Abstract
Epiblast stem cells (EpiSCs) are pluripotent cells derived from post-implantation late epiblasts in vitro. EpiSCs are incapable of contributing to chimerism, indicating that EpiSCs are less pluripotent and represent a later developmental pluripotency state compared with inner cell mass stage murine embryonic stem cells (mESCs). Using a chemical approach, we found that blockage of the TGFβ pathway or inhibition of histone demethylase LSD1 with small molecule inhibitors induced dramatic morphological changes in EpiSCs toward mESC phenotypes with simultaneous activation of inner cell mass-specific gene expression. However, full conversion of EpiSCs to the mESC-like state with chimerism competence could be readily generated only with the combination of LSD1, ALK5, MEK, FGFR, and GSK3 inhibitors. Our results demonstrate that appropriate synergy of epigenetic and signaling modulations could convert cells at the later developmental pluripotency state to the earlier mESC-like pluripotency state, providing new insights into pluripotency regulation.
Keywords: Embryonic Stem Cell, Epigenetics, Mouse, Stem Cell, Transforming Growth Factor β (TGFβ), Conversion, Pluripotency, Small Molecule
Introduction
Conventional ESCs2 are derived from and represent pluripotent cells of the ICM of pre-implantation blastocysts. They can self-renew indefinitely; have the ability to give rise to all cell types in vitro; and most importantly, contribute to an entire animal in vivo, including the germline, when placed back into blastocysts. More recently, a different type of pluripotent cells was derived from post-implantation stage epiblasts, termed EpiSCs (1, 2). Although EpiSCs can self-renew over a long-term period and appear to be pluripotent in vitro as well as in vivo in teratoma assays, in contrast to mESCs, they are incapable of incorporating into the ICM and contributing to chimerism, confirming that EpiSCs are from and represent an advanced/later developmental stage of pluripotency compared with ICM-derived ESCs and suggesting that they cannot be “reprogrammed” back into ICM stage pluripotent cells even in the in vivo environment. Although derived using blastocysts, conventional hESCs seem to correspond very closely to the EpiSCs with respect to many characteristics, including some gene expression, colony morphology (i.e. flat colony), and the signaling responses in self-renewal and differentiation. EpiSCs/hESCs are also functionally and mechanistically distinct from mESCs (which have more compact and domed colony morphology) in many other ways. For example, whereas mESCs self-renew under LIF and BMP condition or under inhibition of MEK and/or FGFR (3), EpiSCs/hESCs appear to be dependent on MAPK, FGF, and TGFβ/activin/Nodal pathway activity for self-renewal and differentiate rapidly when treated with MEK, FGFR, and/or ALK4/5/7 inhibitors (1, 2, 4). In addition, in response to BMP treatment under defined differentiation conditions, mESCs differentiate toward mesoderm lineages, whereas EpiSCs/hESCs generate trophoblasts or primitive endoderm cells (1, 5, 6). These observations strongly support the notion that EpiSCs/hESCs and mESCs represent two distinct pluripotency states: the mESC-like state representing the ICM of pre-implantation blastocysts and the EpiSC/hESC-like state representing the post-implantation epiblasts. This also raised the questions of whether the epiblast state (including conventional hESCs) can be converted back to the ICM state, and more fundamentally and significantly, how this would be achieved in an efficient manner by chemically defined conditions without any genetic manipulations. Because of the distinct difference in their ability to contribute to chimerism from mESCs or mEpiSCs (which would offer a definitive confirmation of the functional conversion of EpiSCs to mESCs), the murine system represents an ideal platform to study the intriguing process and provides a basis for generating perhaps a new type of ICM/mESC-like human pluripotent cell from conventional hESCs.
EXPERIMENTAL PROCEDURES
See the supplemental data for detailed “Experimental Procedures.”
RESULTS
EpiSCs express master pluripotency genes, including Oct4, Sox2, and Nanog. Overexpression of Oct4, Sox2, and Klf4 has been shown to induce reprogramming of murine somatic cells to become germline-competent pluripotent cells. In addition, it has been shown that germline stem cells, which express fewer pluripotency genes (e.g. lack of Nanog expression), can convert to mESC-like cells in culture (7, 8). Furthermore, a non-pluripotent cell type (designated FAB-SC) was recently derived from blastocytes and was shown to generate pluripotent mESC-like cells simply under LIF and BMP condition (9). Moreover, recent studies suggested that subpopulations of cells within mESC colonies exhibit dynamic expression of several key transcription factors (e.g. Nanog, Rex1, and Stella), which makes them continuously fluctuate between different states (e.g. between ESC- and epiblast-like phenotypes) (10–12). These studies raised the possibility that EpiSCs existing in a less “stable” pluripotency state than ICM-derived mESCs may have the ability to transition back to a mESC state “spontaneously” under culture fluctuation in vitro. To test this hypothesis, EpiSCs were trypsinized to single cells, and ∼500 cells were plated under mESC self-renewal conditions based on the notion that “converted” mESC-like cells within EpiSC colonies would be captured/selected and expanded under conditions that promote self-renewal of mESCs but induce differentiation of EpiSCs. We found that EpiSCs differentiated (e.g. cells spread/migrated out of colonies) in the first passage, and no colony could be identified over several passages when the cells were cultured under the conventional mESC growth condition with feeder cells and supplemented with LIF (Fig. 1A, panel A3). Given that the spontaneous conversion from EpiSCs to mESCs might be very inefficient, a stronger and more stringent differential self-renewal-promoting and differentiation-inducing condition might be required to select/capture and expand those “rare” converted mESC-like cells from EpiSCs (e.g. achieving cleaner phenotypic distinction and minimizing the overgrowth of differentiated EpiSCs). On the basis of the differential signaling responses (self-renewal versus differentiation) between mESCs and EpiSCs in the context of FGF and MAPK signaling pathways, as well as the observation that inhibition of MEK-ERK signaling promotes reprogramming of cells toward a more primitive state (13–15), we next treated EpiSCs with a combination of the selective FGFR inhibitor PD173074 (0.1 μm) and MEK inhibitor PD0325901 (0.5 μm) (referred to as 2PD) under regular mESC self-renewal conditions. Under these 2PD/LIF conditions, which promote robust clonal growth of mESCs and inhibit growth of differentiated cells, we observed accelerated differentiation of EpiSCs and decreased growth of the overall cell culture. Most of cells died when they were kept in culture in the 2PD/LIF medium, and no mESC-like colony was identified over serial passages. Similarly, the addition of CHIR99021 (3 μm) to the 2PD/LIF conditions for improved mESC growth/survival did not promote or capture the conversion of EpiSCs to the mESC-like state (Fig. 1A, panel A4). These results suggest that the EpiSCs represent a stable pluripotency state that does not readily convert to the mESC-like state spontaneously under conditions promoting mESC self-renewal. Concurrent and consistent with our studies, it has been recently shown that conversion of EpiSCs to the mESC-like state could be achieved only by overexpression of Klf4 or Nanog in conjunction with the use of chemical inhibitors of MEK and GSK3 (16, 17). Given those challenges, it is critical to identify and devise a pharmacological approach for reprogramming EpiSCs toward the mESC-like state, which may directly provide mechanistic insights into this process and ultimately facilitate converting hESCs to the mESC-like state.
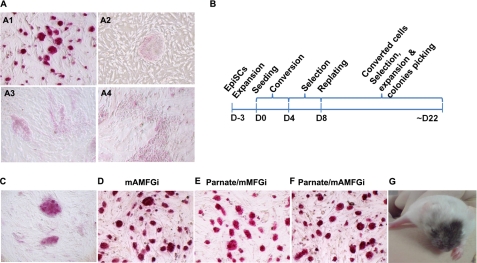
FIGURE 1.
EpiSCs differentiate under mESC growth conditions and convert to the ICM/mESC-like state by treatment with Parnate and inhibitors of ALK4/5/7, MEK, FGFR, and GSK3. A, murine ESCs (R1) grew as compact domed colonies in conventional mESC growth medium supplemented with LIF, and the colonies showed ALP-positive activity (panel A1). EpiSCs grew as large flat colonies in conventional hESC culture medium supplemented with basic FGF, and the colonies showed ALP-negative activity (panel A2). Shown also are EpiSCs differentiated in conventional mESC growth medium supplemented with LIF (panel A3) and with LIF and 0.5 μm MEK inhibitor PD0325901, 0.1 μm FGFR inhibitor PD173074, and 3 μm GSK3 inhibitor CHIR99021 (mMFGi conditions) (panel A4). B, a schematic for the generation of converted cells is shown. EpiSCs were trypsinized to single cells and seeded on feeder cells under the mESC self-renewal condition with supplements of the indicated chemical compounds for ∼4 days (D) to induce conversion, followed by another 4 days of selection. The culture was subsequently replated and further selected and expanded for another 2 weeks, during which time stable clones were picked. C, inhibition of TGFβ signaling by the selective ALK4/5/7 inhibitor A-83-01 (0.5 μm) induced EpiSCs to form more compact and domed colonies that express ALP. D, these colonies could be further stably expanded in mESC growth medium supplemented with LIF and 0.5 μm A-83-01, 0.5 μm PD0325901, 0.1 μm PD173074, and 3 μm CHIR99021 (mAMFGi conditions). E and F, the LSD1 inhibitor Parnate induced EpiSCs to form more compact and domed colonies that express ALP. These colonies could be further stably expanded under mMFGi (E) or mAMFGi (F) conditions. G, stable mESC-like cells converted from EpiSCs under Parnate/mAMFGi conditions contributed to chimerism in adult mice after aggregated embryos were transplanted into pseudo-pregnant mice. The Agouti coat color originated from Parnate/mAMFGi cells.
TGFβ/activin/Nodal activity is dynamically regulated temporally and spatially during mouse embryogenesis and is required during implantation to control the fate of early progenitor cells in epiblasts (18). The derivation of EpiSCs requiring FGF and TGFβ/activin/Nodal pathway activities suggests that TGFβ/activin/Nodal provides an anti-differentiation signal for EpiSCs (1, 2). In addition, it was reported that E-cadherin is expressed in embryos from the one-cell stage and that down-regulation of E-cadherin by signaling facilitates the implantation of blastocysts (19). Moreover, TGFβ/activin/Nodal activities also promote epithelial-mesenchymal transition by down-regulating E-cadherin during gastrulation (20–22). On the basis of these studies, we hypothesized that inhibition of TGFβ/activin/Nodal signaling might promote the process of mesenchymal-epithelial transition and consequently the conversion of EpiSCs to the mESC-like state. A-83-01 is a selective ALK4/5/7 inhibitor that has no cross-inhibitory effect on BMP receptors (23). Consistent with the previous reports, blocking TGFβ/activin/Nodal signaling with 0.5 μm A-83-01 induced rapid differentiation of EpiSCs under EpiSC culture condition supplemented with basic FGF. In dramatic contrast, under mESC culture conditions supplemented with LIF, A-83-01 induced the overall population of EpiSCs to form more compact and domed colonies that resembled mESC colony morphology and expressed ALP (a pluripotency marker highly expressed in mESCs but not in EpiSCs) (Fig. 1C). Another widely used specific ALK4/5/7 inhibitor, SB431542, had a similar effect on EpiSCs (data not shown). When the A-83-01-treated colonies were exposed to 2PD/LIF condition for selection, >50% of the colonies could self-renew and maintain ALP activity, suggesting that the cells acquired some mESC-like properties. These domed colonies were passaged as a whole cell population and further expanded in mESC growth medium supplemented with inhibitors of ALK5, MEK, FGFR, and GSK3. The stable clones were picked up at day ∼22 (termed the mAMFGi condition). These cells could self-renew for a long-term period under the mAMFGi condition; had an indistinguishable mESC colony morphology (Fig. 1D); expressed pluripotency markers such as Oct4, Nanog, and SSEA1; and regained the ICM marker Rex1 (data not shown). However, when these cells were labeled with a constitutively active GFP by lentiviruses and aggregated with morulas, we did not obtain chimeric animals after the resulting embryos were transplanted into mice (supplemental Fig. S1A). These results indicate that inhibition of TGFβ signaling in conjunction with inhibition of MEK, FGFR, and GSK3 has strong reprogramming activity and can promote partial conversion of EpiSCs to the mESC-like state.
Histone modifications, such as acetylation and methylation, have been established to play important roles in gene regulation. It has been demonstrated that Stella is an important gene in mESC germ line competence and is transcriptionally silent in EpiSCs and epiblast-like cells within mESCs. Moreover, histone modification regulates Stella expression in mESCs (2, 11). We hypothesized that a derepression of the silenced gene loci responsible for in vivo pluripotency may promote EpiSCs to overcome the epigenetic restriction/threshold toward the mESC-like state. Consequently, we chose the small molecule Parnate, which has been shown to increase global H3K4 methylation by inhibiting the histone demethylase LSD1, which specifically demethylates mono- and dimethylated histone H3K4 (24). Remarkably, after 4 days of 2 μm Parnate treatment, up to 70–80% of the EpiSCs formed small and compact colonies under the mESC growth condition. When the Parnate-treated cells were then selected with 2PD/LIF, ∼20% of the cells survived the selection as domed and ALP-positive colonies. Those colonies were passaged as a whole cell population and further expanded with inhibitors of MEK, FGFR, and GSK3 (termed the mMFGi condition) or under the mAMFGi condition. Both conditions resulted in stable cell cultures, which were morphologically indistinguishable from mESCs (Fig. 1, E and F). We next examined GFP-labeled Parnate/mMFGi and Parnate/mAMFGi cells in vivo by morula aggregation and transplantation of the resulting embryos. Remarkably, we obtained seven (of nine pups born) adult chimeras from Parnate/mAMFGi cells as determined by coat color and PCR genotyping for the presence of GFP integration in multiple adult tissues (Fig. 1G and supplemental Fig. S1, A and B). Consistently, widespread GFP-positive cells were observed in multiple tissues (i.e. three germ layers, including gonad) of E13.5 embryos from transplantation of the Parnate/mAMFGi cell-aggregated morulas (supplemental Fig. S1, A and C). To examine the germline contribution from Parnate/mAMFGi cells, the GFP/SSEA1 double-positive cells from the gonad were isolated by FACS and confirmed to express the germline markers Blimp1 and Stella by real-time PCR (supplemental Fig. S1D). These data suggest that Parnate/mAMFGi cells converted from EpiSCs regain in vivo pluripotency. In contrast, GFP-positive cells were found only in the yolk sacs of E13.5 embryos recovered from transplantation of Parnate/mMFGi cell-aggregated morulas (supplemental Fig. S1A).
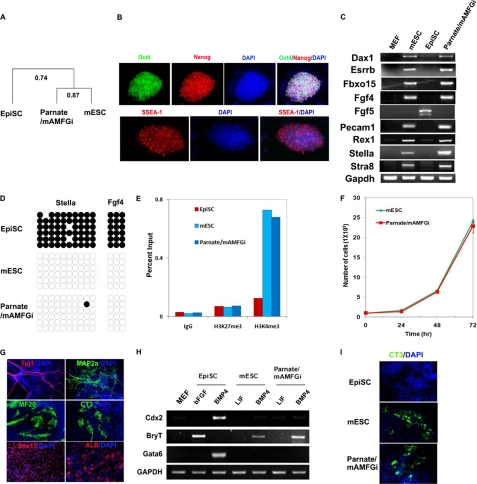
The Parnate/mAMFGi cells were therefore further characterized. Transcriptome analysis demonstrated that the converted Parnate/mAMFGi cells were much more similar to mESCs (Pearson correlation value, 0.87), whereas the original EpiSCs were more distant from mESCs (Pearson correlation value, 0.74) (Fig. 2A), consistent with previous reports. Immunocytochemistry confirmed homogeneous expression of pluripotency-associated markers in long-term expanded Parnate/mAMFGi cells, including Oct4, Nanog, SSEA1, and STELLA (Fig. 2B and supplemental Figs. S2A and S3). In addition, semiquantitative RT-PCR analysis demonstrated restoration of gene expression of specific ICM and germline competence markers (which are expressed in mESCs but absent in EpiSCs) in Parnate/mAMFGi cells, including Dax1, Esrrb, Fbxo15, Fgf4, Pecam1, Rex1, Stella, and Stra8 (Fig. 2C). In contrast, transcripts of genes associated with the epiblast and early germ layers, such as Fgf5 and Brachyury (T), were decreased or undetectable in Parnate/mAMFGi cells (Fig. 2, C and H). To further analyze specific epigenetic changes associated with the conversion, we examined the promoter DNA methylation of Stella and Fgf4, the expression of which is closely associated with ICM properties (11, 25), using bisulfite genomic sequencing. This revealed that the promoter regions of Stella and Fgf4 were largely unmethylated in Parnate/mAMFGi cells and mESCs but were hypermethylated in EpiSCs (Fig. 2D). To further examine the epigenetic state of Stella, which is restricted to the mESC-like state, we performed a ChIP-qPCR analysis of its promoter region in EpiSCs, mESCs, and converted Parnate/mAMFGi cells. We found that the H3K4 and H3K27 methylation pattern of Stella in Parnate/mAMFGi cells is similar to that observed in mESCs but is distinct from that in EpiSCs, confirming that the epigenetic status of Stella in the converted Parnate/mAMFGi cells was switched to the mESC-like status (Fig. 2E).
FIGURE 2.
Molecular and functional characterizations of converted Parnate/mAMFGi cells. A, transcriptome analysis of EpiSCs, mESCs, and Parnate/mAMFGi cells showed that Parnate/mAMFGi cells are much more similar to mESCs than to EpiSCs. Two biological replicates were used for all three cell types. B, immunocytochemistry showed homogeneous expression of the pluripotency markers Oct4 (green), Nanog (red), and SSEA1 (red) in Parnate/mAMFGi cells. C, expression of ICM-specific marker genes (Dax1, Esrrb, Fbxo15, Fgf4, Pecam1, and Rex1), germ line competence-associated marker genes (Stella and Stra8), and an epiblast gene (Fgf5) in mESCs, EpiSCs, and Parnate/mAMFGi cells was analyzed by semiquantitative RT-PCR. GAPDH was used as a control. D, the methylation of Stella and Fgf4 promoters was analyzed by bisulfite genomic sequencing. Open and closed circles indicate unmethylated CpG and methylated CpG, respectively. E, the indicated histone modifications in the Stella locus in various cells were analyzed ChIP-qPCR. Genomic DNAs were immunoprecipitated from feeder-free cultured EpiSCs, mESCs, and Parnate/mAMFGi cells with the antibodies as indicated, followed by qPCR analysis using a primer set specific to the endogenous genomic locus encoding Stella. The levels of histone modifications are represented as a percentage of input. IgG served as a no-antibody control. F, Parnate/mAMFGi cells had similar growth rate as mESCs. mESCs and Parnate/mAMFGi cells were passaged every 3 days, and the cell number was counted every 24 h. G, Parnate/mAMFGi cells effectively differentiated in vitro into cells in the three germ layers, including the characteristic neuronal cells (βIII-tubulin- and MAP2ab-positive), cardiomyocytes (cardiac troponin- and MHC-positive), and endoderm cells (Sox17- or albumin-positive). Nuclei were stained with DAPI. H, BMP4 had differential effects on the induction of mesoderm marker (Brachyury), trophoblast marker (Cdx2), and primitive endoderm marker (Gata6) expression in EpiSCs, mESCs, and Parnate/mAMFGi cells. I, directed cardiomyocyte differentiation under monolayer chemically defined conditions demonstrated that Parnate/mAMFGi cells share a similar differentiation response with mESCs and are different from EpiSCs. Cells were characterized with CT3 staining and beating phenotype.
Parnate/mAMFGi cells were also examined for their in vitro functional properties. They were found to have similar growth rate as mESCs (Fig. 2F). When Parnate/mAMFGi cells were differentiated through embryoid bodies in suspension, they were able to effectively generate cell derivatives in the three primary germ layers as shown by immunocytochemistry, including characteristic neuronal cells (βIII-tubulin- and MAP2ab-positive), beating cardiomyocytes (cardiac troponin- and MHC-positive), and endoderm cells (Sox17- or albumin-positive) (Fig. 2G and supplemental Video S1). Because mESCs and EpiSCs have different responses to signaling inputs (e.g. growth factors) in self-renewal and differentiation, conditions that were developed and work effectively for mESC differentiation may often be inefficient in inducing corresponding differentiation of EpiSCs. One of the advantages of converting EpiSCs to the mESC-like state is that differentiation conditions may be more readily translated from mESC work to EpiSC work. Differential response to BMP4 treatment represents a functional assay to distinguish between mESCs and EpiSCs. Consistent with previous studies (26–28), we found that Parnate/mAMFGi cells were induced to express the mesoderm-specific marker gene Brachyury (T) when treated with BMP4 as mESCs but, under the same conditions, could not give rise to trophectoderm (no induction of the trophoblast marker Cdx2) or primitive endoderm (Gata6) cells as EpiSCs, suggesting a similar in vitro differentiation potential/response of Parnate/mAMFGi cells to mESCs (Fig. 2H). To further demonstrate this, we compared the directed cardiac differentiation of EpiSCs, mESCs, and converted Parnate/mAMFGi cells under monolayer chemically defined conditions. In this stepwise differentiation process, in which BMP activity plays an essential role in the early steps of mesoderm specification, we found that Parnate/mAMFGi cells differentiated into beating cardiomyocytes as efficiently as mESCs but that differentiation of EpiSCs under the same conditions hardly produced cells that expressed appropriate cardiac markers or had the characteristic beating phenotype (Fig. 2I), confirming again that Parnate/mAMFGi cells are functionally similar to mESCs. Moreover, a single-cell survival assay also demonstrated that Parnate/mAMFGi cells clonally expanded as Oct4-positive colonies as efficiently as mESCs under feeder-free and N2/B27 chemically defined conditions, whereas EpiSCs survived poorly from single cells under the same conditions (supplemental Fig. S2B). These data further demonstrate that EpiSCs can be functionally converted to the mESC-like state by pharmacological manipulation that targets epigenetic modifications and differential signaling pathways required by mESCs or EpiSCs.
DISCUSSION
Concurrent with our studies, EpiSC cells have been recently reported to convert to the mESC-like state by overexpression of reprogramming genes (i.e. Klf4) in conjunction with chemical compounds (16, 29). In this study, we devised a chemically defined treatment to convert stable EpiSCs to a mESC-like developmentally earlier pluripotency state without any genetic manipulation. Despite studies providing evidence that epiblast-like cells exist and transition back and forth within colonies of conventional mESCs (11), mESCs and EpiSCs share a substantial set of pluripotency transcription factors (including Oct4, Sox2, and Nanog), and mESCs are more stable in culture, in this study, we found that EpiSCs differentiated rapidly under the conventional mESC culture conditions and that no spontaneously converted mESCs could be readily identified and isolated over serial passages at the population or clonal level. Remarkably, we found that blockage of the TGFβ pathway or inhibition of the H3K4 demethylase LSD1 with small molecule inhibitors induced dramatic morphological changes in EpiSCs toward mESC-like phenotypes with activation of some ICM-specific gene expression. However, full conversion of EpiSCs to the mESC-like state with competence to chimeric contribution could only be readily generated with a combination of inhibitors of LSD1, ALK5, MEK, FGFR, and GSK3. These observations underscore a powerful and direct induction of reprogramming from the developmentally later stage EpiSCs to the ICM stage mESCs by a synergy of signaling and direct epigenetic modulations. They also highlight a significant role for TGFβ pathway inhibition in promoting reprogramming and sustaining pluripotency, further supporting our recent studies in generating chimerism-competent rat pluripotent cells (30). Collectively, our studies provide a proof-of-concept demonstration that pluripotency-restricted EpiSCs can be readily converted to the mESC-like state in the absence of any genetic manipulation by precise pharmacological control of signaling pathways that distinguish the two pluripotency states and an epigenetic target simultaneously and offer a convenient experimental system to further study the mechanism. Such method and concept may also provide an avenue for generating a new type of mESC-like human pluripotent cell.
Supplementary Material
Acknowledgments
We thank Martin Stehling and Claudia Ortmeier (Max Planck Institute for Molecular Biomedicine) for assistance in FACS and real-time RT-PCR analysis. We also thank Dr. Paul Tesar (Case Western Reserve University) for providing EpiSCs.
This work was supported by Fate Therapeutics and the California Institute for Regenerative Medicine (to S. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, Table S1, Video S1, and references.
- ESC
- embryonic stem cell
- ICM
- inner cell mass
- EpiSC
- epiblast stem cell
- mESC
- murine ESC
- hESC
- human ESC
- LIF
- leukemia inhibitory factor
- BMP
- bone morphogenic protein
- FGFR
- fibroblast growth factor receptor
- qPCR
- quantitative polymerase chain reaction.
REFERENCES
- 1.Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 2.Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007) Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 3.Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Wang G., Wang C., Zhao Y., Zhang H., Tan Z., Song Z., Ding M., Deng H. (2007) Differentiation 75, 299–307 [DOI] [PubMed] [Google Scholar]
- 5.Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002) Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 6.D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., Baetge E. E. (2005) Nat. Biotechnol. 23, 1534–1541 [DOI] [PubMed] [Google Scholar]
- 7.Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S., Toyoshima M., Niwa O., Oshimura M., Heike T., Nakahata T., Ishino F., Ogura A., Shinohara T. (2004) Cell 119, 1001–1012 [DOI] [PubMed] [Google Scholar]
- 8.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007) Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- 9.Chou Y. F., Chen H. H., Eijpe M., Yabuuchi A., Chenoweth J. G., Tesar P., Lu J., McKay R. D., Geijsen N. (2008) Cell 135, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A. M., Hamazaki T., Hankowski K. E., Terada N. (2007) Stem Cells 25, 2534–2542 [DOI] [PubMed] [Google Scholar]
- 11.Hayashi K., Lopes S. M., Tang F., Surani M. A. (2008) Cell Stem Cell 3, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. (2008) Development 135, 909–918 [DOI] [PubMed] [Google Scholar]
- 13.Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. (2008) Cell Stem Cell 2, 525–528 [DOI] [PubMed] [Google Scholar]
- 14.Chen S., Takanashi S., Zhang Q., Xiong W., Zhu S., Peters E. C., Ding S., Schultz P. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10482–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008) PLoS Biol. 6, e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. (2009) Development 136, 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. (2009) Cell 138, 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesnard D., Guzman-Ayala M., Constam D. B. (2006) Development 133, 2497–2505 [DOI] [PubMed] [Google Scholar]
- 19.Li Q., Wang J., Armant D. R., Bagchi M. K., Bagchi I. C. (2002) J. Biol. Chem. 277, 46447–46455 [DOI] [PubMed] [Google Scholar]
- 20.Gadue P., Huber T. L., Paddison P. J., Keller G. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16806–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirard C., de la Pompa J. L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S. E., Rossant J., Mak T. W. (1998) Genes Dev. 12, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derynck R., Akhurst R. J. (2007) Nat. Cell Biol. 9, 1000–1004 [DOI] [PubMed] [Google Scholar]
- 23.Tojo M., Hamashima Y., Hanyu A., Kajimoto T., Saitoh M., Miyazono K., Node M., Imamura T. (2005) Cancer Sci. 96, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M. G., Wynder C., Schmidt D. M., McCafferty D. G., Shiekhattar R. (2006) Chem. Biol. 13, 563–567 [DOI] [PubMed] [Google Scholar]
- 25.Imamura M., Miura K., Iwabuchi K., Ichisaka T., Nakagawa M., Lee J., Kanatsu-Shinohara M., Shinohara T., Yamanaka S. (2006) BMC Dev. Biol. 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995) Genes Dev. 9, 2105–2116 [DOI] [PubMed] [Google Scholar]
- 27.Czyz J., Wobus A. (2001) Differentiation 68, 167–174 [DOI] [PubMed] [Google Scholar]
- 28.Beddington R. S., Robertson E. J. (1989) Development 105, 733–737 [DOI] [PubMed] [Google Scholar]
- 29.Hanna J., Markoulaki S., Mitalipova M., Cheng A. W., Cassady J. P., Staerk J., Carey B. W., Lengner C. J., Foreman R., Love J., Gao Q., Kim J., Jaenisch R. (2009) Cell Stem Cell 4, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. (2009) Cell Stem Cell 4, 16–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.