Abstract
Limitations of current steroidal mineralocorticoid receptor (MR) antagonists have stimulated the search for a new generation of molecules. We screened for novel nonsteroidal compounds and identified MR antagonists derived from the chemical class of dihydropyridines. Chemical optimization resulted in BR-4628, which displays high in vitro and in vivo MR potency as well as selectivity with respect to the other steroid hormone receptors and the L-type calcium channel. Biochemical studies demonstrated that BR-4628 forms complexes with MR that do not promote the recruitment of transcriptional co-regulators. Docking experiments, using the crystal structure of the MR ligand-binding domain in an agonist conformation, revealed that BR-4628 accommodates in the MR ligand-binding cavity differently in comparison with the classical steroidal MR antagonists. An alanine scanning mutagenesis approach, based on BR-4628 docking, allowed identifying its anchoring mode within the ligand-binding cavity. Altogether, we propose that BR-4628 is a bulky antagonist that inactivates MR through a passive mechanism. It represents the prototype of a new class of MR antagonists.
Keywords: Drug Action, Drug Design, Heart, Kidney, Steroid Hormone Receptor, Cardiovascular Diseases, Drug Discovery, Mechanism of Action, Steroid Receptors, Antagonist
Introduction
The first and best documented effects of aldosterone are those observed on the kidney distal tubule. In this epithelial tissue, aldosterone promotes sodium reabsorption and potassium secretion, increasing the blood pressure by expanding the extracellular volume (1, 2). These aldosterone effects are mediated by the mineralocorticoid receptor (MR),4 a ligand-activated transcription factor belonging to the nuclear receptor superfamily (3). According to the aldosterone renal effects, inactivating MR mutations provoke salt wasting (4–7), whereas an activating mutation (S810L) has been shown to induce a severe form of early onset hypertension (8). In addition to its renal effects, aldosterone acts in nonepithelial tissues such as brain, vasculature, and heart, in which it has deleterious effects. In the cardiovascular system, aldosterone provokes inflammation and fibrosis, combined with ventricular hypertrophy (9). Whether these adverse effects are mediated by aldosterone and/or cortisol at conditions of inappropriate salt or redox status has not yet been established (10).
Spironolactone and eplerenone, both of which display structural elements of progesterone, have been developed as MR antagonists (11–14). These molecules inhibit aldosterone binding to MR and render it transcriptionally inactive. As a consequence, they prevent the aldosterone-induced sodium reabsorption and are effective in decreasing blood pressure (15, 16). Remarkably, these molecules have been shown to reduce the deleterious aldosterone effects in the cardiovascular system. In the randomized aldactone evaluation study (RALES) and eplerenone post-acute myocardial infarction heart failure efficacy and survival study (EPHESUS), spironolactone and eplerenone, respectively, significantly reduced mortality and morbidity in patients with heart failure (17, 18).
Despite their renal and cardiac benefits, these two steroidal spirolactones suffer from substantial drawbacks that limit their clinical use. Although spironolactone is highly potent, it lacks selectivity toward other members of the oxo-steroid receptor family. Indeed, its prolonged use is associated with sexual side effects that are related to its progestogenic and antiandrogenic activities (19). Eplerenone is characterized by an improved selectivity as compared with spironolactone. However, eplerenone has a low affinity for MR (20) and is less efficient than spironolactone with respect to blood pressure lowering in patients with mild-to-moderate hypertension (21). In addition, both steroidal molecules are not only unable to block the constitutive activity of the mutant MRS810L, a gain-of function mutation that is linked clinically to early onset hypertension in men and gestational hypertension in women, but paradoxically activate this mutant receptor (8, 22). Therefore, the use of spirolactones to treat hypertensive patients carrying the S810L mutation is inappropriate.
The known disadvantages of spirolactones stimulated research for new, selective MR antagonists (23). An ultrahigh throughput screening revealed substituted dihydropyridines (DHP) as novel nonsteroidal MR antagonists. Chemical optimization of these DHP lead compounds resulted in BR-4628. Here we present the in vitro and in vivo pharmacological characterization of BR-4628, together with studies that highlight the binding mode on MR and the molecular reasons for the distinct antagonism.
EXPERIMENTAL PROCEDURES
Compounds
BR-4628 and [3H]BR-4628 were synthesized as described in the supplemental data. Aldosterone and spironolactone were purchased from Sigma. Eplerenone was purified from commercially available tablets (Inspra®). Nitrendipine was obtained by Hantzsch dihydropyridine synthesis using classical published methods (24). 18-Oxo-18-vinylprogesterone (18-vinyl-4-pregnen-3,18,20-trione) was a gift from A. Marquet (Paris, France).
Expression Vectors
The expression vectors pchMR, pchMRN770A, pchMRA773G, pchMRQ776A, pchMRR817A, pchMRM852A, pchMRC942A, and pchMRT945A code for MR, MRN770A, MRA773G, MRQ776A, MRR817A MRM852A, MRC942A, and MRT945A, respectively (25–28). The pchMRS810A, pchMRS810M, and pchMRA773G/S810M coding for MRS810A, MRS810M, and MRA773G/S810M, respectively, were obtained from pchMR using the site-directed mutagenesis procedure (QuikChange; Agilent, Massy, France). The plasmid pFC31Luc contains the murine mammary tumor virus promoter that drives the luciferase gene (29). The coding sequences of the respective LBDs of MR, MRS810L, AR, GR, and PR have been PCR-amplified and fused to the coding sequence of the DNA-binding domain of GAL4 (amino acids 1–147) under the control of a CMV promoter, leading to the expression plasmids pGAL4-MR, pGAL4-MRS810L, pGAL4-GR, pGAL4-AR, and pGAL4-PR. The pVPMR (kindly provided by Dr. G. Pinon) codes for the fusion protein between the VP16 activating domain and the full-length MR. The pGALTIF2 and pGALNcoR vectors, which code for fusion protein between the GAL4 DNA-binding domain and the nuclear activating domain of TIF2 and NCoR, respectively, were kindly provided by Dr. P. Balaguer. The pG5luc (kindly provided by Prof. P. Fuller) contains the luciferase gene driven by a GAL4-responsive promoter.
Generation of Stable Steroid Hormone Receptor Cell Lines
The pGAL4-MR, pGAL4-MRS810L, pGAL4-GR, pGAL4-AR, and pGAL4-PR vectors were transfected into CHO-K1 cells stably expressing a thymidine kinase promoter construct containing five GAL4-binding elements in front of the firefly luciferase gene. For each receptor a stable cell line has been generated by several rounds of limiting dilutions.
Transactivation Assays in Stable Cell Lines
The stable MR, MRS810L, GR, AR, and PR cell lines were cultured at 37 °C and 5% CO2 in DMEM/Ham's F-12 medium with GlutaMAX supplemented with 10% (v/v) inactivated fetal calf serum, 20 mm HEPES, 1.4 mm sodium pyruvate, 1.8 mm sodium bicarbonate, and 1 mg/ml Geneticin. Subconfluent cultures were passaged using Accutase. All of the cell culture reagents were obtained from Invitrogen. The cells were seeded 24 h before testing in Optimem medium containing 2.5% FCS (v/v), 2 mm glutamine, and 10 mm HEPES in 96- or 384-well plates. On the test day compounds were given in eight dilutions to the cells followed by the relevant EC50 concentration of each agonist. After an incubation time of 5–6 h, luciferase activity was determined using a luminescence detecting video camera system. The GraphPad Prism software (version 3.02; GraphPad Software Inc., San Diego, CA) was used for curve fitting and calculation of the IC50 and EC50 values. The IC50 and EC50 values from the luciferase assay were determined in at least three independent experiments performed in duplicate. The IC50 and EC50 values are given as the means ± S.E.
Transactivation Assays in Transiently Transfected Cells
HEK-293T cells were cultured and transfected with the expression vectors pchMR, pchMRN770A, pchMRA773G, pchMRQ776A, pchMRS810A, pchMRS810M, pchMRR817A, pchMRM852A, pchMRC942A, pchMRT945A, or pchMRA773G/S810M, the reporter vector pFC31Luc, and the pcβgal vector according to the method described previously (30). 24 h after transfection, ligands were added, and after a 16-h incubation, the cell extracts were assayed for luciferase and β-galactosidase activities as reported previously (30). The GraphPad Prism software (version 3.02; GraphPad Software Inc., San Diego, CA) was used for curve fitting and calculation of the IC50 and EC50 values. The IC50 values were determined from at least three independent experiments performed in triplicate. The IC50 values are given as the means ± S.E. The EC50 values were calculated from one experiment performed in triplicate.
Mammalian Two-hybrid Assays
HEK 293T cells were transfected with 2 μg of pVPMR, pGALTIF2, or pGALNcoR; 5 μg of pg5luc; and 1 μg of pcβgal. 24 h after transfection, aldosterone (10−10 to 10−8 m) or spironolactone or BR-4628 (10−8 to 10−6 m) were added. Parallel experiments were performed with 10−9 m aldosterone in the presence of 10−8 to 10−6 m spironolactone or BR-4628. After 16 h, the cell extracts were assayed for luciferase and β-galactosidase as reported previously (30).
Animals
Male Wistar rats (Charles River Germany, ∼300 g) were used in the experiment. They were housed with free access to food and water and maintained on a light-dark cycle at 22–24 °C. All of the animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and German legislation on animal welfare.
Model of Acute Natriuresis
A similar protocol as published previously (31) was used. Briefly, after 7 days of acclimatization, male Wistar rats were placed on a low salt diet containing 0.02% (w/v) sodium chloride (S0602-E081; ssniff Spezialdiäten GmbH, Soest, Germany) for 72 h. BR-4628 and spironolactone were administered in 1 ml/kg of vehicle (PEG400, 85.8%; glycerine, 5.3%, water, 8.9%, v/v/v) by oral gavage, and the animals (n = 8/group) were placed in metabolic cages (Tecniplast Deutschland GmbH, Hohenpeissenberg, Germany) for 8 h on water ad libitum. The urine samples were analyzed for volume as well as sodium and potassium concentrations by flame spectroscopy.
Coupled Cell-free Transcription and Translation
The human MR was expressed in vitro in the rabbit reticulocyte lysate system as described previously (27).
BR-4628 Binding Characteristics at Equilibrium
The lysates containing the in vitro expressed MR were diluted 4-fold with TEGWM buffer (20 mm Tris-HCl, 1 mm EDTA, 20 mm sodium tungstate, 1 mm β-mercaptoethanol, and 10% glycerol (v/v), pH 7.4) and incubated with 0.3–300 nm [3H]BR-4628 for 4 h at 4 °C. Bound and unbound ligands were separated by the dextran-charcoal method (27). The change in bound/unbound as a function of the amount bound was analyzed, and the Kd value was calculated as described previously (32). A parallel experiment was performed with an untranslated rabbit reticulocyte lysate.
Kinetic Experiments
The lysates containing the in vitro expressed MR were 4-fold diluted with TEGWM buffer and incubated with 10 nm [3H]BR-4628 for 4 h at 4 °C. One half of the labeled lysate was kept at 4 °C and was used to determine the stability of the [3H]BR-4628·MR complexes, and the other half was incubated with 1 μm BR-4628 for various periods. Bound and free ligands were separated using charcoal-dextran. The findings were corrected for receptor stability and were expressed as percentages of the binding measured at time 0. Dissociation was represented in a semi-logarithmic scale giving a linear manner representation.
Limited Proteolysis Assays
In vitro expressed [35S]MR was incubated for 10 min at 20 °C with or without aldosterone or BR-4628 (10−8 m) and then for 10 min at 20 °C with increasing concentrations of trypsin (0–300 μg/ml). The digestion products were analyzed by SDS-PAGE and autoradiographed.
Protein Modeling, Docking, and Molecular Dynamics Simulations
The x-ray structure of the wild type MR LBD complexed with deoxycorticosterone (Protein Data Bank code 2ABI) (30) served as a model for docking of BR-4628. The size of the binding site was enlarged by modifying several side chain conformations. The program Glide SP (version 3.5) was used to dock BR-4628 into the resulting binding cleft. The complex with the most probable binding pose was energy-minimized using the OPLS-AA/L force field. To check for the stability of hydrogen bonds or alternative contacts, a 4-ns molecular dynamics simulation with 11,522 explicit water molecules using the program Desmond was run (for details, see the supplemental text).
RESULTS
BR-4628 Is a Potent and Selective MR Antagonist
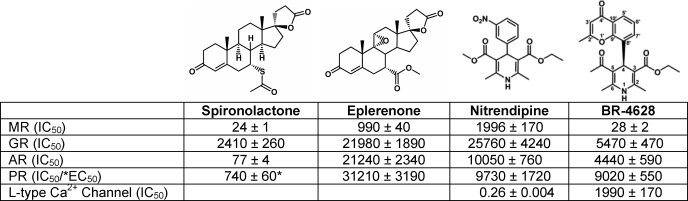
An ultrahigh throughput screening was performed with a cellular transactivation assay based on a recombinant CHO cell line stably expressing the MR LBD. Screening of almost 1,000,000 compounds revealed 677 confirmed primary hits. Approximately 300 screening hits were selected for further evaluation after elimination of toxic, unspecific, and noncompetitive compounds. Among them, a single cluster of ∼100 compounds comprised dihydropyridines (WO2007/025604) (33). This finding was surprising because dihydropyridines constitute the well known class of L-type calcium channel antagonists. Chemical optimization with respect to potency, selectivity, and metabolic stability lead to the synthesis of BR-4628 as a drug candidate (see formula in Table 1).
TABLE 1.
Half-maximally inhibitory/effective concentrations of spironolactone, eplerenone, nitrendipine, and BR-4628 determined for the oxo-steroid receptor and for the L-type calcium channel
The IC50 or EC50 values (nm) are the means ± S.E. of at least four independent experiments, performed in duplicate. The IC50 values at the L-type calcium channel were determined in two independent experiments, performed in duplicate using homogenates of rat cerebral cortex as described (46).
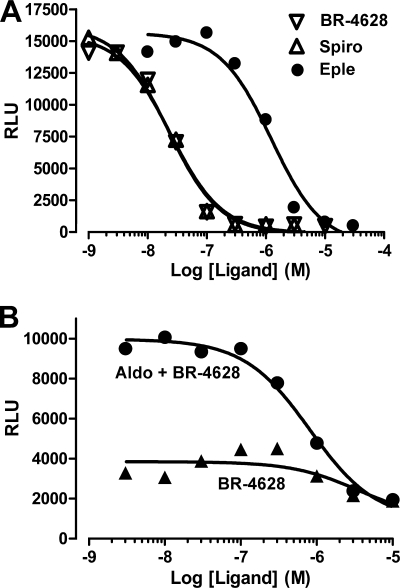
Transactivation assays performed with CHO-K1 cells stably expressing the MR LBD revealed that BR-4628 acts as a full MR antagonist. It inactivates the aldosterone-induced MR-LBD transactivation activity with an IC50 of 27.9 nm, a value comparable with that of spironolactone (24.2 nm) but much lower than that of eplerenone (990 nm) (Fig. 1A and Table 1). Furthermore, its antagonist potency is 2 orders of magnitude higher than that of nitrendipine, the well known dihydropyridine-based calcium channel blocker (Table 1). Spironolactone and eplerenone lose their antagonist feature upon the S810L mutation (8, 22, 30). Thus, we wondered whether BR-4628 still acts as an antagonist when bound to the MRS810L. CHO-K1 cells stably expressing the MRS810L LBD are characterized by a weak constitutive activity (5-fold increase in the base-line luciferase activity; Fig. 1B). Remarkably, BR-4628 inhibits the constitutive MRS810L LBD activity (IC50, 3630 nm) and the aldosterone-induced MRS810L LBD activity (IC50, 813 nm; Fig. 1B) in a dose-dependent manner. Thus, BR-4628 is a potent MR antagonist that retains its antagonist character at the MRS810L mutant.
FIGURE 1.
Representative inhibition curves of aldosterone-induced GAL4-MR-LBD and GAL4-MRS810L-LBD activities in response to antagonists. A, CHO-K1 cells stably expressing the GAL4-MRWT LBD fusion protein and the luciferase reporter gene under the control of a GAL4-responsive element-containing promoter (pFA-luc; Stratagene) were incubated for 6 h with increasing concentrations of BR-4628 (▽), spironolactone (△), or eplerenone ●) in the presence of 10−9 m aldosterone. B, CHO-K1 cells stably expressing the GAL4-MRS810L LBD fusion protein were incubated for 6 h with increasing concentrations of BR-4628 alone (▲) or in the presence of 10−9 m aldosterone (●). The MRWT and MRS810L transactivation activities were determined in duplicate from the respective luciferase activities.
The high sequence similarity among the LBDs of oxo-steroid receptors results in the cross-binding of ligands to the various receptors. To evaluate whether BR-4628 was MR-selective, transactivation assays were performed in CHO-K1 cells stably expressing the LBD of AR, GR, and PR. Spironolactone is a strong AR antagonist (IC50 = ∼77 nm), a weak GR antagonist (IC50 = ∼2.4 μm), and a weak PR agonist (EC50 = ∼740 nm). In contrast, BR-4628, like nitrendipine and eplerenone, is a weak antagonist of AR, GR, and PR, as revealed by IC50 values higher than 4 μm (Table 1). Thus, BR-4628 is at least 160-fold more selective for MR than for AR, whereas spironolactone exhibits only a 3-fold selectivity.
The dihydropyridine-derived nitrendipine is a potent calcium channel blocker used as a antihypertensive agent (34). Binding assays show that BR-4628 has a low calcium channel blocker activity, as revealed by an IC50 value 3 orders of magnitude higher than that of nitrendipine (1.99 μm versus 0.26 nm; Table 1). In summary, we could demonstrate that minor chemical modifications from a classical dihydropyridine calcium antagonist lead to a potent and highly specific MR antagonist.
BR-4628 Is a Potent MR Antagonist in Vivo
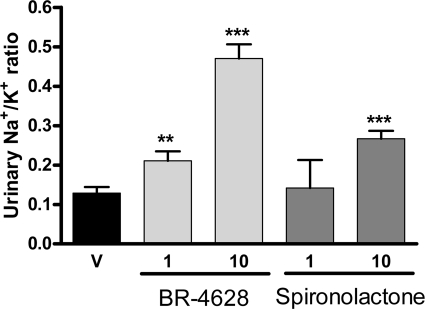
The acute in vivo activity of a MR antagonist can be monitored by measuring its effects on the urinary Na+/K+ ratio. BR-4628 was orally administered to conscious rats by gavage, and the sodium and potassium concentrations were measured in the collected urine. BR-4628 administration increased the urinary sodium/potassium ratio in a dose-dependent manner with a significant effect for a dose as low as 1 mg/kg (Fig. 2). For comparison, spironolactone was found to increase the Na+/K+ at a dose of 10 mg/kg (Fig. 2).
FIGURE 2.
Natriuretic in vivo activity of BR-4628 and spironolactone in conscious rats. The increase of the urinary sodium to potassium ratio after oral application of vehicle (V), BR-4628 (1 and 10 mg/kg), and spironolactone (1 and 10 mg/kg) was determined by flame spectroscopy of urine samples after a collection period of 8 h. Each bar represents the mean value of n = 8 animals ± S.E. *, p < 0.05; **, p < 0.01; and ***, p < 0.005 versus vehicle after calculation using Student's t test.
Docking of BR-4628 within the MR LBD Reveals an Unusual Anchoring Mode
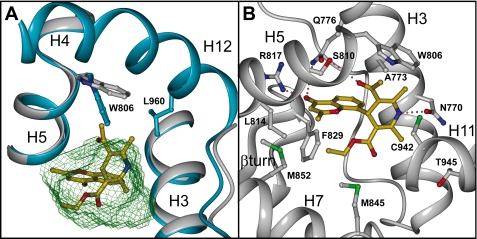
The crystal structures of the LBD of MR and MRS810L complexed with steroidal agonists have been recently solved, allowing their anchoring mode to be studied in detail (28, 30, 35, 36). The nonsteroidal nature of BR-4628 raised the question of its binding mode within the MR binding pocket. To answer this question, a large amount of purified MR·BR-4628 complex was produced. However, despite numerous trials, crystallization of a complex was unsuccessful. Thus, a mutagenesis approach guided by BR-4628 docking within the x-ray structure of the wild type MR LBD (Protein Data Bank code 2ABI) (30) was followed. Initial docking into the agonist conformation of the MR LBD was unsuccessful because the binding pocket is simply too small to accommodate the highly branched BR-4628 (supplemental Fig. S1). Indeed, the 5-acetyl moiety is in close contact with Trp806 and might displace it for accommodation, resulting in a clash with the H12 helix (Fig. 3A). Moreover, the neighboring 6-methyl group of the DHP core clashes with Leu960 located in helix H12 (Fig. 3A). Both of these structural elements suggest that this molecule might have bulky antagonist features by impairing the H12 helix to adopt its agonist position. Therefore, the H12 helix was omitted from further docking experiments and from a constrained 4-ns molecular dynamics simulation (supplemental Video S1 and Fig. S2). The chromenone carbonyl group is anchored to Gln776 and Arg817, the DHP NH group is hydrogen-bonded to the Asn770 carbonyl oxygen, and the carbonyl group of the 5-acetyl moiety forms a hydrogen bond with Ser810 (Fig. 3B). During the course of the 4-ns molecular dynamics simulation, Asn770 is hydrogen-bonded in 92.8% of the conformations, Ser810 is hydrogen-bonded in 65.3%, Arg817 is hydrogen-bonded in 34.5%, and Gln776 is hydrogen-bonded in 0.6%. No hydrogen bond is observed for Cys942; however, the thiol group packs against the core of the DHP ring, forming van der Waal's contacts (average distance from the BR-4628 NH to the sulfur of Cys942 is 3.9 Å). The methyl side chain of Ala773 fits tightly into a notch on the BR-4628 surface, formed by the 6-methyl carbon (3.6 Å), the C5 in the DHP core (3.9 Å), the 5-acetyl carbon (4.0 Å), and the C7′ in the chromenone ring (3.7 Å). Interestingly, in such a position, BR-4628 does not interact with Thr945 (average distance from the BR-4628 NH to Thr945 Cα is 7.4 Å). The 3-ethyl ester fills a lipophilic pocket formed by Leu814, Phe829, Met845, Met852, and Leu938 (Fig. 3B). The methyl substituent on the 4-chromenone ring interacts with close van der Waal's contacts with the side chain of Ser811, Met807, Met852, and Leu938.
FIGURE 3.
Accommodation mode of BR-4628 within the MR LBD. A, picture showing the minimized complex between BR-4628 (gold) and the MR LBD devoid of the H12 helix (gray) superimposed to the structure of the full-length MR LBD (blue). The ligand cavity volume, as calculated with Voidoo (45), is depicted as a green wire surface. B, overall view of the BR-4628 accommodation within the MR LBD devoid of the H12 helix. Only residues that form critical contacts with the ligand are shown. Hydrogen bound between BR-4628 and the polar residues is depicted as dashed red lines. This figure was produced using DINO.
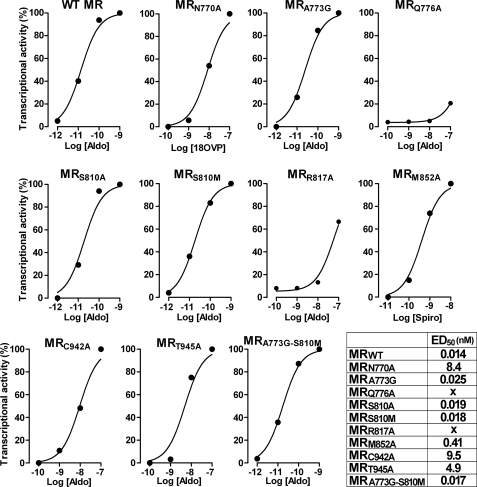
Based on this binding mode, 10 amino acids in the binding site were selected for mutation studies. We focused on Asn770, Gln776, Arg817, and Thr945, known to anchor the steroids (25), and on the Ala773, Ser810, Met852, and Cys942 residues that play a critical role in delimiting the volume of the cavity (25, 26, 28). Each residue was replaced by an alanine, except Ala773, which was replaced by a glycine. All of the mutations were introduced within the full-length MR gene, and the corresponding mutant receptors were transiently expressed in HEK293T cells and tested for their transcriptional activity. Aldosterone activates MRA773G and MRS810A with ED50 values identical to that of MR (0.025 and 0.019 nm, respectively, versus 0.014 nm; Fig. 4). Aldosterone is less efficient in activating MRC942A and MRT945A (ED50, 9.5 and 4.9 nm, respectively). The activity of MRQ776A and MRR817A in response to 10−7 m aldosterone is 20 and 70% that of MR, respectively (Fig. 4). Aldosterone is unable to activate MRN770A and MRM852A (25, 27). Nevertheless, 18-vinyl-4-pregnen-3,18,20-trione and spironolactone activate MRN770A and MRM852A, respectively (ED50, 8.4 and 0.41 nm; Fig. 4). The effect of the mutations on the BR-4628 and spironolactone antagonist potency was analyzed in transfection assays (supplemental Fig. S3 and Table 2). The T945A mutation had no effect on the BR-4628 potency, as revealed by an IC50 identical to that of wild type MR, and confirmed that BR-4628 does not interact with this residue. In sharp contrast, the N770A and A773G mutations have a strong effect, increasing BR-4628 IC50 values by 39- and 25-fold, respectively. From these results it can be proposed that Asn770 and Ala773 are critical for BR-4628 binding. The Q776A, S810A, R817A, M852A, and C942A mutations have intermediate effects, increasing the IC50 of BR-4628 by ∼6–10-fold. This suggests that the amino acids Gln776, Ser810, Arg817, Met852, and Cys942 contribute to the BR-4628 binding. Interestingly, the A773G and S810A mutations have no effect on the spironolactone antagonist potency, whereas these two mutations increase the BR-4628 IC50 value. Moreover, the T945A mutation induces a 6-fold increase in the spironolactone IC50 value, whereas it does not modify BR-4628. Altogether, these results fully agree with the docking experiments showing striking differences between BR-4628 and spironolactone accommodation mode within the ligand-binding pocket of MR.
FIGURE 4.
Transactivation activity of the wild type and mutant MRs. HEK-293T cells transiently expressing the MR, MRN770A, MRA773G, MRQ776A, MRS810A, MRS810M, MRR817A, MRM852A, MRC942A, MRT945A, or MRA773G/S810M were incubated for 16 h with aldosterone (Aldo), 18-vinyl-4-pregnen-3,18,20-trione (18OVP), or spironolactone (Spiro). The cell extracts were assayed for luciferase and β-galactosidase activities as reported previously (30). The GraphPad Prism software was used for curve fitting and calculation of the EC50 values.
TABLE 2.
Half-maximally inhibitory concentrations of BR-4628 and spironolactone for the full-length wild-type and mutant MRs
The IC50 values were calculated using the GraphPad Prism software and are the means ± S.E. of three independent experiments performed in triplicate. The EC50 values for spironolactone were calculated using the Prism software for a representative experiment performed in triplicate.
| BR-4628 |
SPIRO |
||||
|---|---|---|---|---|---|
| IC50 | Fold | IC50 | Fold | EC50 | |
| nm | nm | nm | |||
| MRWT | 34 ± 4 | 1 | 74.0 ± 15 | 1 | |
| MRN770A | 1334 ± 384 | 39.2 | 1313 ± 167 | 17.7 | |
| MRA773G | 958 ± 321 | 28.2 | 84 ± 15 | 1.1 | |
| MRQ776A | 317 ± 87 | 9.3 | 301 ± 77 | 4.1 | |
| MRS810A | 345 ± 21 | 10.1 | 109 ± 24 | 1.5 | |
| MRS810M | 179 ± 30 | 5.3 | 6 | ||
| MRR817A | 323 ± 22 | 9.5 | 331 ± 20 | 4.5 | |
| MRM852A | 270 ± 23 | 7.9 | 0.4 | ||
| MRC942A | 219 ± 21 | 6.4 | 506 ± 55 | 6.8 | |
| MRT945A | 30 ± 5 | 0.9 | 444 ± 43 | 6.0 | |
| MRA773G/S810M | 12100 ± 3000 | 356 | 8.4 | ||
Previous reports have highlighted remarkable differences between the oxo-steroid receptors ligand-binding cavity residues. It concerns the MR Ala773 and Ser810, which correspond to glycine and methionine residues at the corresponding positions of the human AR, GR, and PR (26, 37). Because BR-4628 possesses remarkable selectivity toward MR, we wondered whether these residues are involved in the high MR selectivity. The A773G and S810M single mutations and the A773G/S810M double mutation have no effect on the aldosterone potency to activate MR as illustrated by identical ED50 values (Fig. 4). In sharp contrast, these mutations lead to an increase of BR-4628 IC50 values by 28-, 5-, and 356-fold, respectively (supplemental Fig. S3 and Table 2). These results suggest that the alanine/serine pair at the 773 and 810 positions favor BR-4628 binding to MR.
The MR·BR-4628 Complex Is Unstable and Unable to Recruit Transcriptional Co-regulators
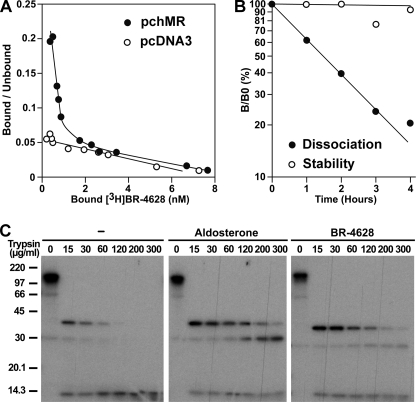
To further characterize the MR·BR-4628 interaction, we synthesized the 3H-labeled BR-4628 (see supplemental data) and characterized its binding properties to the in vitro expressed MR. A dissociation constant at equilibrium (Kd) of 2 nm was measured (Fig. 5A), indicating an affinity for MR in the same order of magnitude as that of aldosterone (25). Interestingly, we observed that, at 4 °C, [3H]BR-4628 dissociates quickly from MR with a half-life time of ∼90 min (Fig. 5B), a value identical to that of the tritiated spirolactone RU26752 (t½ = ∼90 min) (27). We further examined the stability of the MR·BR-4628 complex by measuring the ability of BR-4628 to protect MR against proteolysis. Upon treatment of the unbound [35S]MR for 10 min at 20 °C by 15 μg/ml trypsin, a major fragment of 41 kDa and a minor fragment of 30 kDa both encompassing the LBD (38) were recovered (Fig. 5C). They were completely digested when the trypsin concentration was increased to 120 μg/ml. In contrast, the aldosterone·MR complex was highly resistant to the trypsin action. Although the proteolysis pattern is similar for the MR·BR-4628 complex, the intensity of the 30-kDa fragment is much lower upon BR-4628 treatment (Fig. 5C). This result indicates that BR-4628 is less efficient than aldosterone in protecting the MR against proteolysis, confirming the high instability of the BR-4628·MR complex.
FIGURE 5.
Binding properties of BR-4628 to MR. A, Scatchard plot of the binding of [3H]BR-4628 to MR. The in vitro expressed MR was incubated with [3H]BR-4628 (3 × 10−10 to 3 × 10−7 m) for 4 h at 4 °C. Bound and unbound ligands were separated by the dextran-charcoal method, the evolution of bound/unbound as a function of the amount bound was plotted, and the Kd value was calculated using the ScatMac program (32). A parallel experiment was performed with a rabbit reticulocyte lysate in which no receptor was expressed. B, dissociation kinetics of [3H]BR-4628 from MR. The in vitro expressed MR was incubated with 10−8 m [3H]BR-4628 for 4 h at 4 °C and then incubated for various time with 10−6 m BR-4628. The bound and free ligands were separated by the dextran-charcoal method, and the residual binding was calculated. C, limited proteolysis assays. In vitro expressed [35S]MR was incubated for 10 min at 20 °C with or without 10−8 m aldosterone or BR-4628 and then for 10 min at 20 °C with trypsin (0–300 μg/ml). The digestion products were analyzed by SDS-PAGE and autoradiographed.
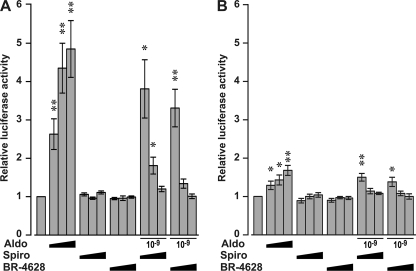
To further characterize the mechanism by which BR-4628 inactivates MR, we evaluated the capacity of MR to recruit transcriptional co-regulators upon BR-4628 binding. Mammalian two-hybrid assays revealed that aldosterone, but not BR-4628 and spironolactone, promoted a dose-dependent binding of the TIF2 to MR (Fig. 6A). Moreover, the two antagonists are able to inhibit the aldosterone-induced MR/TIF2 interaction in a dose-dependent manner. Interestingly, the NCoR is not recruited by MR upon the binding of BR-4628 and spironolactone, whereas a weak interaction is observed after aldosterone binding (Fig. 6B). Furthermore, the two MR antagonists inhibit the binding of NCoR to the MR-aldosterone complex. Thus, despite a high affinity of BR-4628 for MR, the BR-4628·MR complex is highly unstable and unable to recruit transcriptional co-modulators.
FIGURE 6.
Recruitment of transcriptional co-regulators by MR in mammalian two-hybrid assays. HEK293T cells transiently expressing the fusion proteins VP16-MR and the GAL4 DNA-binding domain fused to the receptor interacting domain of TIF2 (A) or NcoR (B) were incubated in triplicate with ethanol, aldosterone (Aldo, 10−10 to 10−8 m), spironolactone (Spiro, 10−8 to 10−6 m), or BR-4628 (10−8 to 10−6 m) alone or with 10−9 m aldosterone in the presence of 10−8 to 10−6 m spironolactone or BR-4628. After harvesting the cells, the luciferase activities were measured and normalized by the values obtained with ethanol. The results are the means ± S.E. of five to six independent experiments. *, p ≤ 0.05; **, p ≤ 0.01.
DISCUSSION
In this study, we performed thorough mutagenesis, structural, biochemical, and pharmacological investigations of a novel dihydropyridine-based MR antagonist, which allowed elucidation of its mechanism of MR inactivation. Transactivation assays using the MR LBD fused to the GAL4 DNA-binding domain revealed that BR-4628 is as potent as spironolactone but significantly more potent than eplerenone (Table 1). A similar inhibitory hierarchy is also observed by using the entire MR, because BR-4628 is characterized by an IC50 of 33.5 nm (Table 2) compared with 50.0 nm for spironolactone and 2000 nm for eplerenone (27). Molecular dynamics and point mutation data suggest that binding of BR-4628 is mediated by hydrogen bonds to Asn770, Gln776, Ser810, and Arg817 and lipophilic contacts to Ala773, Met852, and Cys942. The calculated hydrogen bond probabilities in the molecular dynamics simulation agree well for all point mutation data; the higher the observed H-bond frequency, the more important the amino acid is in the transactivation experiment. Only Gln776 is underestimated in the molecular dynamics simulation. However, even in the published x-ray structures, the distance between the steroidal 3-oxo function and the Gln776 side chain NH is relatively large (3.2–3.3 Å), and an additional H-bond acceptor in a ligand (like the 5-acetyl group in BR-4628) might form water-mediated contacts with Gln776, explaining the importance of this amino acid in our mutational studies.
The second noteworthy feature of BR-4628 is its MR selectivity. It inhibits the transcriptional activity of the other oxo-steroid receptors (AR, GR, and PR) with IC50 values higher than 4 μm, pointing out only residual affinity for these receptors. MR is the unique oxo-steroid receptor, having an alanine residue (Ala773) in the H3 helix and a serine residue (Ser810) in the H5 helix. AR, PR, and GR each harbor a glycine and a methionine at the corresponding positions, respectively. Interestingly, BR-4628 docking within the MR binding pocket underscores that Ser810 forms a hydrogen bond with the C5-acetyl group and that the Ala773 residue is surrounded by the aromatic phenyl moiety of the chromenone, the C5-acetyl, and the C6-methyl group. The dramatic decrease in the BR-4628 inhibitory potency upon the A773G, S810M, and A773G/S810M mutations demonstrates the importance of the interactions between BR-4628 and these residues for its potency and selectivity.
BR-4628 is a dihydropyridine derivative. Classic dihydropyridine-derived molecules, such as nifedipine and nitrendipine, are known to act as potent L-type calcium channel blockers (34). These molecules have been reported to also have a weak MR antagonist activity (39). However, we show here that a classical L-type channel blocker like nitrendipine has no selectivity toward MR and rather nonselectively antagonizes all steroid hormone receptors in the micromolar range (Table 1). In sharp contrast, BR-4628 is a highly potent MR antagonist, with a low L-type calcium channel binding activity in the micromolar range. BR-4628 differs from classical dihydropyrine derivatives mainly by its 2-methylchromenonyl substituent. It is likely that this substituent drastically reduces calcium channel inhibitory activity, and it is responsible for high MR binding affinity, because it rigidifies the active conformation with respect to the MR. The methyl group is involved in close van der Waal's contact with four lipophilic side chains in MR, explaining the pronounced differences in activity between nitrendipine and BR-4628 on MR. Thus, the presence of the 2-methylchromenonyl substituents on the dihydropyridine ring ensures the high MR selectivity versus calcium channel.
Functional and structural data have clearly shown that the inability of progesterone and spironolactone to establish a strong contact with Asn770 is responsible for their antagonistic features (25). BR-4628 strongly interacts with Asn770, raising the question of the mechanism by which it inactivates MR. Two classes of antagonists have been described for steroid receptors. The first one, called “passive,” refers to molecules that are unable to stabilize the receptor in a conformation able to recruit the transcriptional co-regulators. These antagonists are usually small molecules that dissociate quickly from the receptor. This is namely the case for the MR antagonists progesterone and spironolactone, the estrogen receptor β antagonist (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol (40), and the AR antagonist hydroxyflutamide (41). The second class of antagonists, called “active,” comprises molecules that permit the LBD to recruit transcriptional co-repressors. This type of antagonism is observed namely with RU486 for PR and GR and fulvestrant for estrogen receptor α. These molecules are characterized by bulky side chains that impair H12 helix from adopting its active conformation. The docking experiments we performed with BR-4628 clearly show that its 5-acetyl and C6-methyl groups protrude toward the H12 helix, suggesting that BR-4628 behaves as a bulky antagonist. However, BR-4628 does not occupy the binding pocket in the region in which the steroidal D-ring is anchored, namely through Thr945. The inability to establish contacts in this region prevents the formation of a stable complex, as confirmed by the quick dissociation of BR-4628 from MR and the low resistance of the MR·BR-4628 complex against proteolysis. As a consequence, MR is unable to recruit transcriptional co-regulator. Thus, it can be proposed that BR-4628 acts as a bulky-passive antagonist. Similarities of BR-4628 exist with respect to the nonsteroidal AR-antagonist bicalutamide. X-ray structures of point mutated ARs show that bicalutamide only partly fills the binding pocket around the steroidal D-ring and that its sulfonyl moiety clashes with the H12 helix (42, 43), signifying that bicalutamide could also acts as a bulky-passive antagonist. Altogether these findings suggest that the receptor inactivation mode referred as to bulky-passive antagonism that we proposed for MR can be applied to other steroid receptors.
Interestingly, we observed that BR-4628 is able to inhibit the weak MRS810L constitutive activity, thus behaving as an inverse agonist. Moreover, BR-4628, unlike progesterone and spironolactone, still acts as an antagonist when bound to the mutant MRS810L. The presence of a leucine at the 810 position instead of the serine allows the establishment of additional stabilizing contacts with the A-ring of progesterone and spironolactone, triggering the LBD to adopt its active state. In the case of BR-4628, the C5-acetyl group is hydrogen-bonded to Ser810. The replacement of Ser810 by the leucine residue destabilizes the BR-4628·MR complex. The inverse agonist potency of BR-4628 is rather low (IC50, ∼3.6 μm), suggesting that high BR-4628 concentrations would be necessary to inhibit the MRS810L constitutive activity. BR-4628 has also a reduced inhibiting potency when acting through the MRS810L compared with the wild type receptor. However, in the presence of physiological aldosterone concentrations, BR-4628 IC50 at MRS810L is decreased to 0.8 μm. Therefore, its IC50 is still almost 1 order of magnitude lower than that for the other oxo-steroid receptors (4–9 μm), giving the possibility of a selective MRS810L inhibition. Thus, BR-4628 offers a potential for causal treatment of aldosterone-related hypertension and end organ damage in broad patient populations as well as in rare forms of hypertension (including the hypertension associated with the S810L mutation), where available steroidal antagonists are even contraindicated.
Altogether these findings suggest that BR-4628 represents a prototype of an at least third generation MR antagonist (44) that acts through a new bulky-passive mechanism. Several in vivo investigations are underway to evaluate its end organ protective effects in preclinical animal models.
Supplementary Material
Acknowledgments
We thank Verena Jilg and Petra Ammelung for excellent technical assistance. We are also grateful to Dr. P. Balaguer, Prof. P. Fuller, and Dr. G. Pinon for providing us with the vectors for two-hybrid assays.
Portions of these results have been presented at the Congress of the European Society of Cardiology in Vienna in 2007 and at the German Society of Cardiology Congress in 2007 in Mannheim.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Video S1, and Figs. S1–S3.
- MR
- mineralocorticoid receptor
- AR
- androgen receptor
- GR
- glucocorticoid receptor
- PR
- progesterone receptor
- TIF2
- transcriptional intermediary factor 2
- NCoR
- nuclear receptor co-repressor 1
- LBD
- ligand-binding domain
- DHP
- dihydropyridine.
REFERENCES
- 1.Horisberger J. D., Rossier B. C. (1992) Hypertension 19, 221–227 [DOI] [PubMed] [Google Scholar]
- 2.Bonvalet J. P. (1998) Kidney Int. Suppl. 65, S49–S56 [PubMed] [Google Scholar]
- 3.Evans R. M. (1988) Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geller D. S., Rodriguez-Soriano J., Vallo Boado A., Schifter S., Bayer M., Chang S. S., Lifton R. P. (1998) Nat. Genet. 19, 279–281 [DOI] [PubMed] [Google Scholar]
- 5.Sartorato P., Cluzeaud F., Fagart J., Viengchareun S., Lombès M., Zennaro M. C. (2004) Mol. Endocrinol. 18, 2151–2165 [DOI] [PubMed] [Google Scholar]
- 6.Geller D. S., Zhang J., Zennaro M. C., Vallo-Boado A., Rodriguez-Soriano J., Furu L., Haws R., Metzger D., Botelho B., Karaviti L., Haqq A. M., Corey H., Janssens S., Corvol P., Lifton R. P. (2006) J. Am. Soc. Nephrol. 17, 1429–1436 [DOI] [PubMed] [Google Scholar]
- 7.Pujo L., Fagart J., Gary F., Papadimitriou D. T., Claës A., Jeunemaître X., Zennaro M. C. (2007) Hum. Mutat. 28, 33–40 [DOI] [PubMed] [Google Scholar]
- 8.Geller D. S., Farhi A., Pinkerton N., Fradley M., Moritz M., Spitzer A., Meinke G., Tsai F. T., Sigler P. B., Lifton R. P. (2000) Science 289, 119–123 [DOI] [PubMed] [Google Scholar]
- 9.Delcayre C., Silvestre J. S. (1999) Cardiovasc. Res. 43, 7–12 [DOI] [PubMed] [Google Scholar]
- 10.Funder J. W. (2005) J. Steroid Biochem. Mol. Biol. 93, 121–125 [DOI] [PubMed] [Google Scholar]
- 11.Kagawa C. M., Cella J. A., Van Arman C. G. (1957) Science 126, 1015–1016 [DOI] [PubMed] [Google Scholar]
- 12.Liddle G. W. (1957) Science 126, 1016–1018 [DOI] [PubMed] [Google Scholar]
- 13.de Gasparo M., Joss U., Ramjoué H. P., Whitebread S. E., Haenni H., Schenkel L., Kraehenbuehl C., Biollaz M., Grob J., Schmidlin J., et al. (1987) J. Pharmacol. Exp. Ther. 240, 650–656 [PubMed] [Google Scholar]
- 14.de Gasparo M., Whitebread S. E., Preiswerk G., Jeunemaître X., Corvol P., Ménard J. (1989) J. Steroid Biochem. 32, 223–227 [DOI] [PubMed] [Google Scholar]
- 15.Jeunemaitre X., Chatellier G., Kreft-Jais C., Charru A., DeVries C., Plouin P. F., Corvol P., Menard J. (1987) Am. J. Cardiol. 60, 820–825 [DOI] [PubMed] [Google Scholar]
- 16.Croom K. F., Perry C. M. (2005) Am. J. Cardiovasc. Drugs 5, 51–69 [DOI] [PubMed] [Google Scholar]
- 17.Pitt B., Zannad F., Remme W. J., Cody R., Castaigne A., Perez A., Palensky J., Wittes J. (1999) N. Engl. J. Med. 341, 709–717 [DOI] [PubMed] [Google Scholar]
- 18.Pitt B., Remme W., Zannad F., Neaton J., Martinez F., Roniker B., Bittman R., Hurley S., Kleiman J., Gatlin M. (2003) N. Engl. J. Med. 348, 1309–1321 [DOI] [PubMed] [Google Scholar]
- 19.Corvol P., Mahoudeau J. A., Valcke J. C., Ménard J., Bricaire H. (1976) Nouv. Presse Med. 5, 691–694 [PubMed] [Google Scholar]
- 20.Garthwaite S. M., McMahon E. G. (2004) Mol. Cell. Endocrinol. 217, 27–31 [DOI] [PubMed] [Google Scholar]
- 21.Weinberger M. H., Roniker B., Krause S. L., Weiss R. J. (2002) Am. J. Hypertens. 15, 709–716 [DOI] [PubMed] [Google Scholar]
- 22.Hultman M. L., Krasnoperova N. V., Li S., Du S., Xia C., Dietz J. D., Lala D. S., Welsch D. J., Hu X. (2005) Mol. Endocrinol. 19, 1460–1473 [DOI] [PubMed] [Google Scholar]
- 23.Kolkhof P., Bärfacker L., Hillisch A., Haning H., Schäfer S. (2008) in Nuclear Receptors as Drug Targets (Ottow E., Weinmann H. eds) pp. 409–429, Wiley-VCH Verlag, Berlin [Google Scholar]
- 24.Meyer H., Bossert F., Wehinger E., Stoepel K., Vater W. (1981) Arzneimittelforschung 31, 407–409 [PubMed] [Google Scholar]
- 25.Fagart J., Wurtz J. M., Souque A., Hellal-Levy C., Moras D., Rafestin-Oblin M. E. (1998) EMBO J. 17, 3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auzou G., Fagart J., Souque A., Hellal-Lévy C., Wurtz J. M., Moras D., Rafestin-Oblin M. E. (2000) Mol. Pharmacol. 58, 684–691 [DOI] [PubMed] [Google Scholar]
- 27.Fagart J., Seguin C., Pinon G. M., Rafestin-Oblin M. E. (2005) Mol. Pharmacol. 67, 1714–1722 [DOI] [PubMed] [Google Scholar]
- 28.Fagart J., Huyet J., Pinon G. M., Rochel M., Mayer C., Rafestin-Oblin M. E. (2005) Nat. Struct. Mol. Biol. 12, 554–555 [DOI] [PubMed] [Google Scholar]
- 29.Gouilleux F., Sola B., Couette B., Richard-Foy H. (1991) Nucleic Acids Res. 19, 1563–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huyet J., Pinon G. M., Fay M. R., Fagart J., Rafestin-Oblin M. E. (2007) Mol. Pharmacol. 72, 563–571 [DOI] [PubMed] [Google Scholar]
- 31.Brandish P. E., Chen H., Szczerba P., Hershey J. C. (2008) J. Pharmacol. Toxicol. Methods 57, 155–160 [DOI] [PubMed] [Google Scholar]
- 32.Claire M., Rafestin-Oblin M. E., Michaud A., Corvol P., Venot A., Roth-Meyer C., Boisvieux J. F., Mallet A. (1978) FEBS Lett. 88, 295–299 [DOI] [PubMed] [Google Scholar]
- 33.Kuhl A., Kolkhof P., Heckroth H., Schlemmer K. H., Flamme I., Perez S. F., Gielen-Haertwig H., Grosser R., Ergüden J. K., Lang D. (2007) 4-CHROMENONYL-l,4-DIHYDROPYRIDINES AND THEIR USE. WO2007/025604 [Google Scholar]
- 34.Meredith P. A., Elliott H. L. (2004) J. Hypertens. 22, 1641–1648 [DOI] [PubMed] [Google Scholar]
- 35.Bledsoe R. K., Madauss K. P., Holt J. A., Apolito C. J., Lambert M. H., Pearce K. H., Stanley T. B., Stewart E. L., Trump R. P., Willson T. M., Williams S. P. (2005) J. Biol. Chem. 280, 31283–31293 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Suino K., Daugherty J., Xu H. E. (2005) Mol. Cell 19, 367–380 [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Geller D. S. (2008) J. Steroid Biochem. Mol. Biol. 109, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couette B., Jalaguier S., Hellal-Levy C., Lupo B., Fagart J., Auzou G., Rafestin-Oblin M. E. (1998) Mol. Endocrinol. 12, 855–863 [DOI] [PubMed] [Google Scholar]
- 39.Dietz J. D., Du S., Bolten C. W., Payne M. A., Xia C., Blinn J. R., Funder J. W., Hu X. (2008) Hypertension 51, 742–748 [DOI] [PubMed] [Google Scholar]
- 40.Shiau A. K., Barstad D., Radek J. T., Meyers M. J., Nettles K. W., Katzenellenbogen B. S., Katzenellenbogen J. A., Agard D. A., Greene G. L. (2002) Nat. Struct. Biol. 9, 359–364 [DOI] [PubMed] [Google Scholar]
- 41.Söderholm A. A., Lehtovuori P. T., Nyrönen T. H. (2005) J. Med. Chem. 48, 917–925 [DOI] [PubMed] [Google Scholar]
- 42.Bohl C. E., Gao W., Miller D. D., Bell C. E., Dalton J. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6201–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohl C. E., Miller D. D., Chen J., Bell C. E., Dalton J. T. (2005) J. Biol. Chem. 280, 37747–37754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Funder J. W. (2005) Nat. Clin. Pract. Endocrinol. Metab. 1, 4–5 [DOI] [PubMed] [Google Scholar]
- 45.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 46.Ehlert F. J., Roeske W. R., Itoga E., Yamamura H. I. (1982) Life Sci. 30, 2191–2202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.