Abstract
1,4-Dihydroxy-2-naphthoyl coenzyme A (DHNA-CoA) synthase is a typical crotonase-fold protein catalyzing an intramolecular Claisen condensation in the menaquinone biosynthetic pathway. We have characterized this enzyme from Escherichia coli and found that it is activated by bicarbonate in a concentration-dependent manner. The bicarbonate binding site has been identified in the crystal structure of a virtually identical ortholog (96.8% sequence identity) from Salmonella typhimurium through comparison with a bicarbonate-insensitive orthologue. Kinetic properties of the enzyme and its site-directed mutants of the bicarbonate binding site indicate that the exogenous bicarbonate anion is essential to the enzyme activity. With this essential catalytic role, the simple bicarbonate anion is an enzyme cofactor, which is usually a small organic molecule derived from vitamins, a metal ion, or a metal-containing polyatomic anionic complex. This finding leads to classification of the DHNA-CoA synthases into two evolutionarily conserved subfamilies: type I enzymes that are bicarbonate-dependent and contain a conserved glycine at the bicarbonate binding site; and type II enzymes that are bicarbonate-independent and contain a conserved aspartate at the position similar to the enzyme-bound bicarbonate. In addition, the unique location of the enzyme-bound bicarbonate allows it to be proposed as a catalytic base responsible for abstraction of the α-proton of the thioester substrate in the enzymatic reaction, suggesting a unified catalytic mechanism for all DHNA-CoA synthases.
Keywords: Bacterial Metabolism, Enzyme Mechanisms, Quinones, Respiratory Chain, Vitamins and Cofactors, Bicarbonate, Crotonase-fold Enzymes
Introduction
Menaquinone is a lipid soluble naphthoquinone that plays important biological roles. In many bacterial microorganisms, menaquinone serves as an electron transporter in the respiratory chain and is essential for their survival (1, 2). In humans and animals, menaquinone is a vitamin (K2) and is used as an enzyme cofactor involved in post-translational glutamate γ-carboxylation of proteins in blood coagulation (3, 4), bone metabolism (5), and calcification of arteries and other soft tissues (6). This naphthoquinone is synthesized from the chorismate of the shikimate pathway in microorganisms (1, 2, 7, 8), whereas it has to be acquired from diet or intestinal microflora in humans or animals (9, 10). Due to its absence in humans and animals, menaquinone biosynthesis has been an attractive target for development of antibiotics against a number of important microbial pathogens, such as Mycobacterium tuberculosis and Staphylococcus aureus (11, 12).
Menaquinone biosynthesis has been most extensively studied in the facultative anaerobe Escherichia coli that uses the naphthoquinone (MK-8) and its demethylated congener (demethylmenaquinone, DMK-8) as an obligatory electron transporter under anaerobic conditions when the electron acceptor is fumarate, dimethyl sulfoxide, or trimethylamine N-oxide (13, 14). The naphthoquinone level is significantly up-regulated during the aerobiosis-to-anaerobiosis transition (15–17), indicating that menaquinone biosynthesis is subject to aeration control. However, the O2-dependent Arc, Fnr, and AppY transcriptional regulatory systems have been demonstrated not to regulate the biosynthesis of menaquinone by gene knock-out experiments (18, 19). In addition, global transcription suppression by chloramphenicol has no effect on the anaerobiosis-induced menaquinone biosynthesis, suggesting that the biosynthesis is controlled post-transcriptionally via enzymatic activity modulation (19). So far, no enzyme has been identified to be involved in such a regulation system.
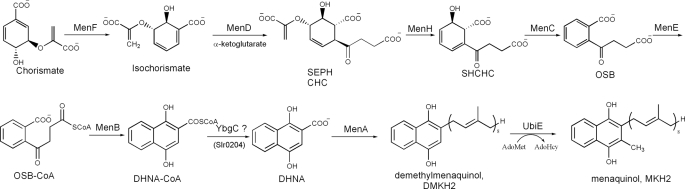
The classical menaquinone biosynthetic pathway (Fig. 1) in E. coli has been better understood through two recent findings. A new intermediate, (1R, 2S, 5S, 6S)-2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate (SEPHCHC), has been identified as the true MenD product (20, 21). MenH, which was previously proposed to be responsible for the hydrolysis of 1,4-dihydroxynaphthoyl-CoA (DHNA-CoA),2 has been reassigned as a (1R, 6R)-2-succinyl-6-hydroxyl-2, 4-cyclohexadiene-1-carboxylate (SHCHC) synthase that adopts an α/β-hydrolase fold and uses its serine-histidine-aspartate triad to catalyze a proton transfer reaction (22, 23). As a result of these findings, this classical pathway has been revised to involve nine enzymatic reactions starting from chorismate with only eight known enzymes (MenA–F, MenH, and UbiE in Fig. 1). It is found to operate in a large set of bacterial microorganisms by homolog search for the known men cluster genes (24). In addition, biosynthesis of phylloquinones has been found to follow this bacterial pathway with all the men cluster genes identified in plants and cyanobacteria (25–27). More recently, a hot dog-fold thioesterase (Slr0204) has been shown to catalyze DHNA-CoA hydrolysis in the phylloquinone biosynthesis of Synechocystis (28), but the catalytic properties of its bacterial homologs (YbgC in E. coli) have not been reported. As an alternative to this classical menaquinone/phylloquinone pathway, a new route from chorismate to menaquinone has been found in a subset of bacteria such as Helicobacter pylori and Campylobacter jejuni, which use futalosine as an intermediate (29, 30).
FIGURE 1.
The biosynthesis of menaquinone (vitamin K2) in E. coli. SEPHCHC, (1R, 2S, 5S, 6S)-2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate; DMKH2, demethylmenaquinol; MKH2, menaquinol; AdoMet, S-adenosyl methionine; AdoHcy, S-adenosyl homocysteine; Slr0204, a hot dog thioesterase committed to catalyzing DHNA-CoA hydrolysis in Synechocystis; YbgC, a homolog of Synechocystis Slr0204 in E. coli.
DHNA-CoA synthase, or MenB (EC 4.1.3.36), is the sixth enzyme in the classical menaquinone pathway and converts o-succinylbenzoyl-CoA (OSB-CoA) to DHNA-CoA through an intramolecular Claisen condensation. Three-dimensional structures have been solved for MenB proteins from M. tuberculosis (MtbMenB) (31, 32), S. aureus (SaMenB) (33), Salmonella typhimurium (StMenB, Protein Data Bank (PDB) ID: 3HO2), and Geobacillus kaustophilus HTA 426 (GkMenB, PDB ID: 2IEX) either in an apo form or in complex with a substrate analog or the product. They all display a typical crotonase family fold as an (α3)2 hexamer with the distinct feature that two of its three C-terminal helices cross the trimer-trimer interface and form a flexible part of the active site within the opposing trimer, as shown for MtbMenB. Using the crystal structure of MtbMenB in complex with acetoacetyl-CoA, the active site residues involved in substrate binding and catalysis have been identified and confirmed through site-directed mutagenesis (31). Surprisingly, a catalytically essential aspartate (Asp-185) in MtbMenB, which occupies a position similar to the highly conserved acidic residue responsible for proton abstraction from the α-carbon of the coenzyme A thioester substrates in enoyl-CoA hydratases (crotonase) and enoyl-CoA isomerases (34–40), is only conserved among a subset of the MenB proteins. In multiple sequence alignment, many MenB proteins contain a glycine at this position (31). Partly due to this lack of sequence conservation, the essential Asp-185 in MtbMenB is proposed to promote the α-proton abstraction by a substrate carboxylate in initiation of the intramolecular Claisen condensation, together with another catalytically essential acidic residue, Asp-192 (31).
In this study, we have characterized the MenB enzyme from E. coli (EcMenB) and found that its catalytic activity is dependent on an exogenous bicarbonate anion. Interestingly, the crystal structure of StMenB, which is virtually identical to EcMenB because of high sequence identity (96.8%), contains a bicarbonate anion at a position similar to the side-chain carboxylate of the essential Asp-185 residue in MtbMenB. Site-directed mutagenic studies show that the bound bicarbonate anion likely plays the same essential catalytic role of the MtbMenB Asp-185. In accordance with these findings, MenB proteins are divided into two evolutionarily conserved groups with distinct bicarbonate dependence. These findings have not only demonstrated that the simple bicarbonate anion is able to serve as an enzyme cofactor but have also provided new insights into the catalytic mechanism of the menaquinone biosynthetic enzyme. In addition, these results also identify EcMenB as the only enzyme subjected to external control in the E. coli biosynthesis of menaquinone, which may be involved in the proposed regulation of the pathway by post-transcriptional modulation of enzymatic activity.
EXPERIMENTAL PROCEDURES
Materials
Chemical reagents, including NaHCO3, NH4HCO3, KHCO3, DHNA, chorismate, α-ketoglutarate, thiamine diphosphate, coenzyme A, adenosine 5′-triphosphate, isopropyl β-d-thiogalactopyranoside (IPTG), buffers, and other salts, were purchased from Sigma.
Expression and Purification of Enzymes
Expression and purification of the menaquinone biosynthetic enzymes MenC and MenE from E. coli followed the reported procedures (20). The menB genes were amplified from the genomic DNA isolated from E. coli XL1 Blue (Stratagene), Bacillus subtilis 1A 698 (Bacillus Genetic Stock Center), and Mycobacterium smegmatis (ATCC 23037D) and were subcloned into pET32a (Novagen) for overexpression as untagged proteins. The menB gene from a clinical strain of M. tuberculosis was amplified from the genomic DNA sample provided by Professor Pak-Leung Ho of the Hong Kong University and subcloned into a pET28a vector (Novagen) for overexpression as a hexahistidine-tagged protein. The menB gene from S. aureus was cut from pET15b-TEV-menB (a generous gift from Prof. William N. Hunter of the University of Dundee, Scotland, UK) (33) with NdeI and BamHI and subcloned into pET32a (Novagen) for overexpression as an untagged protein. The subcloned genes were confirmed by restriction analysis and full-length DNA sequencing. The oligodeoxynucleotides used in the gene amplification are listed in supplemental Table S1.
The recombinant His6-tagged proteins were expressed in E. coli BL21 (DE3) harboring the menB-containing plasmids in Luria broth containing 0.1 mm IPTG at 18 °C for 16 h and purified to greater than 95% in purity (SDS-PAGE, supplemental Fig. S1) by a combination of metal-chelating chromatography and gel filtration chromatography. The recombinant untagged proteins were expressed similarly and purified to more than 90% purity by a combination of fractionation by ammonium sulfate precipitation, DEAE chromatography, and size-exclusion chromatography using Sephacryl S-200 beads (GE Healthcare). The purified proteins were quantified by a Coomassie Blue protein assay kit (Pierce) and stored in 50 mm sodium phosphate buffer (pH 7.0) containing 10% glycerol at −20 °C until use.
Site-directed Mutagenesis
The QuikChange® site-directed mutagenesis kit (Stratagene) was used to introduce point mutations (single and double mutants) into the menB genes from E. coli and M. tuberculosis. The plasmids obtained above for the expression of the corresponding MenB proteins were used as the template in the mutagenic reactions. Oligodeoxynucleotides carrying the mutated sequences are listed in supplemental Table S1. All the mutant constructs were verified by full-length DNA sequencing using a T7 promoter primer and a T7 terminator primer.
MenB Activity Assays
A reported method (31) was slightly modified for the assay of the MenB activity. To avoid interference of the decomposition of OSB-CoA to OSB spirodilactone, OSB-CoA was prepared in situ using a coupled reaction with MenC and MenE from SHCHC. Preparation of SHCHC from chorismate has been described previously (22). In a typical assay, a buffered solution containing 50 mm sodium phosphate (pH 7.0), 1–30 μm SHCHC, 200 μm ATP, 200 μm CoA-SH, and 10 mm MgCl2 was added to MenC to a final concentration of 1 μm and incubated 15 min at room temperature (23 °C) for complete conversion of SHCHC to OSB (verified by HPLC and UV-visible spectroscopy). MenE was then added to the reaction mixture to a final concentration of 0.5 μm to completely convert OSB to OSB-CoA within 8 min. Subsequently, the DHNA-CoA synthase at a varied concentration was added to the resulting reaction mixture to determine the initial reaction rate by measuring the increase of absorbance at 392 nm, which is characteristic of DHNA-CoA.
For the assays of the activation of MenB by bicarbonate and the effect of various small anions on the activity of MenB, the phosphate buffer (200 mm) was prepared with fresh Milli-Q water and purged with nitrogen gas. The SHCHC concentration used in the assay was 30 μm, and the other reagent concentrations were the same as the MenB activity assay. The titration of NaHCO3 and the addition of the other anions to the reaction system did not significantly change the pH value. The maximum pH change was 0.1 when the final concentration of NaHCO3 was 50 mm.
Circular Dichroism
Equilibrium circular dichroism (CD) spectra were measured using a JASCO J-810 spectropolarimeter (JASCO, Tokyo, Japan) with a 0.1-cm path length cuvette. All measurements were carried out at room temperature. All the spectra were averaged from three scans from 190 to 290 nm at an interval of 0.1 nm. The measured ellipticity θ (in degrees) is converted to the residue ellipticity, [θ], using the following equation: [θ] = θ × 100 × Mr/(C × d × NA) deg·cm2·dmol−1, where θ is the measured ellipticity in degrees, C is the protein concentration in mg/ml, d is the path length in cm, Mr is the protein molecular weight, and NA is the number of amino acid residues in the protein. The protein concentration was about 0.4 mg/ml in the measurements. The far-ultraviolet CD spectra of the MenB proteins and their mutants are given in supplemental Fig. S2.
Structural Analysis
All structures were generated using PyMOL 1.0 (41) from the M. tuberculosis MenB (PDB code 1Q51) and the S. typhimurium MenB (PDB code 3HO2) using SWISS-MODEL (42). Structural comparison was performed with Coot (43).
Sequence Alignment and Phylogenic Analysis
The MenB sequences used in the sequence alignment and phylogenetic analysis are from GenBankTM using the previous criteria of sequence homology (BLAST E < 10−20 relative to EcMenB) and proximity to the other menaquinone pathway genes (24). The accession numbers of the sequences used and the species abbreviations are listed in supplemental Table S2. These sequences were automatically aligned using ClustalX (Version 2.0) (44). The phylogenetic trees were constructed by the Neighbor program in PHYLIP 3.67 under the Jones-Taylor-Thornton evolutionary model and were edited using the TreeExplorer software.
RESULTS
Bicarbonate Activation of the EcMenB Activity
The E. coli MenB protein was expressed both as an untagged protein (EcMenB) containing the wild-type sequence and as a protein with a hexahistidine tag (His6-EcMenB). Due to high overproduction in E. coli, the untagged protein was purified to homogeneity with a combination of ammonium sulfate precipitation, anion-exchange chromatography, and size-exclusion chromatography. The tagged protein has a high tendency to precipitate in low salt buffers and is soluble in solutions containing one molar sodium chloride, whereas the untagged protein is soluble in almost any buffer. Despite this difference in solubility, both forms of MenB exhibit overlapping far-ultraviolet circular dichroism spectra and are present in solutions as a hexameric protein as shown by analytical ultracentrifugation, demonstrating that they have the same secondary and quaternary structures. Nevertheless, His6-EcMenB is totally inactive, whereas EcMenB demonstrates a low level of DHNA-CoA synthase activity (Km = 2.8 ± 0.3 μm, kcat = 0.1–1.0 min−1). No DHNA synthase activity was detected for either form of the enzyme.
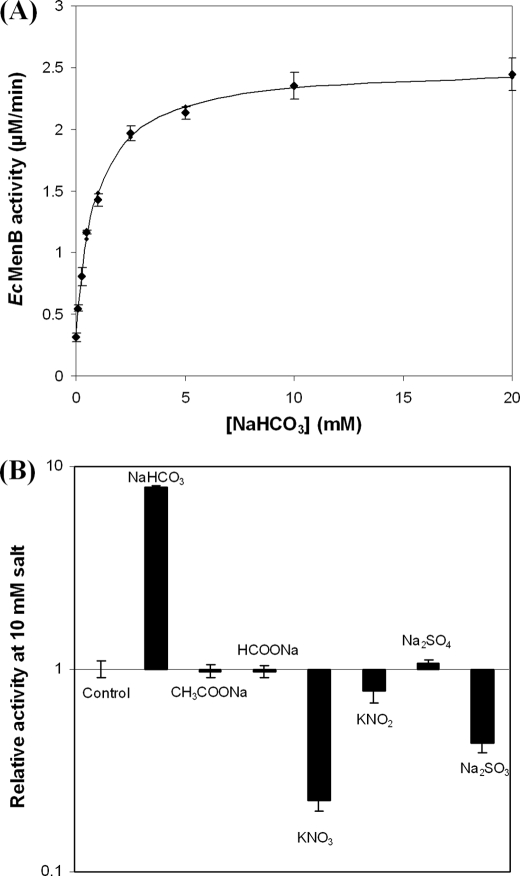
It was noticed that EcMenB exhibited a variable level of the DHNA-CoA synthase activity, which was affected by whether the buffers used in the protein purification and enzyme assay were freshly prepared. Realizing that the activity variability may be caused by solvated CO2 in the buffers, bicarbonate was purposely added into the assay buffer to find that the EcMenB activity was stimulated at least 8-fold in a concentration-dependent manner. The activity versus the [HCO3−] relationship fits well with a saturation model with an apparent effective concentration (EC50) of 0.73 mm and a background level of 0.12 mm bicarbonate in the buffer, suggesting that bicarbonate binding is essential to the enzyme activity (Fig. 2A). This activation effect was neither due to altered pH nor due to the metal ion because the pH change was negligible (<0.03) under the employed reaction conditions and EcMenB was stimulated equally well by NaHCO3, KHCO3, or NH4HCO3 (data not shown).
FIGURE 2.
Bicarbonate activation of the E. coli DHNA-CoA synthase (EcMenB). A, EcMenB activity titrated with bicarbonate; the solid line is the curve-fitting result using a saturation model (KD = 0.72 mm) under the assumption that there is a low background level of bicarbonate in the buffer. B, effects of various small anions on the activity of EcMenB. The concentration of the anions is 10 mm in B. Error bars indicate S.E.
Various simple anions were tested for their effects on the EcMenB activity. It was found that sulfite and nitrate inhibited rather than activated the synthase (Fig. 2B). This inhibition of the enzyme activity was reversed by the addition of bicarbonate, indicating that these simple inorganic anions compete for the same binding site in the enzyme. Further kinetic studies show that nitrate is a competitive inhibitor for bicarbonate binding with Ki = 2.0 mm (supplemental Fig. S3). Most other anions such as chloride, sulfate, formate, or acetate have no effect on the enzyme. EcMenB recycled from the inhibition or activation experiments by ultrafiltration removal of the anions was found equally sensitive to either activation or inhibition by the anions like the freshly prepared protein, indicating that none of the anions was tightly bound. These results show that bicarbonate specifically activates EcMenB through reversible binding.
Active Site and Bicarbonate Binding Site in EcMenB/StMenB
It was noted that among all structurally determined MenB proteins, only StMenB contains a bicarbonate anion in its crystal structure. Because EcMenB and StMenB share 96.8% sequence identity and are thus expected to have an essentially identical three-dimensional structure, the bound bicarbonate in the crystal structure is likely related to the observed bicarbonate dependence of EcMenB.
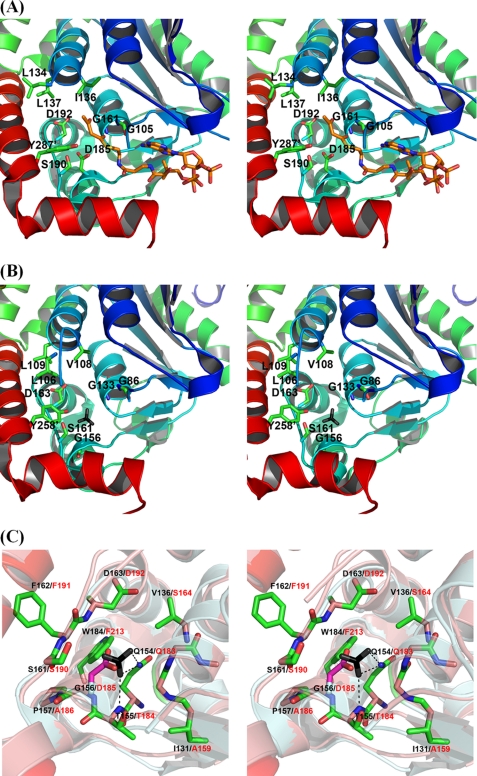
The StMenB structure was compared with the MtbMenB crystal structure, which is best characterized among all available MenB structures (31, 32). In a structure alignment, MtbMenB has an additional N-terminal extension and three insertions between the A0 and A1 strands (residues 42–46), the A2 and B2 strands (residue 90–99), and the B2 strand and H2B helix (residues 120–128), which are mostly disordered without electron densities in the crystal structure (see Ref. 32 for definition of the secondary structures). The remaining major part of the two crystal structures superimposes very well with a root-mean-square deviation of 1.06 Å for 245 aligned Cα positions. As shown in Fig. 3A, nine active site residues of MtbMenB have been identified in a deep cleft between two subunits through the help of a bound substrate analog, acetoacetyl-CoA, and site-directed mutagenesis (31). In comparison, equivalents of eight of these crucial amino acid residues are present in the StMenB active site (Fig. 3B), which are believed to play the same catalytic roles of the corresponding MtbMenB residues despite the difference in orientation of the phenyl ring of two corresponding tyrosine residues. These residues include: 1) Leu-106, Val-108, and Leu-109, forming a hydrophobic patch for recognition and binding of the nonpolar moiety of the aromatic ring of the OSB-CoA substrate; 2) Gly-86 and Gly-133, whose backbone amides form an oxyanion hole for stabilization of the enolate intermediate in the Claisen condensation; 3) Tyr-258 from a different subunit and Ser-161 that serve as hydrogen bond donors in substrate recognition and general acids in the catalysis; and 4) Asp-163, equivalent to the essential MtbMenB Asp-192 in catalysis.
FIGURE 3.
The active site and bicarbonate binding site in DHNA-CoA synthases. A, stereo view of the active site of MtbMenB in complex with acetoacetyl-CoA. The carbon atoms in the thioester ligand are represented in rainbow. B, stereo view of the active site of StMenB. The exogenous bicarbonate is represented in black for the carbon atom and gray for the oxygen atoms. C, stereo view of the bicarbonate binding site and its surroundings in the overlaid structures of MtbMenB and StMenB. The essential side-chain carboxylate of Asp-185 in MtbMenB and the exogenous bicarbonate in StMenB occupy a similar position in the superimposed structures. MtbMenB and StMenB are superimposed with PyMOL 1.0 (41) with the former in pale cyan and the latter in salmon pink. Asp-185 in MtbMenB is represented in stick with pink carbon atoms, whereas all other amino acid residues in stick presentation are from StMenB, which are labeled in black with their corresponding residues in MtbMenB labeled in red. The dashed lines in C denote probable hydrogen-bonding interaction. In A and B, the primed residues are from a different subunit.
Despite the extensive similarities, there is also a major difference between the active site pockets of StMenB and MtbMenB; the catalytically essential MtbMenB Asp-185 has no equivalent in StMenB, which contains a glycine (Gly-156) at the corresponding position. However, this location is where the exogenous bicarbonate is bound in StMenB. The bound anion is close to Gly-156 and occupies the position of the side-chain carboxylate of Asp-185 in MtbMenB when the two crystal structures are superimposed (Fig. 3C). Despite the difference in orientation of their carbonyl planes, the coincidence of the bound anion and the chemically similar active site carboxylate is a strong indication that the bound bicarbonate in StMenB may play the same catalytic role, like the essential side-chain carboxylate of Asp-185 in MtbMenB. Taking this into consideration, the two active sites are believed to be catalytically equivalent.
Except for the difference at the positions of StMenB Gly-156/MtbMenB Asp-185, the two proteins contain similar amino acid residues at other corresponding positions surrounding the bound exogenous bicarbonate. As shown in Fig. 3C, two oxygen atoms of the bicarbonate anion are within a hydrogen-bonding distance from the side-chain amide of a conserved glutamine (StMenB Gln-154 and MtbMenB Gln-183) and the backbone amide of a conserved threonine (StMenB Thr-155 and MtbMenB Thr-184). The third oxygen atom of the exogenous bicarbonate is unliganded and points toward the empty active site cavity, which corresponds to the active site in the MtbMenB structure. Another StMenB amino acid residue in close proximity of the bound anion is Trp-184, which corresponds to a phenylalanine (Phe-213) in the aligned MtbMenB structure. Another difference between the two proteins is found at a position farther away than the tryptophan residue from the bound bicarbonate, where a valine (Val-138) is present in StMenB and a serine (Ser-164) is present in MtbMenB. This residue is unlikely in direct interaction with the exogenous bicarbonate because of the long distance. Due to the high sequence identity, EcMenB is believed to be the same as StMenB to bind a bicarbonate anion in a likely identical binding site.
Mutagenesis of the Amino Acid Residues at the Bicarbonate Binding Site
The amino acid residues at the EcMenB bicarbonate binding site are examined for their effects on the binding of bicarbonate and the DHNA-CoA synthase activity. The Gly-156 residue close to the bound bicarbonate anion is changed into asparagine or aspartate by site-directed mutagenesis. These mutations cause a complete loss of the DHNA-CoA synthase activity, suggesting that the asparagine or aspartate side chains in these mutants prevent bicarbonate binding by spatial occlusion and thus providing strong support for the dependence of the enzyme activity on the exogenous bicarbonate. The inactivity of the G156D mutant indicates that the side-chain carboxylate of the introduced aspartate cannot play the catalytic role like the bound bicarbonate in the wild-type enzyme or the Asp-185 side-chain carboxylate in MtbMenB, despite their occupation of a similar position in the protein structure.
Two additional amino acid residues at the EcMenB binding site were studied. The side-chain amide in direct hydrogen-bonding contact with the bound bicarbonate in Gln-154 was removed by mutating the amino acid residue into alanine (Q154A), whereas the side chain in close proximity of the exogenous anion in Trp-184 was mutated to phenylalanine (W184F). The Q154A mutation reduced the specific DHNA-CoA synthase activity by 15-fold when compared with the wild-type enzyme, whereas its affinity for bicarbonate was reduced by 36-fold by activity titration (Table 1). Similarly, the W184F mutation diminished the specific activity by 530-fold and reduced the bicarbonate affinity by 20-fold (Table 1). These results show that both Gln-164 and Trp-184 make significant contribution to the binding of bicarbonate and provide further experimental support for the dependence of the DHNA-CoA synthase activity of EcMenB on the anionic ligand.
TABLE 1.
Activity and bicarbonate binding constants of DHNA-CoA synthases and their mutants
| Protein | Specific activitya | Apparent Kdb |
|---|---|---|
| min−1 | mm | |
| EcMenB | ||
| Wild type | 1.24 ± 0.06 | 0.73 ± 0.12 |
| Q154A | 0.078 ± 0.005 | 22.0 ± 5.6 |
| G156N | 0 | NDc |
| G156D | 0 | NDc |
| W184F | 0.0023 ± 0.003 | 12.5 ± 4.0 |
| G156D + W184F | 0 | NDc |
| MtbMenB | ||
| Wild type | 12.3 ± 0.6 | >100 mm |
| D185G | 0 | NDc |
| D185G + F213W | 0 | NDc |
a The enzyme activity was determined in the presence of 20 mm NaHCO3.
b The apparent Kd was determined by fitting the bicarbonate titration curve of the enzyme activity using a growth model under the assumption that there is a low background level of bicarbonate in the buffer.
c ND, not determined.
These mutagenic analyses confirm that EcMenD contains a bicarbonate binding site similar to that found in the StMenD crystal structure. They also provide compelling evidence that the bound bicarbonate anion is essential to the DHNA-CoA synthase activity, consistent with the saturating activation curve of the enzyme activity by the anion (Fig. 2A). The retention of a low level activity for wild-type EcMenB in the absence of externally added bicarbonate might be a result of the combined effect of its high affinity for the anion and a low level of bicarbonate formed from solvation of air CO2 in the reaction buffer, which is not possible to remove completely according to previous experience (45). The buffer-solvated carbon dioxide might also be the source of bicarbonate found in the structure of StMenB, whose crystallization was achieved in buffers without added bicarbonate.
Two DHNA-CoA Synthase Subfamilies with Different Bicarbonate Dependence
The essential catalytic role for the exogenous bicarbonate in EcMenB and its occupation of a position similar to the essential side-chain carboxylate of the MtbMenB Asp-185 indicate that the DHNA-CoA synthase activity is strictly dependent on an acidic group at this location. As such, MenB proteins without an acidic residue at this critical position should be sensitive to bicarbonate activation, like EcMenB. In contrast, for those MenB proteins containing an acidic residue at this location, like MtbMenB, they should not respond to activation by bicarbonate.
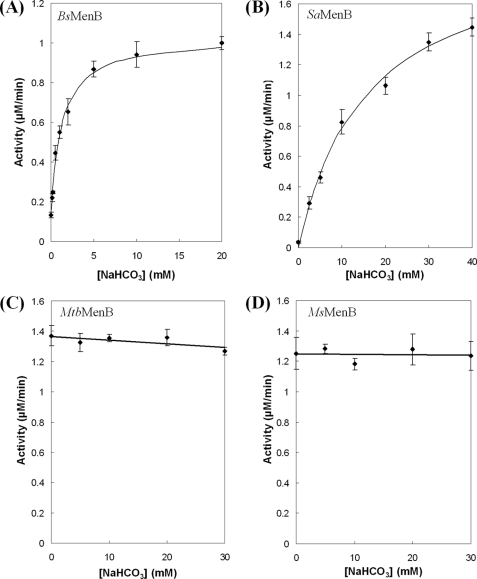
To test these predictions, four additional DHNA-CoA synthase orthologs from S. aureus (SaMenB), B. subtilis (BsMenB), M. tuberculosis (MtbMenB), and M. smegmatis (MsMenB) were obtained and subjected to activation by bicarbonate. The first two enzymes contain a glycine at the position corresponding to the EcMenB Gly-156 or the MtbMenB Asp-185 in a sequence alignment, whereas the latter two enzymes contain an aspartate residue at this location. As shown in Fig. 4, neither MtbMenB nor MsMenB is affected by the externally added bicarbonate, whereas both SaMenB and BsMenB, like EcMenB, are readily activated by bicarbonate with a varied EC50. The lower bicarbonate affinity of SaMenB may underlie the absence of the anion in its crystallographic structure (33). Indeed, these results strongly support that there exist two distinct types of DHNA-CoA synthases, whose different sensitivity to bicarbonate activation can be predicted by whether an acidic residue is available at a position similar to Asp-185 in MtbMenB or Gly-156 in EcMenB.
FIGURE 4.
Effects of bicarbonate on the activity of DHNA-CoA synthases from different bacterial species. A, BsMenB; B, SaMenB; C, MtbMenB; D, MsMenB. Solid lines in A and B represent the curve-fitting results with a saturation model under the assumption that there is a low background level of bicarbonate in the buffer. Error bars indicate S.E.
Convertibility between the Two Different Types of DHNA-CoA Synthases
According to the requirement of an acidic acid at the position similar to Gly-156 in EcMenB or Asp-185 for the activity of DHNA-CoA synthases, introducing an aspartate to position 156 of EcMenB should result in an active enzyme similar to MtbMenB. However, the EcMenB G156D mutant is completely inactive (Table 1), suggesting that the activity of the enzyme and its bicarbonate sensitivity are not entirely determined by one amino acid residue. To further test whether the bicarbonate sensitivity can be changed, a second mutation, W184F, was introduced to the EcMenB G156D mutant to make the active site more similar to that of MtbMenB. However, the double mutations also resulted in a completely inactive mutant, although all the active site residues match those in MtbMenB. Similarly, mutation of MtbMenB Asp-185 to glycine with or without additional mutation of Phe-213 to tryptophan also failed to transform the synthase into a bicarbonate-activatable enzyme, although the active site residues and the main bicarbonate binding site residues match those of EcMenB. In both cases, a complete inactive mutant without the ability to bind bicarbonate was obtained (Table 1). These results demonstrate that there is a substantial barrier between the two DHNA-CoA synthase subfamilies, which cannot be overcome by simple remodeling of the residues at the active site and the bicarbonate binding site.
DISCUSSION
We have found that bicarbonate activates EcMenB and controls its catalytic activity in a concentration-dependent manner. The saturating mode of activation suggests that the bound bicarbonate is essential to the enzyme activity. This is strongly supported by the severe loss of the DHNA-CoA synthesis activity for the site-directed mutants of the bicarbonate binding site amino acid residues. Structural comparison and site-directed mutagenesis support that the bicarbonate anion plays the same catalytic role as the side-chain carboxylate group of the essential Asp-185 in MtbMenB. In addition, the bicarbonate binding site has been identified to mainly consist of the hydrophobic tryptophan side chain (Trp-184), the side-chain amide of Gln-154, and the backbone amide of Thr-155, of which the latter two functional groups form hydrogen bonds with two oxygen atoms of the anionic ligand. With the essential catalytic role, bicarbonate is an enzyme cofactor, which is usually a small organic molecule derived from vitamins, a metal ion, or a metal-containing polyatomic complex.
The essential cofactor role of the EcMenB-bound bicarbonate appears to be distinct from the various biochemical roles played by this inorganic ion in other enzyme systems. In previous investigations, bicarbonate was found to affect the activities of photosystem II (46–48), nucleotidyl cyclases (49–51), and amine oxidase (52) without a clear molecular basis. It has also been found to function as a nonessential general base in dizinc leucine aminopeptidases (53) and to modulate β-carbonic anhydrases as an allosteric regulator (54). In addition, bicarbonate is known to affect bacterial growth without a clear molecular mechanism (55, 56). Together with these unique biochemical effects, the observed capability of bicarbonate to serve as an enzyme cofactor allows us to better appreciate the catalytic potential of this simple anion in diverse biochemical and cellular processes.
The finding of the catalytically essential bicarbonate in EcMenB/StMenD, which is likely equivalent to the essential side-chain carboxylate of the MtbMenB Asp-185, sheds new light on the catalytic mechanism of DHNA-CoA synthases. These synthases are typical crotonase-fold enzymes, whose catalysis involves an oxyanion hole-stabilized enolate intermediate generated from abstraction of α-proton from the acyl-CoA substrates by a conserved catalytic base (35–40, 57). However, the catalytically essential Asp-185 of MtbMenB, located at a position similar to the conserved catalytic base Glu-164 in enoyl-CoA hydratases, was not considered to be the catalytic base responsible for α-proton abstraction in DHNA-CoA synthesis (31). Instead, a carboxylate group of the OSB-CoA substrate was proposed to be responsible for the critical α-proton abstraction that initiates the intramolecular Claisen condensation. This was in large part due to the absence of an acidic residue similar to MtbMenB Asp-185 in a large subset of MenB proteins. Now that it is known that a bound bicarbonate likely plays the catalytic role of an acidic residue in this subset of MenB enzymes such as EcMenB, SaMenB, and BsMenB, there is a conserved acidic group among all DHNA-CoA synthases that structurally aligns with the conserved catalytic base of other members of the crotonase-fold superfamily. We propose that this conserved acidic group is a catalytic base responsible for abstraction of the α-proton in OSB-CoA to initiate the intramolecular Claisen condensation in DHNA-CoA synthesis. This is supported by the close distance of either the bound bicarbonate in EcMenB/StMenB or the side-chain carboxylate of the essential Asp-185 in MtbMenB to the α-proton of acetoacetyl-CoA in the superimposed structures. In the meantime, this proposal is also consistent with the observed inhibitory effects of nitrate and sulfite (Fig. 2B), which bind to the bicarbonate binding site but cannot serve as a catalytic base because of their lower basicity.
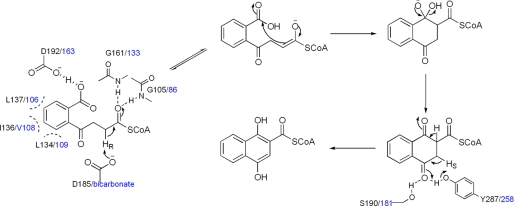
To be consistent with the proposed catalytic role for the enzyme-bound bicarbonate or equivalent acidic groups, we also propose that the other essential acidic residue in the enzyme active site, namely Asp-192 in MtbMenB or Asp-163 in EcMenB, affects the acidity of the carboxylate group of the substrate and promotes its protonation to facilitate the intramolecular Claisen condensation in DHNA-CoA synthesis. Moreover, all the remaining active site residues are considered to have the same catalytic functions as proposed previously (31). Taken together, a convergent catalytic mechanism is proposed for EcMenB and MtbMenB (Fig. 5). The stereochemistry in different proton abstraction steps is consistent with the results from a previous isotope-labeling/exchange study of the menaquinone biosynthesis (58).
FIGURE 5.
The proposed convergent mechanism for the reactions catalyzed by EcMenB and MtbMenB. The reaction scheme is similar to that described in Ref. 31. The active site residues are only shown at selected reaction steps. The residues from MtbMenB are labeled in black, whereas the residues and the bound bicarbonate ligand from EcMenB are given in blue.
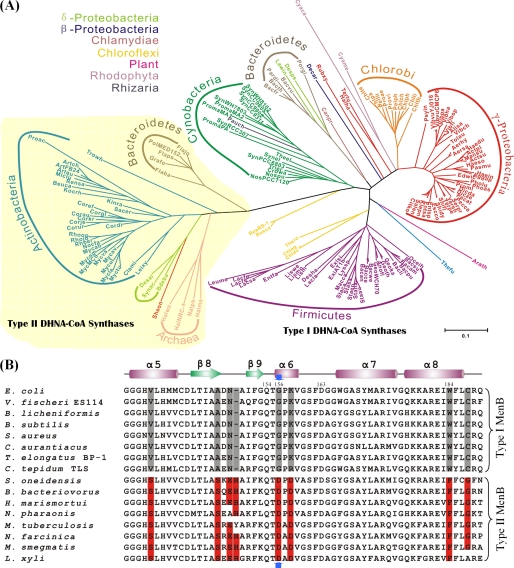
The role of the EcMenB-bound bicarbonate and the side-chain carboxylate of the Asp-185 in MtbMenB as a likely catalytic base is consistent with the existence of two distinct subfamilies of DHNA-CoA synthases. Members of the first subfamily, called type I enzymes, contain a nonacidic residue at the position similar to Gly-156 of EcMenB or Asp-185 of MtbMenB and have to base their catalytic activity on an exogenous bicarbonate such as EcMenB. In contrast, members of the second subfamily, called type II enzymes, contain a bicarbonate-equivalent acidic residue at the same location and are insensitive to activation by bicarbonate, such as MtbMenB. This classification is strongly supported by the distinct dependence on bicarbonate for the randomly selected four MenB orthologs (Fig. 4). In addition, it is also supported by the fact that all of the 163 MenB sequences identified from GenBank according to the previously set criteria (24) fall into these two categories. In a multiple sequence alignment, the type I enzymes contain a strictly conserved glycine at the position similar to Gly-156 of EcMenB, whereas the type II enzymes contain a strictly conserved aspartate at this position. Consistently, the type I enzymes (118 out of 163 sequences) form an evolutionarily related cluster in the phylogenic tree and are widely found in Bacteroidetes, Chlamydiae, Chloroflexi, Chlorobi, γ-Proteobacteria, Firmicutes, Cyanobacteria, Rhodophyta, Rhizaria, and plants (Fig. 6A). In the meantime, the type II enzymes form another distinct evolutionary cluster in the phylogenic analysis and are mainly found in Archaea, Bacteroidetes, Actinobacteria, and δ-Proteobacteria.
FIGURE 6.
Distribution and distinctive sequence conservation of type I and type II DHNA-CoA synthases. A, type I and type II DHNA-CoA synthases form distinct clusters in phylogenic analysis. The Bayesian phylogenetic tree of the proteins in the MenB family was generated from 163 sequences, whose abbreviations are listed in the supplemental material with their gene identification numbers. Proteins are colored according to phylum, and arcs indicate the main subfamilies. The type II DHNA-CoA synthases are shaded in pale yellow. B, type-specific conservation of eight amino acid residues in close proximity of the essential Asp-185 in MtbMenB. The sequences are selected to represent the gene diversity of the 163 identified MenB proteins in the phylogenetic analysis. The distinctively conserved residues are shaded in gray for type I enzymes and in red for type II enzymes. The blue arrows point to Gly-156 in EcMenB and Asp-185 in MtbMenB. A few EcMenB residues mentioned “under ”Results“ are numbered in the alignment. V. fischeri, Vibrio fischeri; B. licheniformis, Bacillus licheniformis; C. aurantiacus, Chloroflexus aurantiacus; T. elongatus, Thermosynechococcus elongatus; C. tepidum, Chlorobium tepidum; S. oneidensis, Shewanella oneidensis; B. bacteriovorus, Bdellovibrio bacteriovorus; H. marismortui, Haloarcula marismortui; N. pharaonis, Natronomonas pharaonis; N. farcinica, Nocardia farcinica; L. xyli, Leifsonia xyli.
Although the DHNA-CoA synthases can be classified on the basis of the difference at only one active site residue, the failure to overcome the evolutionary barrier between the two synthase subfamilies by simple active site remodeling indicates that the proper positioning and functioning of the conserved acidic group, either the bound bicarbonate in type I enzymes or the conserved aspartate in type II enzymes, is affected by structural elements beyond the active site. In this connection, it has been noted that the type II MtbMenB contains an additional N-terminal extension and three insertions the A0 and A1 strands, the A2 and B2 strands, and the B2 strand and the H2B helix in comparison with the type I StMenB structure. Interestingly, these additional sequences are a conserved feature in all type II enzymes, whereas none of the type I enzymes contain extra amino acid residues at any of the three locations. In addition, examinations of the multiple sequence alignment of the MenB proteins find that each subfamily contains a set of conserved, type-specific amino acid residues at seven positions close to the location of the essential acidic residue (Asp-185) in MtbMenB (Fig. 6B). These additional structural differences may have a critical effect on positioning and functioning of the conserved acidic group through long range electrostatic interactions and causing subtle conformational or even dynamic changes in the protein structure, thereby increasing the evolutionary barrier between the two enzyme subfamilies.
Although the exogenous bicarbonate plays an essential catalytic role in the EcMenB-catalyzed reaction, this role can be fulfilled by a side-chain carboxylate as seen for the type II DHNA-CoA synthases. It is not clear why these synthases are evolved into two evolutionary groups with distinct bicarbonate dependence. However, it is noticeable that the bicarbonate-activatable EcMenB is the only enzyme subjected to external control in the menaquinone biosynthetic pathway of E. coli, which may be linked to the proposed regulation of the pathway by post-transcriptional modulation of enzymatic activity (19). Due to this potential connection, it may be worthwhile to investigate the correlation of menaquinone/phylloquinone biosynthesis to intracellular bicarbonate level in other species utilizing the type I DHNA-CoA synthases.
Supplementary Material
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region of the People's Republic of China (Grant GRF601209).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
- DHNA-CoA
- 1,4-dihydroxy-2-naphthoyl coenzyme A
- SHCHC
- (1R, 6R)-2-succinyl-6-hydroxyl-2, 4-cyclohexadiene-1-carboxylate
- OSB
- o-succinylbenzoyl-CoA
- BsMenB
- MenB from B. subtilis
- EcMenB
- MenB from E. coli
- GkMenB
- MenB from G. kaustophilus HTA 426
- MsMenB
- MenB from M. smegmatis
- MtbMenB
- MenB from M. tuberculosis
- SaMenB
- MenB from S. aureus
- StMenB
- MenB from S. typhimurium.
REFERENCES
- 1.Meganathan R. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C., Curtiss R., 3rd, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. eds) 2nd Ed., Vol. 1, pp 642–656, American Society for Microbiology, Washington, DC [Google Scholar]
- 2.Meganathan R. (2001) Vitam. Horm. 61, 173–218 [DOI] [PubMed] [Google Scholar]
- 3.Berkner K. L. (2005) Annu. Rev. Nutr. 25, 127–149 [DOI] [PubMed] [Google Scholar]
- 4.Dowd P., Ham S. W., Naganathan S., Hershline R. (1995) Annu. Rev. Nutr. 15, 419–440 [DOI] [PubMed] [Google Scholar]
- 5.Bugel S. (2008) Vitamin K, pp 393–416, Elsevier Academic Press Inc, San Diego [Google Scholar]
- 6.Shanahan C. M., Proudfoot D., Farzaneh-Far A., Weissberg P. L. (1998) Crit. Rev. Eukaryot. Gene Expr. 8, 357–375 [DOI] [PubMed] [Google Scholar]
- 7.Bentley R. (1990) Crit. Rev. Biochem. Mol. Biol. 25, 307–384 [DOI] [PubMed] [Google Scholar]
- 8.Dosselaere F., Vanderleyden J. (2001) Crit. Rev. Microbiol. 27, 75–131 [DOI] [PubMed] [Google Scholar]
- 9.Fernandez F., Collins M. D., Hill M. J. (1985) Biochem. Soc. Trans. 13, 223–224 [Google Scholar]
- 10.Suttie J. W. (1995) Annu. Rev. Nutr. 15, 399–417 [DOI] [PubMed] [Google Scholar]
- 11.Kurosu M., Narayanasamy P., Biswas K., Dhiman R., Crick D. C. (2007) J. Med. Chem. 50, 3973–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X., Zhang H., Tonge P. J., Tan D. S. (2008) Bioorg. Med. Chem. Lett. 18, 5963–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wissenbach U., Kröger A., Unden G. (1990) Arch. Microbiol. 154, 60–66 [DOI] [PubMed] [Google Scholar]
- 14.Wissenbach U., Ternes D., Unden G. (1992) Arch. Microbiol. 158, 68–73 [DOI] [PubMed] [Google Scholar]
- 15.Polglase W. J., Pun W. T., Withaar J. (1966) Biochim. Biophys. Acta 118, 425–426 [DOI] [PubMed] [Google Scholar]
- 16.Jones R. W., Garland P. B. (1982) Function of Quinones in Energy Conserving Systems (Trumpover B. C. ed) pp. 465–476, Academic Press, New York [Google Scholar]
- 17.Bekker M., Kramer G., Hartog A. F., Wagner M. J., de Koster C. G., Hellingwerf K. J., Teixeira de Mattos M. J. (2007) Microbiology 153, 1974–1980 [DOI] [PubMed] [Google Scholar]
- 18.Unden G. (1988) Arch. Microbiol. 150, 499–503 [DOI] [PubMed] [Google Scholar]
- 19.Shestopalov A. I., Bogachev A. V., Murtazina R. A., Viryasov M. B., Skulachev V. P. (1997) FEBS Lett. 404, 272–274 [DOI] [PubMed] [Google Scholar]
- 20.Jiang M., Cao Y., Guo Z. F., Chen M., Chen X., Guo Z. (2007) Biochemistry 46, 10979–10989 [DOI] [PubMed] [Google Scholar]
- 21.Jiang M., Chen M., Cao Y., Yang Y., Sze K. H., Chen X., Guo Z. (2007) Org. Lett. 9, 4765–4767 [DOI] [PubMed] [Google Scholar]
- 22.Jiang M., Chen X., Guo Z. F., Cao Y., Chen M., Guo Z. (2008) Biochemistry 47, 3426–3434 [DOI] [PubMed] [Google Scholar]
- 23.Jiang M., Chen X., Wu X. H., Chen M., Wu Y. D., Guo Z. (2009) Biochemistry 48, 6921–6931 [DOI] [PubMed] [Google Scholar]
- 24.Glasner M. E., Fayazmanesh N., Chiang R. A., Sakai A., Jacobson M. P., Gerlt J. A., Babbitt P. C. (2006) J. Mol. Biol. 360, 228–250 [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre-Legendre L., Rappaport F., Finazzi G., Ceol M., Grivet C., Hopfgartner G., Rochaix J. D. (2007) J. Biol. Chem. 282, 13250–13263 [DOI] [PubMed] [Google Scholar]
- 26.Gross J., Meurer J., Bhattacharya D. (2008) BMC Evol. Biol. 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J. P. (2008) Plant Physiol. 147, 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widhalm J. R., van Oostende C., Furt F., Basset G. J. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5599–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiratsuka T., Furihata K., Ishikawa J., Yamashita H., Itoh N., Seto H., Dairi T. (2008) Science 321, 1670–1673 [DOI] [PubMed] [Google Scholar]
- 30.Seto H., Jinnai Y., Hiratsuka T., Fukawa M., Furihata K., Itoh N., Dairi T. (2008) J. Am. Chem. Soc. 130, 5614–5615 [DOI] [PubMed] [Google Scholar]
- 31.Truglio J. J., Theis K., Feng Y., Gajda R., Machutta C., Tonge P. J., Kisker C. (2003) J. Biol. Chem. 278, 42352–42360 [DOI] [PubMed] [Google Scholar]
- 32.Johnston J. M., Arcus V. L., Baker E. N. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 1199–1206 [DOI] [PubMed] [Google Scholar]
- 33.Ulaganathan V., Agacan M. F., Buetow L., Tulloch L. B., Hunter W. N. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Ordine R. L., Bahnson B. J., Tonge P. J., Anderson V. E. (1994) Biochemistry 33, 14733–14742 [DOI] [PubMed] [Google Scholar]
- 35.Müller-Newen G., Janssen U., Stoffel W. (1995) Eur. J. Biochem. 228, 68–73 [DOI] [PubMed] [Google Scholar]
- 36.Engel C. K., Mathieu M., Zeelen J. P., Hiltunen J. K., Wierenga R. K. (1996) EMBO J. 15, 5135–5145 [PMC free article] [PubMed] [Google Scholar]
- 37.Engel C. K., Kiema T. R., Hiltunen J. K., Wierenga R. K. (1998) J. Mol. Biol. 275, 847–859 [DOI] [PubMed] [Google Scholar]
- 38.Modis Y., Filppula S. A., Novikov D. K., Norledge B., Hiltunen J. K., Wierenga R. K. (1998) Structure 6, 957–970 [DOI] [PubMed] [Google Scholar]
- 39.Kiema T. R., Engel C. K., Schmitz W., Filppula S. A., Wierenga R. K., Hiltunen J. K. (1999) Biochemistry 38, 2991–2999 [DOI] [PubMed] [Google Scholar]
- 40.Mursula A. M., van Aalten D. M., Hiltunen J. K., Wierenga R. K. (2001) J. Mol. Biol. 309, 845–853 [DOI] [PubMed] [Google Scholar]
- 41.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 42.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2978 [DOI] [PubMed] [Google Scholar]
- 45.Shevela D., Klimov V., Messinger J. (2007) Photosynth. Res. 94, 247–264 [DOI] [PubMed] [Google Scholar]
- 46.Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 47.Shevela D., Su J. H., Klimov V., Messinger J. (2008) Biochim. Biophys. Acta 1777, 532–539 [DOI] [PubMed] [Google Scholar]
- 48.Ulas G., Olack G., Brudvig G. W. (2008) Biochemistry 47, 3073–3075 [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Cann M. J., Litvin T. N., Iourgenko V., Sinclair M. L., Levin L. R., Buck J. (2000) Science 289, 625–628 [DOI] [PubMed] [Google Scholar]
- 50.Steegborn C., Litvin T. N., Levin L. R., Buck J., Wu H. (2005) Nat. Struct. Mol. Biol. 12, 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun L., Wang H., Hu J., Han J., Matsunami H., Luo M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2041–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez-Guillamon M., Bolea I., Solé M., Boada M., Tipton K. F., Unzeta M. (2007) J. Biochem. 142, 571–576 [DOI] [PubMed] [Google Scholar]
- 53.Sträter N., Sun L., Kantrowitz E. R., Lipscomb W. N. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11151–11155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cronk J. D., Rowlett R. S., Zhang K. Y., Tu C., Endrizzi J. A., Lee J., Gareiss P. C., Preiss J. R. (2006) Biochemistry 45, 4351–4361 [DOI] [PubMed] [Google Scholar]
- 55.Repaske R., Clayton M. A. (1978) J. Bacteriol. 135, 1162–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watsuji T. O., Kato T., Ueda K., Beppu T. (2006) Biosci. Biotechnol. Biochem. 70, 753–756 [DOI] [PubMed] [Google Scholar]
- 57.Holden H. M., Benning M. M., Haller T., Gerlt J. A. (2001) Acc. Chem. Res. 34, 145–157 [DOI] [PubMed] [Google Scholar]
- 58.Igbavboa U., Leistner E. (1990) Eur. J. Biochem. 192, 441–449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.