three systems (I–III), covalent attachment of the heme moiety occurs on the positive side of energy-transducing membranes (i.e. bacterial periplasm, mitochondrial intermembrane space, and thylakoid lumen) and requires minimally the following: 1) synthesis and transport across at least one biological membrane of apocytochromes c and heme, 2) reduction and maintenance of the heme ferrous iron and the CXXCH sulfhydryls prior to 3) formation of thioether bonds between the heme and apocytochromes c (2–5).
System II, the assembly pathway investigated in this study, was originally discovered in the green alga Chlamydomonas reinhardtii through genetic studies of ccs mutants (ccs for cytochrome c synthesis) that display a specific defect in membrane-bound cytochrome f and soluble cytochrome c6, two thylakoid lumen resident c-type cytochromes functioning in photosynthesis (11). All the ccs mutants exhibit a photosynthetic deficiency and fail to accumulate holoforms of both cytochrome f and c6 because the heme attachment reaction, which is the only common step in the biogenesis of both proteins, is compromised (11–13). At least six loci, plastid ccsA and nuclear CCS1 to CCS5, were uncovered through genetic analysis, and it is likely that saturating mutagenesis screens will reveal additional loci controlling the assembly process (11, 13–15). CcsA and CCS1, the defining components of this pathway, are polytopic membrane proteins that were postulated to function together as a heme relay unit from stroma to lumen (16–18). Recently, the Helicobacter pylori CcsBA protein, a naturally occurring fusion of the CCS1 and CcsA orthologs, was proposed to work as a heme channel that also exhibits the heme ligation activity (19, 20).
The involvement of redox chemistry in system II was first established via genetics in bacterial models. Components of this pathway include a thiol/disulfide membrane transporter of the CcdA/DsbD family (21–23) whose implication in cytochrome c maturation was first demonstrated in the bacteria Bacillus subtilis and Bordetella pertussis (24–26). Another redox factor of the system II thiol reduction pathway is the membrane-anchored periplasm-facing thioredoxin-like ResA/CcsX required for the assembly of multiple bacterial cytochromes c (27–29). The placement of CcdA/DsbD and ResA/CcsX in a thiol reduction pathway is inferred from the finding that ccda/dsbD and resA/ccsx mutants can be rescued by exogenous thiols (24–27, 29, 30). The current thinking is that exogenous thiols substitute for the reducing activity of the redox factors and maintain the apocytochrome c CXXCH motif in a reduced form (i.e. sulfhydryls). The proposed model is that electrons are transferred from NADPH in the cytoplasm to the apocytochrome c CXXCH target in the periplasmic space through disulfide-dithiol exchange reactions involving sequentially a cytosolic NADPH-dependent thioredoxin reductase, a thioredoxin, CcdA/DsbD, and ResA/CcsX (31, 32). This view is supported by the finding that a recombinant form of ResA is active in thiol-disulfide exchange in an in vitro assay (29).
By analogy to the bacterial membrane, provision of reducing equivalents to the thylakoid lumen is also expected in the context of plastid cytochrome c assembly. The occurrence of CcdA orthologs encoded in the plastid genomes of some photosynthetic eukaryotes suggests that plastids might have inherited a bacteria-like thiol reduction pathway (15, 33). In the plant Arabidopsis thaliana, the CCDA protein localizes at the thylakoid membrane, and loss-of-function ccdA mutations result in a photosynthetic deficient phenotype because of a defect in cytochrome b6f complex assembly (15, 34). A possible redox partner for CCDA is the thylakoid membrane-bound lumen-facing HCF164 thioredoxin-like protein. This is suggested by the fact that loss of HCF164 function in Arabidopsis also leads to a cytochrome b6f deficiency (28). If experimental investigations in Arabidopsis support the implication of the thiol-disulfide transporter CCDA and the thioredoxin-like HCF164 in the biogenesis of the cytochrome b6f complex, they do not elucidate whether heme attachment to apocytochrome f is compromised by ccda or hcf164 mutations because the heme attachment reaction has not been directly monitored in the vascular plant system (15, 28). However, in Chlamydomonas, cytochrome f and cytochrome c6 are synthesized in different compartments (plastid versus cytosol), and the only common step in biogenesis is the heme attachment. Therefore, ccs mutants that are deficient in both proteins are most likely defective at heme attachment. Indeed, pulse-chase analyses in the ccs mutants revealed the synthesis of apo-form but not holoform of cytochrome f and c6, indicative of a block in conversion (11–13).
In this study, we report the identification of another CCS locus, CCS5, controlling plastid c-type cytochrome assembly in C. reinhardtii. The CCS5 gene encodes the ortholog of Arabidopsis thioredoxin-like HCF164. The placement of CCS5 in the thiol reduction pathway for plastid cytochrome c assembly is supported by the following findings: (a) the ccs5 mutant is rescued by exogenous thiols; (b) the soluble domain of CCS5 interacts with apo-forms of cytochrome f and c6 in a yeast two-hybrid assay; and (c) a recombinant form of CCS5 is able to reduce a disulfide bond in apocytochrome f CXXCH motif. We discuss the operation of thiol-disulfide chemistry on the lumenal side of the thylakoid membrane.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Strains were grown at 22–25 °C in TAP liquid or solid medium (35) with or without copper supplementation under dim light (25 μmol/m2/s) for ccs strains or under standard illumination for wild type strains (125–500 μmol/m2/s) as described in Ref. 11.
Insertional Mutagenesis and Identification of the ccs5 Mutant
The arg7 cw15A mt− strain was used as a recipient strain for insertional mutagenesis. 2 μg of pARG7.8Φ3 DNA (36) was used to perform glass bead transformation (37), and 1.6 × 104 arginine prototrophic transformants were selected under dim light. Insertional mutants were screened on a fluorescence video imaging system (38) at the University of Geneva. Candidate mutants displaying a rapid fluorescence induction that failed to relax, a characteristic of a block in electron transfer on the reducing side of PSII, were further analyzed by immunoblotting for holocytochrome f accumulation.
Molecular Genetic Analysis of the T78 Strain
The T78 mutant was crossed to a wild type 137 derivative strain to generate T78.3+. Tetrad analysis of T78.3+ by wild type CC621 (NO−) crosses showed that the CCS phenotype segregated as a monogenic trait along with the Φ flag present in the integrated ARG7Φ insertional marker (36). The Φ flag was detected by diagnostic PCR using the phi1 (5′-GTCAGATATGGACCTTGCT-3′) and phi2 (5′-CTTCTGCGTCATGGAAGCG-3′) primers and genomic DNAs as templates. Spores T78.5a− and T78.5d+ come from the same tetrad in the cross of T78.3+ to wild type. T78.15b− originated from an incomplete tetrad and was retained for its ability to mate well. For the DTT-dependent rescue, T78.15b− was crossed to a cw− wild type strain, and over 50 ccs5 spores were tested for DTT rescue. For our analysis, we retained PH9–25 that showed the best DTT-dependent rescue of the photosynthetic growth.
Complementation analyses for test of allelism were done as described previously (35). Fluorescence induction kinetics were analyzed on the lawn of meiotic zygotes resulting from the cross of ccs5 mutant strains (T78.5d+ and T78.5a−) to strains carrying the ccs1-ac206 (ac206.11−), ccs1-4 (CF951.15+), ccs2-1 (CF139.3−), ccs2-3 (CF521.1a−), ccs2-4 (CF779.B1−), ccs2-5 (CF854.141−), ccs3-F18 (F18.a2−), and ccs4-F2D8 (F2D8–9−) alleles. The in vivo chlorophyll fluorescence induction kinetics of dark-adapted colonies or diploid zygotes were recorded with a homemade fluorescence imaging system (Dr. D. Kramer, Washington State University, Pullman) or with the FluorCam 700 MF (Photon Systems Instruments, Ltd.). The colonies were illuminated with the homemade system for 2 s with 30 μmol/m2/s of red light (640 nm) from light-emitting diodes (HLMP C116, Hewlett Packard), and the emitted fluorescence was captured immediately by a CCD camera (Cohu, 2122–1000) in conjunction with a PIXCI-SV4 imaging board and XCIP software (EPIX Inc., Buffalo Grove, IL). For the FluorCam, illumination was 3 s under actinic light of 60 μmol/m2/s. Emitted fluorescence was captured by the camera whose sensitivity and shutter were set up at 80% and 1:500, respectively.
Molecular Complementation of the ccs5 Mutant Strain
Electroporation was used as a transformation procedure (39), and the T78.15b− strain was chosen as a recipient strain. For transformation experiments, an aphVIII PCR fragment generated with primers aph8-R (5′-CTGGGTACCCGCTTCAAATA-3′) and aph8-F2 (5′-TCAGGCAGACGGGCAGGTG-3′) and pSL18 (40) as a template was used. The aphVIII PCR fragment was used in a co-transformation experiment with a CCS5 PCR fragment encompassing the entire ORF and 150 bp upstream of the ATG and 180 bp downstream of the stop codon. The CCS5 PCR fragment was amplified from a wild type genomic DNA using primers HCF164-18 (5′-CGAAACACCAAGGTTCTGAGAGC-3′) and HCF164-28 (5′-GCTTGTCAGATGTGCCCAAGC-3′).
RNA Preparation and RT-PCR Experiments
RNAs were extracted as described previously (41) and reverse-transcribed into cDNAs with MMLV reverse-transcriptase or the 5′-rapid amplification of cDNA ends kit from Invitrogen. An additional exon at the 5′ end of the CCS5 gene was identified via PCR using primers HCF164-6 (5′-AGTTGGCGTAGAACTCCACCAG-3′) and HCF164-24 (5′-GCAGACGAGTGTGTCAACCTATC-3′) and cDNAs as a template. The CCS5 cDNA was synthesized with HCF164-30 (5′-GCCAATTCCCGCATCGCTTCC-3′) as a CCS5 mRNA-specific primer and then used as a template in a semi-nested PCR. Primers HCF164-36 (5′-GACTCCAGTGTGGCCAGCGTGG-3′) and HCF164-32 (5′-GTTTCGCCAAGTTACATATTG-3′) were chosen in the first PCR, and primers HCF164-37 (5′-CCTGGGAAGGTAGCTGCTCAGC-3′) and HCF164-32 were used in the second PCR. The start codon was identified by sequencing of the PCR product. A stop codon in the same reading frame was found 41 nucleotides upstream. The CCS5 sequence has been deposited in GenBankTM under accession number ADC32800.
Protein Preparation and Analysis
Supernatant and pellet fractions were obtained by freeze-thaw fractionation. Fractions were electrophoretically separated, and plastid c-type cytochromes were revealed by immunodetection or a heme-staining procedure (11). Polyclonal antisera raised against C. reinhardtii cytochrome c6, cytochrome f GST-fusion protein, CCS5 (this work), OEE1, CF1, and plastocyanin were used for immunodetection by alkaline phosphatase-conjugated secondary antibodies. Pulse-chase experiments to monitor the synthesis of apocytochrome f were performed as described previously (11, 13, 42).
Expression and Purification of the Soluble Form of CCS5
The DNA sequence encoding the soluble domain of Chlamydomonas CCS5 was codon-optimized for expression in Escherichia coli using the DNA2.0 Gene Designer software (43). The optimized sequence was synthesized (Mr Gene, Germany) and cloned in pSK plasmid. The resulting plasmid (pMK-RQ) was used as a template in a PCR with NdeI and XhoI engineered oligonucleotides as primers (HCF164 O/F Nde1, 5′-GAAGGAGATATACATATGGGTGCTCCAACTCTGGCT-3′, and HCF164 O/R XhoI, 5′-GTGGTGGTGGTGCTCGAGTGCGTGGTCACGTGGCATC-3′). The PCR product was cloned into the NdeI/XhoI sites of the hexahistidinyl tag vector pET24b (Novagen) using the In-FusionTM cloning kit (Clontech), resulting in the pET24b/HCF164opt plasmid. For expression of the recombinant His6-tagged protein, 1 liter of LB broth (with 30 μg/ml kanamycin) of E. coli strain BL21(DE3) (Novagen) carrying pET24b/HCF164opt was grown from a 20-ml LB broth (with 30 μg/ml kanamycin) overnight starter culture. To induce the recombinant protein, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm at A600 = 0.4, and the culture was further grown for 3 h at 37 °C. Cells were then harvested by centrifugation at 4,000 × g for 20 min at 4 °C, and the pellet was stored at −20 °C. Batch purification of the His6-tagged protein was performed under native condition using the nickel-nitrilotriacetic acid resin (Qiagen). Recombinant CCS5 was used as an antigen to raise a polyclonal antibody (Covance). Immunoaffinity purification of the anti-CCS5 polyclonal was performed using Affi-Gel 10 and 15 according to the instructions of the manufacturer (Bio-Rad).
Disulfide Reductase Assay
The recombinant soluble domain of wild type CCS5 was assayed for disulfide reductase activity by using insulin properties. As insulin reduction proceeds, a precipitate is formed from the free B chain, which can be monitored at 650 nm for the change in turbidity (28, 44). The reaction mixture was prepared in a stirred and thermostated (25 °C) cuvette with a final volume of 1 ml containing 100 mm sodium phosphate (pH 7.0), 2 mm EDTA, 1 mg/ml bovine insulin. The reaction was initiated by adding 330 μm DTT. The nonenzymatic reduction of insulin by DTT only and the reduction of insulin by 10 μm recombinant CCS5 without DTT served as negative controls. As a positive control, 2 μm thioredoxin from Spirulina sp. (Sigma) was used.
Yeast Two-hybrid Experiments
The soluble domain of Chlamydomonas CCS5 (Gly206 to Ala254, see Fig. 4A) was used as bait, and the corresponding sequence was cloned via In-FusionTM technique (Clontech) as a PCR fragment at the NdeI/SalI sites of the pGBKT7 vector. The sequence corresponding to the soluble domain of wild type CCS5 was PCR-amplified using PHCF164BD-F (5′-GATCTCAGAGGAGGACCTGCAAGGTGCTCCAACTCTGGCTACTC-3′) and PHCF164BD-R (5′-TGCGGCCGCTGCAGGTCGAGCTGCGTGGTCACGTGGCATCG-3′) as primers and pET24b/HCF164opt as a template. Plasmids expressing the mutant forms of CCS5 (WCXXS and WSXXS) were constructed via QuickChange II site-directed mutagenesis kit (Stratagene). Chlamydomonas apocytochrome f without the membrane anchor (Tyr32 to Arg281) was used as prey, and the corresponding sequence was cloned via In-FusionTM as a PCR fragment at the NdeI/XhoI of the pGADT7 vector. The sequence corresponding to apocytochrome f was PCR-amplified using PcytfAD-F (5′-CGACGTACCAGATTACGCTCAATACCCTGTATTTGCACAAC-3′) and PcytfAD-R (5′-CGATTCATCTGCAGCTCGAGCACGAGCAGGGTTTTGTAATAC-3′) as primers and Chlamydomonas wild type genomic DNA as a template. Chlamydomonas apocytochrome c6 (Ala59 to stop) was used as prey, and the corresponding sequence was cloned via In-FusionTM as a PCR fragment at the NdeI/XhoI of the pGADT7 vector. The sequence corresponding to apocytochrome c6 was PCR-amplified using Pcytc6optAD-forward (5′-CGACGTACCAGATTACGCTCAAGCGGATCTGGCCCTGGGCGCACAGG-3′) and Pcytc6optAD-reverse (5′-CGATTCATCTGCAGCTCGAGCGTATTTCCACGCAGCATCGGTAGC-3′) as primers and the pMA/cytc6 plasmid carrying the codon-optimized cytochrome c6 encoding DNA as a template. The yeast strain PJ69-4A was used as a reporter (45), and β-galactosidase activity was measured as described previously (46).
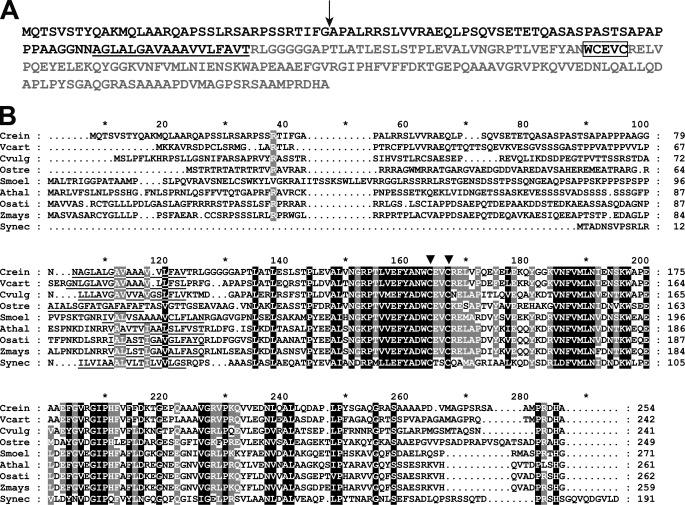
FIGURE 4.
CCS5 is a thioredoxin-like protein conserved in the green lineage. A, amino acid sequence of Chlamydomonas CCS5. The amino acid sequence of Chlamydomonas CCS5 protein was deduced from assembly of a full-length cDNA. The cleavage site of the plastid targeting sequence predicted by PredSL (65) is indicated by a black arrow, and the putative transmembrane domain is underlined. The hydrophilic C-terminal domain predicted to be facing the lumenal side of the thylakoid membrane is highlighted in light gray. The thioredoxin motif with active cysteines is boxed. B, alignment of CCS5/HCF164-like proteins. Sequences of CCS5/HCF164 from C. reinhardtii (Crein), Volvox carteri (Vcart, 80757), Chlorella vulgaris (Cvulg, 79381), Ostreococcus RCC809 (Ostre, 68525), Selaginella moellendorffii (Smoel, 159329), A. thaliana (Athal, CAC19858), Oriza sativa (Osati, NP_001051387), Zea mays (Zmays, NP_001152226), and TxlA from Synechococcus sp. PCC 7942 (Synec, U05044) were aligned using the ClustalW algorithm (Blosum62 scoring matrix) in Bioedit. The alignment was edited using the GeneDoc multiple alignment editor. Amino acids strictly conserved in all sequences are shaded in black and those conserved in the majority of the sequences (7 of 9) are shaded gray. Asterisk marks residues 10, 30, 50, 70, etc. The putative membrane anchor is underlined. Two downward arrowheads indicate the cysteines in the WCXXC motif.
In Vitro Redox Assay
The DNA sequence encoding the soluble domain of Chlamydomonas apocytochrome f without the membrane anchor was codon-optimized for expression in E. coli using the DNA2.0 Gene Designer software (43). The optimized sequence was synthesized (Mr Gene, Germany). The resulting plasmid (pMK-CYTF) was used as a template in a PCR with NdeI- and XhoI-engineered oligonucleotides as primers (Solcytfopt-forward, 5′-CTTTAAGAAGGAGATATACATATGTACCCGGTTTTCGCTCAGCAGAACTA-3′, and Solcytfopt-reverse, 5′-CGGATCTCAGTGGTGGTGGTGGTGGTGACGTGCCGGGTTCTGCAGGACGA-3′). The PCR product was cloned into the NdeI/XhoI sites of the hexahistidinyl tag vector pET24b (Novagen) using the In-FusionTM cloning kit (Clontech), resulting in the pET24b/CYTFsol plasmid. Purification of apocytochrome f was as described for soluble CCS5 except that isopropyl 1-thio-β-d-galactopyranoside concentration was 0.4 mm and induced cells were harvested at A600 = 0.6. Soluble CCS5 in 25 mm Tris-HCl (pH 7.5) was reduced by 200 μm DTT during 1 h on ice. DTT was eliminated by buffer exchange using the Amicon Centriprep system (Ultracel-10 membrane, Millipore). Air-oxidized apocytochrome f (8 μm) in 25 mm Tris-HCl (pH 7.5) was incubated for 60 min at 25 °C in the presence or absence of reduced soluble CCS5 (16 μm). After incubation, proteins were precipitated with trichloroacetic acid (final 5%), washed with ice-cold acetone, and then dissolved in buffer containing 50 mm Tris-HCl (pH 6.8), 2% SDS, 10 mm 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS).6 After 1 h 30 min of incubation, reduced (AMS derivative) and oxidized forms of cytochrome f and CCS5 were separated by 15% nonreducing SDS-PAGE and visualized by Coomassie staining.
RESULTS
CCS5, a New Locus Controlling Plastid Cytochrome c Assembly
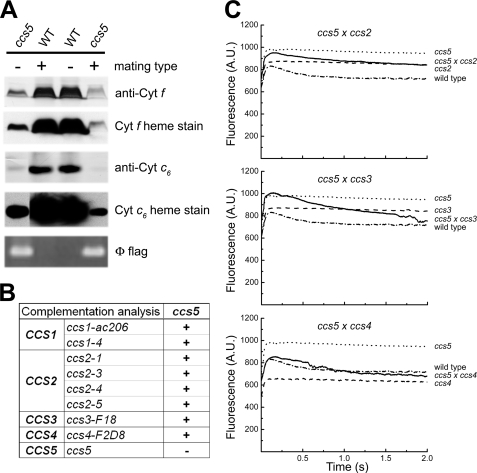
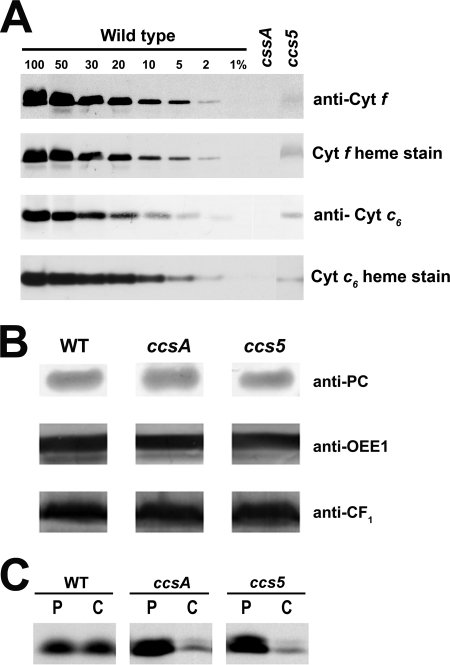
The fact that the Chlamydomonas CCS3 and CCS4 loci are only defined by a single allele suggests that mutagenesis screens for plastid cytochrome c-deficient mutants are not saturated (13). To identify additional CCS loci controlling plastid c-type cytochrome assembly, an insertional mutagenesis with the ARG7Φ marker (36) was performed and led to the isolation of 80 photosynthesis-minus mutants that displayed a fluorescence kinetic characteristic of a deficiency in the cytochrome b6f complex (see Figs. 2C and 5A for example). This fluorescence signature is similar to the high chlorophyll fluorescence phenotype (hcf) described for Arabidopsis mutants with defects in the photosynthetic light reactions (47). Among the 80 insertional mutants, one strain (T78) exhibited the features of a c-type cytochrome-deficient mutant (ccs). T78 accumulated low levels of holoforms of cytochrome f (<5%) and c6 (<5%) similarly to a ccsA mutant (Fig. 1A), but it was unaffected for the accumulation of lumen resident proteins such as plastocyanin and OEE1 or other photosynthesis proteins such as ATP synthase at the thylakoid membrane (Fig. 1B). This localized the defect to cytochrome assembly and ruled out a general block in the maturation of lumenal proteins. RNA blot analyses confirmed that the expression of the petA and CYC6 genes encoding apocytochrome f and apocytochrome c6, respectively, is unaltered in the T78 strain (data not shown). Pulse-chase experiments showed that apocytochrome f is synthesized in T78 but rapidly degraded, comparably to the ccsA mutant (Fig. 1C) and all of the ccs strains (11–13). This suggests that T78 is blocked at the step of heme attachment in the plastid lumen. Tetrad analysis of T78 crossed to a wild type strain revealed that the cytochrome f and c6 deficiency co-segregated as a single Mendelian trait (Fig. 2A). Analysis of a total of 25 mutant progeny showed a tight linkage between the cytochrome c deficiency and the Φ molecular tag in the ARG7 Φ insertional marker (p > 0.05) (Fig. 2A). Based on the genetic analyses, we concluded that the mutation in T78 segregated as a single CCS locus that is tagged with the ARG7Φ marker. To determine whether the ccs mutation in the T78 strain defines an allele of an existing or new CCS locus, we undertook an allelism test by performing complementation analysis. Complementation can be assessed by testing the fluorescence rise and decay kinetics of diploid zygotes (48, 49). We reasoned that if the ccs mutation in T78 defines a new CCS locus, we should observe complementation of the CCS phenotype in zygotes resulting from the cross of strains carrying the ccs mutation isolated in T78 by the genetically defined ccs mutants. All zygotes resulting from the cross of the ccs mutation to strains carrying ccs1-ac206, ccs1-4, ccs2-1, ccs2-3, ccs2-4, ccs2-5, ccs3-F18, and ccs4-F2D8 alleles exhibited fluorescence curves with a decay phase indicating that the b6f function was restored (Fig. 2, B and C). These results established that the affected CCS locus in T78 is distinct from CCS1, CCS2, CCS3, and CCS4. Moreover, based on the fact that the CCS1 locus was found to be intact by Southern blot analysis in the T78 strain, we confirmed that this strain does not carry a molecular lesion in the CCS1 gene (data not shown). Therefore, we concluded that the T78 mutant defines yet a fifth nuclear CCS locus, and the ccs allele has been named ccs5-1::ARG7.
FIGURE 2.
T78 mutant defines a new CCS locus (CCS5). A, monogenic Mendelian segregation of the ccs mutation in T78. Two spores displaying the photosynthetic deficient phenotype (ccs5) (left, T78.5a− and right, T78.5d+) and two wild type spores (WT) from the same tetrad were used to analyze accumulation of plastid cytochromes (cyt) c by heme stain and immunoblotting. The tetrad originates from a cross of the T78 insertional mutant to a wild type strain (CC621). The mating type mt+ or mt− of each spore is indicated. Segregation of the Φ flag in the progeny was followed by PCR amplification from total genomic DNA samples of the molecular tag using diagnostic primers. B, complementation of ccs5 mutant with strains carrying alleles of the CCS1, CCS2, CCS3, and CCS4 loci. Diploid zygotes were obtained by crossing ccs5 mutant strains by strains carrying the ccs1-ac206, ccs1-4, ccs2-1, ccs2-3, ccs2-4, ccs2-5, ccs3-F18, and ccs4-F2D8. “+” indicate a positive complementation based on the presence of the decay phase in fluorescence induction experiments. “−” indicates a negative complementation. C, fluorescence kinetics indicate restoration of cytochrome b6f in the diploid zygotes. Fluorescence induction and decay kinetics of the ccs5xccs2-5, ccs5xccs3-F18, and ccs5xccs4-F2D8 diploid zygotes, respective parental strains (see B), and wild type strain (arg7cw15A). The fluorescence transients were measured using a homemade device. The fluorescence is in arbitrary units (A.U.) and recorded over a 2-s illumination period. The rise and plateau curve for the ccs mutants is the signature of a specific block in electron transfer at the level of cytochrome b6f complex because of its impaired assembly in the absence of membrane-bound holocytochrome f. When the energy absorbed by the chlorophyll cannot be utilized because of a block in photosynthetic electron transfer through cytochrome b6f, an increase in the chlorophyll fluorescence is observed. Note the absence of the decay phase corresponding to reoxidation of the quinone pool (the primary electron acceptor of photosystem II) by the cytochrome b6f complex. In the complemented diploid zygote, the presence of the decay phase indicates restoration of the b6f complex function.
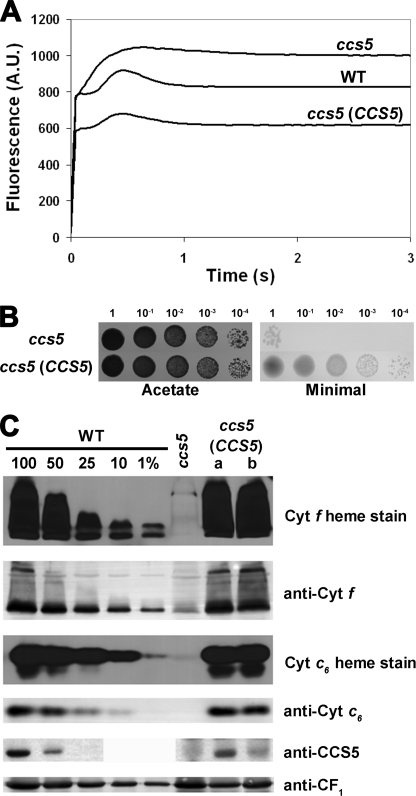
FIGURE 5.
The thioredoxin-like encoding gene complements the ccs5 mutant. For ABC, the ccs5 mutant (T78.15b−) carries the PmR cassette, and the CCS5 -complemented transformants result from the co-transformation of the PmR cassette and the CCS5 gene. A, fluorescence kinetics indicate restoration of cytochrome b6f in the complemented ccs5 mutant. Fluorescence induction and decay kinetics of the ccs5, wild type, and ccs5 complemented with the wild type CCS5 gene. One representative transformant (lane a) is shown. Fluorescence transients were measured on colonies grown for 2 days on solid TAP medium after a short dark adaptation using FluorCam 700 from Photon Systems Instruments. The fluorescence is in arbitrary units (A.U.) and recorded over a 3-s illumination period. B, the CCS5 gene restores phototrophic growth to the ccs5 mutant. 10-Fold dilution series of ccs5 and a representative ccs5 transformant (lane a) carrying the CCS5 gene have been plated on acetate (heterotrophic conditions, 20 μmol/m2/s of light) and minimal medium (phototrophic conditions, 500 μmol/m2/s of light) and incubated for 1 week at 25 °C. C, plastid c-type cytochromes (cyt) accumulation is restored in the CCS5-complemented ccs5 strain. Wild type (CC124), ccs5, and two independent ccs5 transformants (lanes a and b) carrying the CCS5 gene were analyzed for cytochrome f and cytochrome c6 accumulation via heme stain and immunoblot analyses. Samples corresponding to 18 μg of chlorophyll were separated in 12% SDS-acrylamide gel to detect cytochrome f, CCS5, and CF1 of the ATPase that serves as a loading control. Samples corresponding to 16 μg of chlorophyll were separated in 15% native acrylamide gel to detect cytochrome c6. For an estimation of the protein abundance in the ccs5 complemented strain, dilutions of the wild type sample were loaded on the gel. Gels were transferred to polyvinylidene difluoride membranes before heme staining by chemiluminescence and immunodetection with antisera against Chlamydomonas cytochrome f, cytochrome c6, CCS5, or CF1.
FIGURE 1.
T78 displays a dual deficiency in holocytochrome f and c6. A and B, accumulation of plastid c-type cytochromes in the T78 mutant. Soluble and membrane fractions of copper-deficient cultures were analyzed for the accumulation of cytochrome f (anti-cyt f and cyt f heme stain) and cytochrome c6 (anti-cyt c6 and cyt c6 heme stain), respectively. All the samples were found to be copper-deficient based on the absence or very low abundance of immunoreactive holoplastocyanin (data not shown). Samples of wild type strains (WT), T78 mutant (ccs5), and B6 (ccsA) corresponding to 5 μg of chlorophyll were separated in SDS-containing acrylamide (12%) gels or native acrylamide gels (15%) to detect cytochrome f and cytochrome c6, respectively. For an estimation of the cytochrome abundance, dilutions of the wild type sample (arg7cw15A for cytochrome f and CC125 for cytochrome c6) were loaded on the gel. Gels were then transferred to polyvinylidene difluoride membranes following electrophoresis before heme staining by chemiluminescence and immunodecoration with antisera against Chlamydomonas cytochrome f and cytochrome c6 in A and with antisera against plastocyanin, OEE1, ATPase CF1 in B. C, in vivo synthesis and degradation of apocytochrome f in the ccs5 mutant. The synthesis of cytochrome f was assessed by immunoprecipitation of anti-cytochrome f-reactive polypeptides from solubilized acetone extracts of wild type (WT), B6 (ccsA), and T78 (ccs5) cells labeled for 10 min with Na235SO4 (P indicates pulse). The labeled cells were sampled 40 min later after further incubation in the presence of unlabeled sulfate and chloramphenicol to assess the fate of the newly synthesized protein (C indicates chase). Immunoprecipitates were resolved by denaturing gel electrophoresis followed by fluorography.
CCS5 Gene Product Functions in Redox Chemistry
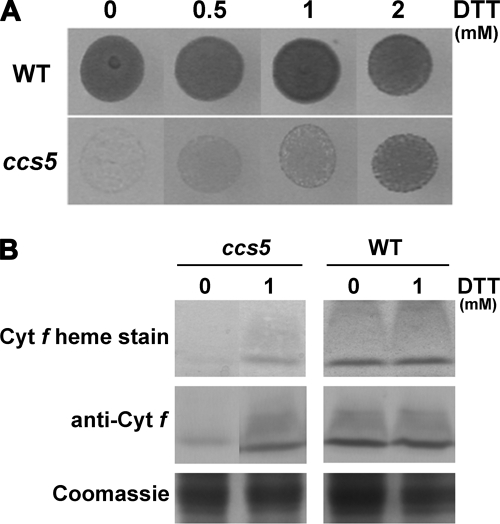
In bacteria, loss of cytochrome c assembly redox factors with a proposed reducing activity can be by-passed by provision of exogenously reduced thiols in the medium (24–27, 30). Therefore, as a preliminary functional assessment of the products of the genetically defined CCS loci, we tested the ability of exogenous thiols (2-mercaptoethane sulfonate, DTT, and cysteine) to rescue the phototrophic growth of all ccs strains. Among the different compounds tested, we found that exogenous thiols such as DTT and 2-mercaptoethane sulfonate could rescue the photosynthetic deficiency of the ccs5 mutant (Fig. 3A and data not shown). Photosynthetic competence in the chemically rescued colonies correlates with accumulation of holocytochrome f, which is strictly required for photosynthesis (Fig. 3B). Note that cytochrome c6 is only required for photosynthesis in copper-deficient conditions where it acts as a substitute for plastocyanin, a copper-containing protein. For experimental convenience, we usually monitor photosynthetic growth in copper-replete conditions where cytochrome f is the only c-type cytochrome required for photosynthesis. As expected, fluorescence rise and decay kinetics confirmed that DTT restores the function of cytochrome b6f (data not shown). The DTT-dependent rescue of the ccs5 mutant suggests that the gene product acts as a reductant in the cytochrome c assembly process.
FIGURE 3.
ccs5 mutant is rescued by exogenous thiols. A, DTT-dependent photosynthetic rescue of the ccs5 mutant. Equal numbers of wild type (CC124) and ccs5 mutant (PH9–25) cells were spotted on minimal medium supplemented with DTT as indicated and incubated at 25 °C at a light intensity of 500 μmol/m2/s for 12 days. B, DTT-dependent restoration of holocytochrome f assembly in the ccs5 mutant. In gel cytochrome (cyt) f heme staining (41) and anti-cytochrome f immunoblot analyses were performed on total protein extracts from the ccs5 mutant (PH9–25) and wild type (WT) strains. Cells were grown heterotrophically (acetate, low light) in the absence or presence of 1 mm DTT. Coomassie Blue staining is used to show equal loading. Note that cells grown heterotrophically showed the best rescue with 1 mm DTT.
CCS5 Encodes a Thioredoxin-like Protein
To identify the CCS5 gene, we searched the Chlamydomonas genome for candidate proteins based on the occurrence of the following: 1) a redox-active domain (e.g. CXXC), and 2) an algorithm-predicted targeting sequence to the plastid. Our approach was greatly facilitated by the detailed annotation of the Chlamydomonas genome (50). Several candidate proteins were retrieved, and we focused our attention on a thioredoxin-like protein because of its sequence similarity to HCF164, a lumen-located membrane-anchored thioredoxin-like protein involved in cytochrome b6f complex biogenesis in Arabidopsis (28, 51). Because the ccs5 mutant is an insertional mutant, we reasoned that the physical integrity of the gene that encodes the HCF164-like protein should be altered in the mutant strain if ccs5 is deficient for this protein. PCR-based scanning of the locus confirmed that up to 3 kb of the corresponding locus, including the entire coding sequence, is deleted in the ccs5 strain (data not shown). This alteration in the ccs5 mutant presumably occurred upon insertion of the ARG7 insertional marker, a common occurrence in Chlamydomonas (52). In addition, quantitative PCR experiments showed that the transcript is detected in wild type but not in the ccs5 strain (data not shown). We concluded that ccs5 is a null mutant of the gene encoding the HCF164-like protein. Because only a partial EST sequence was available in the data base, we sought to confirm the 5′ end of the CCS5 gene model and determine the initiation codon by performing RT-PCR experiments. A full-length cDNA was assembled and shown to encode a 254-amino acid protein with a predicted plastid targeting sequence (Fig. 4A). The CCS5 protein is 35% identical to Arabidopsis HCF164. From similarity to HCF164, CCS5 is predicted to be anchored to the membrane by a single hydrophobic stretch with the redox-active motif exposed to the thylakoid lumen (Fig. 4A). Orthologs of CCS5/HCF164 are found in the green lineage, and they all display a similar topology with an N-terminal membrane anchor and the redox-active domain in the C-terminal part of the protein (Fig. 4B).
To confirm that the ccs5 mutation affects the CCS5 gene, we performed complementation experiments. The ccs5 mutant strain was co-transformed with two PCR products corresponding to the aphVIII selectable marker conferring paromomycin resistance (PmR) and a 2.5-kb fragment containing the entire CCS5 gene. PmR transformants were selected and screened for restoration of photosynthesis on the basis of fluorescence. Out of the 211 PmR transformants that we screened, 23 showed wild type-like fluorescence, suggesting that b6f and hence cytochrome f assembly was restored (Fig. 5A). Consistently, the transformants displaying a wild type-like fluorescence rise and decay kinetics were able to grow photoautotrophically and accumulate wild type level of holocytochrome f and c6 (Fig. 5, B and C). We also showed that CCS5 protein accumulation was restored in the complemented transformants (Fig. 5C). However, the two independent transformants that we analyzed did not show wild type level of CCS5 protein despite the fact that cytochrome f and c6 accumulated to wild type level. The different level of accumulation of CCS5 might reflect the differential expressions of the transforming DNA at the integration site in the chromosome. This suggests that CCS5 is probably not limiting for the assembly process. As expected, all of the 68 PmR transformants that were only transformed with the aphVIII PCR product exhibited a b6f-minus fluorescence rise and decay kinetics (Fig. 5A).
CCS5 Displays Disulfide Reductase Activity
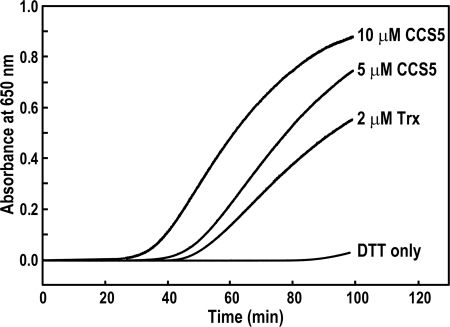
The DTT-dependent rescue of the ccs5 mutant and the fact that CCS5 displays a thioredoxin-like motif speaks to a reducing activity for this protein in the assembly process of holocytochrome c. One hypothesis is that CCS5 is required to reduce the cysteines in the CXXCH motif prior to the heme attachment reaction. In such a working model, CCS5 would act as a disulfide reductase. To demonstrate such an activity, we purified the soluble domain of CCS5 (Fig. 4A) as a recombinant protein and tested its disulfide reductase activity in the insulin reduction assay (28, 44, 51). This is a quantitative assay that measures the rate of insulin reduction as turbidity formation from the precipitation of the free insulin B chain released after reduction of a disulfide bond. As shown on Fig. 6, CCS5 is active in disulfide reduction, similarly to the stromal thioredoxin from the alga Spirulina sp. that is known to display disulfide reductase activity (44). We concluded that CCS5 exhibits disulfide reductase activity.
FIGURE 6.
Recombinant soluble CCS5 displays disulfide reductase activity. The disulfide reductase activity of recombinant CCS5 was measured by using the insulin reduction assay with 5 and 10 μm protein as described under “Experimental Procedures.” As a positive control, 2 μm thioredoxin (Trx) from Spirulina sp. were used. The nonenzymatic assay with DTT only served as a negative control. No reduction of insulin was observed with 10 μm recombinant CCS5 in the absence of DTT (data not shown).
CCS5 Interacts with Apocytochrome f and c6
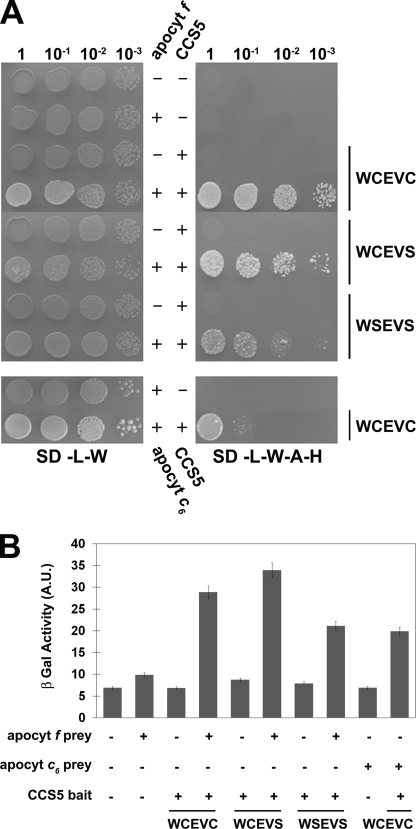
To provide evidence that plastid apo-forms of cytochrome c are the relevant targets of action of CCS5 in vivo, we aimed to demonstrate interaction between the thioredoxin-like domain of CCS5 and the domain of apocytochrome f and c6 carrying the CXXCH heme-binding site. We first chose to evidence such an interaction using apocytochrome f as a prey and CCS5 as a bait in a GAL4-based two-hybrid system. As shown in Fig. 7, CCS5 and apocytochrome f are able to interact based on the recovery of GAL4-dependent adenine/histidine prototrophies and β-galactosidase activities in the yeast reporter strain. Next, we addressed the importance of the cysteines residues in CCS5 for such an interaction. In thioredoxin proteins, the first cysteine residue of the WCXXC motif attacks the disulfide in a target and forms a mixed disulfide that is resolved by the second cysteine of the motif (53). Mutation of the resolving cysteine or both cysteines in the WCXXCH did not abolish interaction between the thioredoxin-like domain and apocytochrome f (Fig. 7). Compatible with its function in c-type cytochrome assembly, CCS5 also interacts with apocytochrome c6 in the yeast two-hybrid assay (Fig. 7). Similarly to our finding with apocytochrome f, the cysteines of the thioredoxin motif in CCS5 are not required for such an interaction (data not shown). We concluded that the cysteines of the thioredoxin motif are not absolutely required for CCS5 to interact with apocytochrome f and c6, its relevant targets of action in vivo.
FIGURE 7.
CCS5 interacts with apocytochrome f and c6 in a yeast two-hybrid assay. The soluble domain of Chlamydomonas wild type (WCEVC) or cysteine-less CCS5 (WCEVS and WSEVS) constitutes the bait and is expressed as a fusion with the GAL4 DNA binding domain from the TRP1-based pGBKT7 vector. The soluble domain of Chlamydomonas apocytochrome (apocyt) f or c6 constitutes the prey and is expressed as a fusion with the GAL4 activation domain from the LEU2-based pGADT7 vector. The yeast PJ69-4A reporter strain was co-transformed with various combinations of two-hybrid plasmids. + refers to the presence of the bait or prey expressing constructs, and − refers to the presence of pGBKT7 or pGADT7 vectors in the yeast transformants. A, GAL4-dependent adenine and histidine prototrophies indicate interaction between apocytochrome f/c6 and CCS5. The yeast transformants were tested for adenine/histidine prototrophies that depend upon reconstitution of an active GAL4. 10-Fold dilution series of one representative transformant for each combination were plated on solid medium lacking leucine and tryptophan (SD-L-W) or lacking leucine, tryptophan, adenine, and histidine (SD-L-W-A-H) and incubated at 28 °C for 2 or 4 days, respectively. B, GAL4-dependent β-galactosidase activities indicate interaction between apocytochrome f/c6 and CCS5. The yeast PJ69-4A transformants were tested for β-galactosidase activity that depends upon reconstitution of an active GAL4. 2-Nitrophenyl β-d-galactopyranoside was used as a substrate. β-Galactosidase (β-Gal) activity is expressed in arbitrary units (A.U.). The values displayed are the average β-galactosidase activities from three individual transformants with standard deviation indicated by error bars.
In Vitro Reduction of Apocytochrome f CXXCH by CCS5
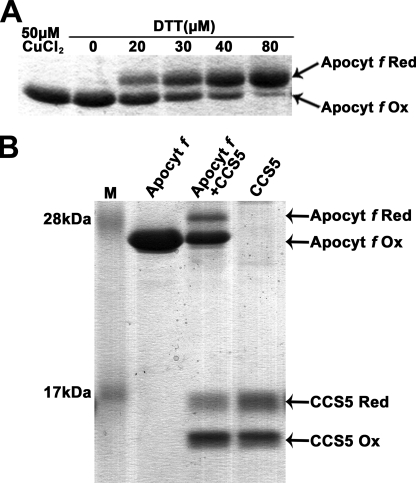
The fact that CCS5 exhibits disulfide reductase activity and interacts with apocytochrome f prompted us to develop an in vitro assay to test if apo-forms of plastid cytochromes c can be acted upon by CCS5. Despite several attempts, we were unable to express apocytochrome c6 as a recombinant protein. Hence, this substrate could not be tested in our in vitro assay. However, we were able to express and purify a recombinant form of apocytochrome f that does not contain the membrane anchor and carries no other cysteines than the ones present in the CXXCH motif. We first tested if the disulfide-bonded CXXCH motif in apocytochrome f could be chemically reduced by action of a reductant. As shown in Fig. 8A, oxidized apocytochrome f can be converted to its reduced form in a dose-dependent manner by the action of DTT. Next, we tested if reduced CCS5 can catalyze the reduction of oxidized apocytochrome f. DTT treatment of CCS5 resulted in full reduction of the protein (data not shown). However, upon DTT removal, we noticed that a fraction of reduced CCS5 is converted back to its oxidized form (Fig. 8B, 4th lane). Incubation of CCS5 with the apocytochrome substrate resulted in the conversion of oxidized to reduced apocytochrome f (Fig. 8B, 3rd lane). As expected, if oxidized apocytochrome f is reduced via CCS5-mediated thiol-disulfide exchange, we noted a conversion of reduced CCS5 to its oxidized form (Fig. 8, 3rd lane).
FIGURE 8.
Reduction of apocytochrome f CXXC disulfide by recombinant CCS5. 4-Acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid-treated samples were separated by nonreducing SDS-PAGE (15%) and visualized by Coomassie Brilliant Blue R-250 staining. The position of reduced (Apocyt f Red) and oxidized (Apocyt f Ox) forms of apocytochrome f and reduced (CCS5 Red) and oxidized (CCS5 Ox) forms of CCS5 are indicated by arrows. AMS is an alkylating molecule and treatment of exposed thiols in apocytochrome f and CCS5 with AMS results in an increase in molecular mass of the alkylated molecules that can be detected by SDS-PAGE. Only reduced apocytochrome f or CCS5 reacts with AMS. A, DTT-dependent reduction of oxidized apocytochrome f CXXCH. Recombinant apocytochrome f was oxidized by incubation with 50 μm CuCl2. Air-oxidized apocytochrome f was reduced by incubation with increasing concentrations of DTT during 1 h. B, CCS5-dependent reduction of oxidized apocytochrome f CXXCH. Prestained protein ladder (Fermentas) was used in lane M. Lane Apocyt f, oxidized apocytochrome f. Lane Apocyt f + CCS5, oxidized apocytochrome f (10 μm) was treated with recombinant reduced CCS5 (16 μm) as described under “Experimental Procedures.” Lane CCS5, DTT-reduced CCS5 after DTT removal. Quantification using the ImageJ software indicates that 22% of oxidized apocytochrome f (2.2 μm) becomes reduced, whereas 14% of reduced CCS5 (2.24 μm) is converted to its oxidized form.
Quantification of the oxidized and reduced species indicates that one molecule of oxidized apocytochrome f reacted with one molecule of reduced CCS5 enzyme. Extended incubation time (from 1 to 3 h) or a change in pH (4 instead of 7.5) did not increase the fraction of oxidized apocytochrome f that is converted to reduced by the activity of CCS5 (data not shown). We concluded that the disulfide between the cysteines of apocytochrome f heme binding is reduced by the activity of CCS5. Hence, CCS5 acts as a CXXCH disulfide reductase.
DISCUSSION
In this study, we report the identification of the C. reinhardtii ccs5 mutant that displays a specific block in the assembly of cytochrome f and c6, two plastid c-type cytochromes. We show the following: 1) the ccs5 mutant can be chemically rescued by exogenous thiols; 2) the CCS5 gene encodes a thioredoxin-like protein with similarity to Arabidopsis HCF164, a protein involved in the assembly of the cytochrome b6f complex; 3) the CCS5 protein displays disulfide reductase activity; 4) it interacts with apocytochrome f and c6; and 5) it is able to reduce an intramolecular disulfide at the CXXCH motif in the apocytochrome f target.
CCS5/HCF164 Is a CXXCH Disulfide Reductase Involved in Plastid c-Type Cytochrome Assembly
Through this work, we have identified CCS5, the algal ortholog of HCF164 that was previously shown to be required for cytochrome b6f assembly in Arabidopsis (28, 51). Chlamydomonas CCS5 and Arabidopsis HCF164 are similar proteins (35% identity), and it is expected that the algal protein also localizes to the thylakoid membrane with the redox domain facing the lumen similarly to its vascular plant counterpart (Fig. 4A) (22, 29). CCS5/HCF164 is very well conserved from unicellular algae to vascular plants. Remarkably, the WCXXC redox motif is strictly conserved in all orthologs (Fig. 4B). CCS5/HCF164 displays some similarity to Synechococcus sp. TxlA that was shown to be required for photosynthesis. However, it is not clear if TxlA is the cyanobacterial counterpart of plastid CCS5/HCF164 because the photosynthetic defects due to mutations in the txlA gene were not examined in detail (54).
The implication of Arabidopsis HCF164 in a thiol-reducing pathway for cytochrome c assembly was not ascertained because a specific defect in the maturation of holocytochrome f could not be demonstrated (28). Note that in land plants like Arabidopsis, cytochrome f is the only c-type cytochrome that is required for photosynthesis. Cytochrome c6, the substitute for plastocyanin in copper-deficient conditions is not present in land plants but only in green algae and cyanobacteria (55, 56). Based on the fact that the ccs5 mutant displays a dual deficiency in the assembly of plastid cytochromes c, namely cytochrome f and cytochrome c6, we have demonstrated the placement of CCS5/HCF164 in the cytochrome c assembly pathway (Figs. 1, 2, and 5). The DTT-dependent rescue of the ccs5 mutant indicates that the CCS5/HCF164 protein acts as a reductant in the heme attachment reaction to the CXXCH motif (Fig. 3). Indeed, recombinant CCS5 displays a disulfide reductase activity in the insulin assay, an indication that disulfides are the targets of CCS5 activity in vivo (Fig. 6). Moreover, we have shown that apocytochrome f and c6 can physically interact with CCS5 and that apocytochrome f heme-binding site can be acted upon by CCS5 and converted from oxidized to reduced form (Figs. 7 and 8). This is the first demonstration that a cytochrome c assembly factor involved in redox chemistry carries this biochemical activity. The view that CCS5/HCF164 is only active as an apocytochrome disulfide reductase may be oversimplified, and it is also conceivable that the protein exerts some chaperoning activity on its apocytochrome c targets. Recent work on CcmG, the CCS5/HCF164 counterpart in bacterial system I, points to an apocytochrome c chaperone function that is distinct from the redox activity (57). Such function could be required to present the reduced apocytochrome c for efficient ligation to the heme co-factor. The structure of ResA, the bacterial system II equivalent of CCS5/HCF164, shows that some residues could serve as a binding interface for apocytochrome c substrates when the protein is in the reduced conformation (58, 59). The fact that the cysteines in CCS5 are not strictly required for interaction with its apocytochrome c targets in a yeast two-hybrid assay is compatible with a possible chaperone function for this assembly factor (Fig. 7).
Both heme and the cysteine sulfhydryls need to be reduced for stereospecific attachment of heme to the apocytochrome c (2–5). It is unlikely that CCS5 is also involved in the reduction of heme based on the fact that disulfide/dithiol exchange is a two-electron process, while hemes can only receive one electron at a time. However, this possibility cannot be formally excluded if two hemes are being reduced at the same time or if another electron acceptor is involved.
What Are Other Candidate Targets of CCS5/HCF164 Activity in the Thylakoid Lumen?
Thiol-trapping experiments using Arabidopsis HCF164 that carries a mutation at the second cysteine in the thioredoxin motif have identified cytochrome f as a target of Arabidopsis HCF164 (51). This is consistent with our finding that cytochrome f assembly is deficient in the ccs5 mutant and that the disulfide in apocytochrome f heme-binding site is the target of CCS5 reducing activity. Based on the fact that ccs5/hcf164 mutants display a specific defect in cytochrome b6f (data not shown) (28), it is expected that the relevant targets of CCS5/HCF164 control b6f biogenesis and/or activity. An additional target identified by thiol trapping is the Rieske protein, a structural subunit of the cytochrome b6f complex (51). The relevance of this interaction is unclear, but it is possible that the disulfide bond in the Rieske protein (60) needs to be reduced under certain conditions to regulate the activity of the cytochrome b6f complex. The functionality of this disulfide in the b6f complex remains unexplored. One possible additional target is the STT7/STN7 kinase, which interacts with cytochrome b6f and is required for state transitions (40, 61), an adaptation of the photosynthetic machinery to changes in light intensities that involves the cytochrome b6f complex (62). STT7/STN7 is a transmembrane thylakoid protein with a conserved pair of cysteines exposed to the lumen. Alteration of the conserved cysteines inactivates the kinase activity suggesting that the disulfide-bonded form of STT7/STN7 is the active form (61). Because state transitions are a reversible process, it is possible that CCS5/HCF164 mediates the reduction of the disulfide in a redox situation where the kinase requires to be inactive (61). PS1-N, a structural subunit of photosystem I, was also identified in the thiol-trapping experiments and shown to be acted upon in an in vitro assay by HCF164 (51). The significance of this finding is obscure because loss of HCF164 does not seem to impact photosystem I function (28). However, it is possible to envision that PSI-N undergoes thiol-disulfide regulation under certain conditions and that CCS5/HCF164 acts as the transducer of reducing power. Other cysteine-containing proteins in the lumen such as violaxanthin de-epoxidase might also be regulated by CCS5/HCF164 reducing activity. Violaxanthin de-epoxidase is an enzyme involved in photoprotection whose activity is inhibited under reducing conditions (63, 64).
Operation of a Multicomponent Trans-thylakoid Thiol Reduction Pathway
If CCS5/HCF164 acts as a reductant in the thylakoid lumen, from where does the reducing power come? By analogy to the bacterial membrane, a transmembrane thiol-disulfide relay from stroma to lumen must operate at the thylakoid membrane. It is very likely that CCDA in the plastid is involved in such a pathway (15, 33). The cytochrome b6f-deficient phenotype due to loss-of-function mutations in the CCDA gene is compatible with such a proposed activity, but it does not indicate whether 1) the cytochrome b6f-deficient phenotype is due to loss of cytochrome f assembly and 2) whether redox chemistry is required in the context of plastid cytochrome c assembly in vivo. Chlamydomonas remains the best model system to confirm the placement of CCDA in the cytochrome c assembly pathway because of the availability of ccs mutants that are specifically blocked in the conversion of plastid apocytochromes c to their corresponding holoforms (11, 13–15). However, no CCS locus was found to be affected for the CCDA gene in any of the genetically defined CCS loci (15). Hence, the involvement of CCDA in cytochrome c assembly, although likely, is still hypothetical at this point. The reductant of CCDA on the stromal side is also not known, but stromal thioredoxins are possible electron donors (33). Recombinant spinach thioredoxin-m was shown to be able to reduce CCDA and HCF164 in an in organello assay, an indication that it might be recruited as a reductant on the stroma to convey redox power across the thylakoid membrane (34, 51). The fact that the complete absence of CCS5/HCF164 does not abolish c-type cytochrome assembly suggests that there is functional redundancy for the provision of reducing equivalents to apocytochrome f and c6 (Fig. 1) (28). It is likely that one of the genetically defined CCS locuses is functionally redundant with CCS5/HCF164. We hypothesize that the CCS4 gene product is functionally redundant with CCS5/HCF164 based on the fact that the ccs4 mutant is not completely deficient in holocytochrome f (13) and is also rescued by exogenous thiols (15). The CCS4 locus must encode a novel redox factor because it does not correspond to the CCDA gene (15).
Acknowledgments
We thank Dr. J.-D. Rochaix for hosting Dr Dreyfuss in his laboratory at the University of Geneva. We also thank Dr. D. Knaff for input on the development of the redox assay.
This work was supported, in whole or in part, by National Institutes of Health Grant GM48350. This work was also supported by Muscular Dystrophy Association Grant 4727, National Science Foundation Grant MCB-0920062 (to P. P. H.), and National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2004-35318-14953 (to S. S. M.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) ADC32800.
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid.
REFERENCES
- 1.Thöny-Meyer L. (1997) Microbiol. Mol. Biol. Rev. 61, 337–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamel P., Corvest V., Giegé P., Bonnard G. (2009) Biochim. Biophys. Acta 1793, 125–138 [DOI] [PubMed] [Google Scholar]
- 3.Ferguson S. J., Stevens J. M., Allen J. W., Robertson I. B. (2008) Biochim. Biophys. Acta 1777, 980–984 [DOI] [PubMed] [Google Scholar]
- 4.Giegé P., Grienenberger J. M., Bonnard G. (2008) Mitochondrion 8, 61–73 [DOI] [PubMed] [Google Scholar]
- 5.Kranz R. G., Richard-Fogal C., Taylor J. S., Frawley E. R. (2009) Microbiol. Mol. Biol. Rev. 73, 510–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnard G., Corvest V., Meyer E. H., Hamel P. (2010) Antioxid. Redox. Signal., in press [DOI] [PubMed] [Google Scholar]
- 7.Sanders C., Turkarslan S., Lee D. W., Daldal F. (2010) Trends Microbiol. 18, 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuras R., de Vitry C., Choquet Y., Girard-Bascou J., Culler D., Büschlen S., Merchant S., Wollman F. A. (1997) J. Biol. Chem. 272, 32427–32435 [DOI] [PubMed] [Google Scholar]
- 9.Kuras R., Saint-Marcoux D., Wollman F. A., de Vitry C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyska D., Paradies S., Meierhoff K., Westhoff P. (2007) Plant Cell Physiol. 48, 1737–1746 [DOI] [PubMed] [Google Scholar]
- 11.Howe G., Merchant S. (1992) EMBO J. 11, 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe G., Mets L., Merchant S. (1995) Mol. Gen. Genet. 246, 156–165 [DOI] [PubMed] [Google Scholar]
- 13.Xie Z., Culler D., Dreyfuss B. W., Kuras R., Wollman F. A., Girard-Bascou J., Merchant S. (1998) Genetics 148, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreyfuss B. W., Merchant S. (1999) in Proceedings of the 11th International Congress on Photosynthesis, Budapest, Hungary, August 17–22, 1998 (Pusztai J., Garab G. eds) Vol. IV, pp. 3139–3142, Kluwer Academic Publishers, The Netherlands [Google Scholar]
- 15.Page M. L., Hamel P. P., Gabilly S. T., Zegzouti H., Perea J. V., Alonso J. M., Ecker J. R., Theg S. M., Christensen S. K., Merchant S. (2004) J. Biol. Chem. 279, 32474–32482 [DOI] [PubMed] [Google Scholar]
- 16.Goldman B. S., Beck D. L., Monika E. M., Kranz R. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamel P. P., Dreyfuss B. W., Xie Z., Gabilly S. T., Merchant S. (2003) J. Biol. Chem. 278, 2593–2603 [DOI] [PubMed] [Google Scholar]
- 18.Dreyfuss B. W., Hamel P. P., Nakamoto S. S., Merchant S. (2003) J. Biol. Chem. 278, 2604–2613 [DOI] [PubMed] [Google Scholar]
- 19.Feissner R. E., Richard-Fogal C. L., Frawley E. R., Loughman J. A., Earley K. W., Kranz R. G. (2006) Mol. Microbiol. 60, 563–577 [DOI] [PubMed] [Google Scholar]
- 20.Frawley E. R., Kranz R. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimball R. A., Martin L., Saier M. H., Jr. (2003) J. Mol. Microbiol. Biotechnol. 5, 133–149 [DOI] [PubMed] [Google Scholar]
- 22.Stirnimann C. U., Grütter M. G., Glockshuber R., Capitani G. (2006) Cell. Mol. Life Sci. 63, 1642–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porat A., Cho S. H., Beckwith J. (2004) Res. Microbiol. 155, 617–622 [DOI] [PubMed] [Google Scholar]
- 24.Deshmukh M., Brasseur G., Daldal F. (2000) Mol. Microbiol. 35, 123–138 [DOI] [PubMed] [Google Scholar]
- 25.Bardischewsky F., Friedrich C. G. (2001) J. Bacteriol. 183, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erlendsson L. S., Hederstedt L. (2002) J. Bacteriol. 184, 1423–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beckett C. S., Loughman J. A., Karberg K. A., Donato G. M., Goldman W. E., Kranz R. G. (2000) Mol. Microbiol. 38, 465–481 [DOI] [PubMed] [Google Scholar]
- 28.Lennartz K., Plücken H., Seidler A., Westhoff P., Bechtold N., Meierhoff K. (2001) Plant Cell 13, 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erlendsson L. S., Acheson R. M., Hederstedt L., Le Brun N. E. (2003) J. Biol. Chem. 278, 17852–17858 [DOI] [PubMed] [Google Scholar]
- 30.Feissner R. E., Beckett C. S., Loughman J. A., Kranz R. G. (2005) J. Bacteriol. 187, 3941–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortenberg R., Beckwith J. (2003) Antioxid. Redox. Signal. 5, 403–411 [DOI] [PubMed] [Google Scholar]
- 32.Möller M., Hederstedt L. (2006) Antioxid. Redox. Signal. 8, 823–833 [DOI] [PubMed] [Google Scholar]
- 33.Nakamoto S. S., Hamel P., Merchant S. (2000) Biochimie 82, 603–614 [DOI] [PubMed] [Google Scholar]
- 34.Motohashi K., Hisabori T. (2010) Antioxid. Redox Signal., in press [DOI] [PubMed] [Google Scholar]
- 35.Harris E. H. (1989) in The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, pp. 25–50, Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- 36.Gumpel N. J., Rochaix J. D., Purton S. (1994) Curr. Genet. 26, 438–442 [DOI] [PubMed] [Google Scholar]
- 37.Kindle K. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenton J. M., Crofts A. R. (1990) Photosynth. Res. 26, 59–66 [DOI] [PubMed] [Google Scholar]
- 39.Shimogawara K., Fujiwara S., Grossman A., Usuda H. (1998) Genetics 148, 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Depège N., Bellafiore S., Rochaix J. D. (2003) Science 299, 1572–1575 [DOI] [PubMed] [Google Scholar]
- 41.Quinn J. M., Merchant S. (1998) Methods Enzymol. 297, 263–279 [DOI] [PubMed] [Google Scholar]
- 42.Howe G., Merchant S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villalobos A., Ness J. E., Gustafsson C., Minshull J., Govindarajan S. (2006) BMC Bioinformatics 7, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmgren A. (1979) J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 45.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer E. H., Giegé P., Gelhaye E., Rayapuram N., Ahuja U., Thöny-Meyer L., Grienenberger J. M., Bonnard G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miles D. (1982) in Methods in Chloroplast Molecular Biology (Edelman M., Hallick R. B., Chua N.-H., eds) pp. 75–107, Elsevier Biomedical Press, Amsterdam [Google Scholar]
- 48.Bennoun P., Masson A., Delosme M. (1980) Genetics 95, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldschmidt-Clermont M., Girard-Bascou J., Choquet Y., Rochaix J. D. (1990) Mol. Gen. Genet. 223, 417–425 [DOI] [PubMed] [Google Scholar]
- 50.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motohashi K., Hisabori T. (2006) J. Biol. Chem. 281, 35039–35047 [DOI] [PubMed] [Google Scholar]
- 52.León R., Fernández E. (2007) Adv. Exp. Med. Biol. 616, 1–11 [DOI] [PubMed] [Google Scholar]
- 53.Meyer Y., Buchanan B. B., Vignols F., Reichheld J. P. (2009) Annu. Rev. Genet. 43, 335–367 [DOI] [PubMed] [Google Scholar]
- 54.Collier J. L., Grossman A. R. (1995) J. Bacteriol. 177, 3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerfeld C. A., Krogmann D. W. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 397–425 [DOI] [PubMed] [Google Scholar]
- 56.Bialek W., Nelson M., Tamiola K., Kallas T., Szczepaniak A. (2008) Biochemistry 47, 5515–5522 [DOI] [PubMed] [Google Scholar]
- 57.Turkarslan S., Sanders C., Ekici S., Daldal F. (2008) Mol. Microbiol. 70, 652–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colbert C. L., Wu Q., Erbel P. J., Gardner K. H., Deisenhofer J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4410–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crow A., Acheson R. M., Le Brun N. E., Oubrie A. (2004) J. Biol. Chem. 279, 23654–23660 [DOI] [PubMed] [Google Scholar]
- 60.Carrell C. J., Zhang H., Cramer W. A., Smith J. L. (1997) Structure 5, 1613–1625 [DOI] [PubMed] [Google Scholar]
- 61.Lemeille S., Willig A., Depège-Fargeix N., Delessert C., Bassi R., Rochaix J. D. (2009) PLoS Biol. 7, e1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rochaix J. D. (2009) in Organellar and Metabolic Processes: The Chlamydomonas Sourcebook (Stern D. B. ed) 2nd Ed., pp. 819–845, Vol. 2, Elsevier [Google Scholar]
- 63.Bilger W., Björkman O. (1990) Photosynth. Res. 25, 173–185 [DOI] [PubMed] [Google Scholar]
- 64.Niyogi K. K., Grossman A. R., Björkman O. (1998) Plant Cell 10, 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petsalaki E. I., Bagos P. G., Litou Z. I., Hamodrakas S. J. (2006) Genomics Proteomics Bioinformatics 4, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]